Abstract
In this review, we compare four assays that are currently used to measure HIV integration and discuss their strengths and weaknesses. We then outline advances that have been made toward development of a more robust, more sensitive, quantitative HIV integration assay suitable for clinical use. The assay that we have developed uses repetitive-sampling Alu-gag PCR. The detailed protocol describes our assay step-by-step, the creation of an integration standard cell line and accompanying standard curve, as well as the quantitation of integration and calculation of associated error estimates. Finally, we speculate on fundamental, unresolved issues in HIV latency that can be addressed by measuring HIV integration.
Keywords: HIV, integration, latent infection, Alu-gag PCR, quantitative real-time PCR, HAART
1. Introduction
Integration is a central step in the HIV life cycle and is defined as the insertion of the HIV reverse transcript into the host cellular DNA. Integration is required for efficient spreading infection (1–9) and so is an important step to measure. Measurements of integration in vitro continue to enhance our understanding of basic retrovirology and how cells restrict HIV integration (10–13). Measuring integration in vivo has also enhanced our understanding of latent HIV infection as it has been demonstrated that resting CD4+ T cells contain HIV, but fail to produce infectious virus unless stimulated (14). Thus, such latently infected cells are resistant to antiretroviral therapy (15, 16).
With the advent of more sensitive assays for HIV integration, it may be useful to measure the level of integrated HIV DNA in various cellular subsets within HIV infected individuals especially in combination with other viral intermediates (17, 18). Because of the limitations of prior HIV integration assays, there is very little data on the level of HIV integration in various CD4+ T subsets (19) and less information regarding the level of integration in CD4+ non-T cell subsets. Monitoring integrated DNA within subsets over time could demonstrate, for example, if HIV is cleared from short lived CD4+ non-T cells (20) in the presence of antiretroviral therapy. By combining measurements of total DNA and integrated DNA (18), it may even be possible to indirectly determine the relative level of replication within different subsets. One recent study provides evidence that the half-life of integrated HIV DNA is greater than the half-life of unintegrated HIV DNA (18). This would suggest that cells with higher levels of total HIV DNA relative to integrated HIV DNA experienced more rounds of replication. Thus, this approach might reveal if HIV replication persists within specific cellular subsets for patients on HAART.
1.1 Overview of existing integration assays
There are three main hurdles to measuring integration well. One is distinguishing integrated from unintegrated DNA since not all reverse transcripts integrate (14). Thus, one cannot measure total DNA as a surrogate marker for integrated DNA. Measuring integration in vivo presents two additional hurdles. These challenges include enhancing sensitivity since the level of integration is low in vivo (14) and detecting all variants of the integrated population since it is known that HIV has a relatively high mutation rate (21–24).
Apart from the method that we use to measure integration, Alu-gag PCR (25–29), three other methods have been used to assay integration: gel separation, inverse PCR, and linker ligation PCR. In the first method, integration in HIV infected individuals is measured by first using gel separation methods to segregate genomic DNA from episomal DNA (18, 30) and then measuring the amount of HIV DNA within the genomic DNA by routine quantitative real-time (kinetic) PCR. However, the separation method is too laborious for large numbers of clinical samples and it is unclear how effectively this method separates episomal DNA from genomic DNA. The two other methods, inverse PCR (14, 31, 32) and linker ligation PCR (33), are conceptually similar to Alu-gag PCR, the method we prefer. They distinguish integrated from unintegrated DNA by designing primers that only allow exponential amplification of HIV DNA when it is integrated. These methods tend to use two PCR steps and approach quantitation via endpoint dilution analysis. In all three methods, inverse, linker ligation, and Alu-gag PCR, one primer is HIV-specific and binds to both integrated and unintegrated DNA. By design, the second primer only binds to the same target DNA template in the correct orientation if HIV has integrated. In this way, only integrated HIV is exponentially amplified. In all three methods, primer extension (or linear amplification) of unintegrated HIV DNA occurs. In all three techniques, only amplicons that contain HIV DNA sequences are detected.
Exponential amplification of human chromosomal DNA is a limitation in both linker ligation and Alu-gag PCR as it results in limiting substrate. Thus, it limits the sensitivity of the assays. However, the amplified human chromosomal DNA that lacks HIV DNA is not detected. While inverse PCR avoids amplification of chromosomal DNA, it is significantly more labor intensive than the other PCR approaches, making it less attractive for widespread clinical use.
Detecting all HIV variants is one of the biggest challenges when measuring integration in vivo. Inverse and linker ligation PCR are most affected by this problem. In both assays, a restriction enzyme digestion step is included. Given the relatively high mutation rate, the restriction enzymes used in these two methods will at some frequency fail to digest some of the HIV sequences in the DNA sample at the predicted site(s). These mutated sequences would often be undetected (or under certain circumstances detected at a much lower efficiency). For example, we found that approximately 5% of the sequences in the Los Alamos database (34) were mutated in a commonly used restriction enzyme site (unpublished observation).
Applications of inverse PCR (14, 31, 35) and Alu-gag PCR (25) to the measurement of integration in HIV-infected individuals led to groundbreaking discoveries and provided the first demonstration that integrated DNA existed in resting CD4+ T cells in infected individuals (14, 25, 31). However, these methods were laborious and had weaknesses that prevented their widespread application to clinical samples. For example, end point PCR is a semiquantitative technique and thus is not amenable to robust quantitation. Both methods lacked a rigorous integration standard. In addition, the Alu method failed to account for events lying too far away from an Alu site to be detected. Neither method provided rigorous background controls to account for unintegrated DNA.
Given the above limitations, several groups tried to improve the quantitation of integration by adapting the endpoint Alu-gag based PCR assay approach as first described by Chun et al (14, 25, 31, 36). These modified assays (26, 27, 29, 37) added a kinetic PCR step, and provided an integration standard as well as a background gag only control to enhance quantitation and control for false positives. Nonetheless, these attempts to improve the original assays resulted in inferior sensitivity.
We recently developed an Alu-gag PCR based assay for HIV integration that has the sensitivity of the endpoint PCR assay, but also provides robust quantitation. We chose to develop the Alu-gag PCR method because it is the least labor intensive, least susceptible to lack of detection (i.e., does not require digestion with a restriction endonuclease), and thus the most amenable to adaptation for clinical use. We overcame the sensitivity limit by incorporating a repetitive sampling step (38) and by additional modifications that enhance sensitivity by forgoing a large dynamic range (39). Repetitive sampling also provides a means to calculate confidence intervals (39). Thus, our new assay should provide a measurement that will allow hypothesis testing to determine, for example, if a treatment regimen reduces the level of integrated DNA within a cellular subset.
Using this approach, we recently showed that patients on HAART have lower levels of integration than patients off HAART (39). This suggests that monitoring patients over time might show that integration is decreased with HAART. In addition, we have demonstrated routine detection of as few as ~0.5 provirus in 10,000 cellular genomes.
Assays to measure HIV integration have improved in a stepwise fashion over several years. Below we present the assay we currently perform. We also mention future directions that could lead to further improvements in assay sensitivity.
2. Description of method
2.1 Overview
Our two-step, Alu-gag PCR assay for detection and quantitation of integrated HIV DNA is depicted schematically in Figure 1. After isolating total DNA from HIV-infected cells, the first PCR is performed with one primer that anneals to Alu, and the other that anneals to gag. Alu is a repeat element in the human genome that occurs approximately every 5,000 base pairs (40, 41). Gag is an HIV gene that encodes for the structural components of the virion particle (42). The second PCR detects HIV-specific products by using primers to the R and U5 regions within the HIV long terminal repeat (LTR).
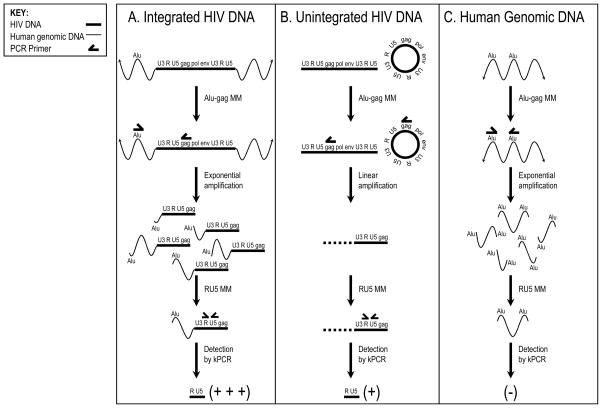
Fig. 1.
Alu-gag PCR integration assay overview. The assay is performed on isolated DNA. First, the Alu-gag master mixture (MM) is added, which contains primers that bind to Alu and gag. Then, the first PCR round is performed, exponentially amplifying integrated HIV DNA, as Alu and gag primers anneal to opposite strands of the same template DNA (panel A). (B) Unintegrated HIV DNA (both circular and linear forms) is only amplified linearly, because the gag primer anneals to only one strand of the HIV template. (C) Alu-Alu segments of human genomic DNA are also exponentially amplified when the elements are sufficiently close together and in opposite orientation. The Alu-Alu amplicons will not be detected, however; only amplicons that contain HIV DNA are detected because a second nested kinetic PCR (kPCR) is performed using a forward primer to R and a reverse primer to U5 and a molecular beacon complementary to the intervening sequence of the HIV LTR. Integrated HIV DNA (panel A) is amplified exponentially and thus more efficiently than unintegrated DNA (panel B). To measure the background signal or the signal expected from unintegrated DNA, a control reaction using only the gag (and not the Alu) primer is included in the first PCR (not shown). This is done in every assay to define the background. The signals from Alu-gag and gag-only are compared to make sure the Alu—gag signal is stronger, and thus that the sample is positive for integration. Then a correlation is developed that relates the kinetic PCR signals to proviral level, which is used to quantify the level of integration in the samples.
In the case of integrated HIV DNA (Fig. 1A), the Alu primer serves as an anchor in the human genome and the gag primer serves as an anchor in the HIV genome. Binding sites for these two primers will be present in the DNA target template only when HIV has integrated into the human genome. Furthermore, when the primer binding sites are close enough and aligned correctly, the region between them can be amplified exponentially. The first PCR is followed by a second, HIV-specific, real-time or kinetic PCR step to quantify the level of Alu-gag amplicons produced in the first round of PCR. Quantification is achieved by comparing the resultant signals to those obtained with an integration standard (IS).
In contrast to integrated DNA, which is selectively and exponentially amplified in this PCR strategy, unintegrated HIV DNA, whether linear or circular, is only linearly amplified (Fig. 1B). This is because, although the Alu and gag primers bind to their cognate sites, the primers are not bound to the same DNA template. In this way, integrated HIV DNA (Fig. 1A) is always preferentially amplified in comparison to unintegrated HIV DNA (panel B). This, in turn, results in a stronger signal from Alu-gag amplicons than gag-only amplicons in the second PCR step. Not explicitly shown in the schematic is a control PCR performed with only the gag primer added. This serves as a background measurement and approximates the signal expected from unintegrated HIV DNA.
Finally, human genomic DNA is not detected by the second PCR step (Fig. 1C). Although the Alu primers bind to the same DNA target template resulting in exponential amplification, these Alu-Alu amplicons are not detected by the second PCR step because it is HIV-specific.
2.2 Detailed protocols
2.2.1 Preparation of an IS
HIV integration occurs at numerous positions throughout the host cell genome. It is therefore essential to prepare the assay standard that best mimics the natural polyclonality of integration during infection. Using our assay, when integration occurs nearby an Alu site, amplification occurs more efficiently than when HIV integrates further away from an Alu site. With a polyclonal standard we are able to compensate for this heterogeneity in our proviral estimates. To prepare the IS, we used a retroviral construct (kind gift of Bob Siliciano) that is engineered to only allow a single round of infection (11). In this engineered virus, HIV env is deleted and replaced with a cassette that encodes for green fluorescent protein (GFP) followed by an IRES and hygromycin resistance gene. Since this construct only allows a single round of infection, over time we expect that unintegrated DNA will be diluted to undetectable levels while integrated HIV DNA will remain stably associated with the cell as it doubles.
2.2.1.1 Validation of our IS
We performed several steps to validate our IS sample. In agreement with our expectations, we demonstrated that, by one week post-infection, only integrated DNA could be detected by probing a southern blot of the IS with HIV specific sequences, which showed that HIV DNA co-localized with chromosomal DNA after gel electrophoresis of undigested genomic DNA (not shown, but described in (37)). Before preparing the IS DNA, we purified GFP positive cells by fluorescence-activated cell sorting and detected approximately ~1 copy of HIV DNA/cell using single round HIV-specific kinetic PCR. We also determined that the integration insertion sites in the prepared IS sample were, as predicted, polyclonal by performing a southern blot on the Alu-gag amplicons with an HIV-specific probe. This demonstrated a range of amplicon lengths, reflecting the range of integration site distances to Alu elements within the infected cell population. These last three steps are not necessary, but we performed them to validate our assay conceptually.
2.2.1.2 Method for IS sample preparation
Prepare single-round transducing virus by transfecting 293T cells with HIV ΔenvhygromycinGFP and pVSV-G (pHIT) plasmid DNAs using the standard CaPO4 transfection protocol as described (11). We use 29 μg pHIV ΔenvhygromycinGFP and 1μg pVSV-G for a 10 cm plate.
Harvest viral supernatants 48 h after transfection.
Infect CEMss cells with the viral supernatant by spinoculation as described (43). Briefly, resuspend cells at 5×106/mL of viral supernatant and spin in one well of a 6 well plate at 1200 g for 2 h at 25 °C. Wash the cells once after spinoculation.
Add 300 μg/mL hygromycin to cells 3 d after infection.
Add 600 μg/mL hygromycin to cells 5 d after infection.
Culture the cells for 4 weeks.
Prepare genomic DNA from infected cells using the Qiagen Blood and Cell culture maxi prep (we isolate DNA from 100 million cells).
Perform HIV-specific kinetic PCR (the RU5 assay; section 2.2.2.1.2 below) on the isolated IS DNA. Include in separate wells in this reaction several known dilutions of an HIV copy standard (e.g., genomic DNA isolated from 8E5 cells (44), which harbor a single integrated provirus, or known copies of a plasmid molecular clone).
Collect cycle threshold (Ct) data. To do this, a horizontal line called the fluorescence threshold is placed above the background noise and near the middle of the exponential amplification region using the kinetic PCR software. The Ct is the cycle number where the PCR amplification curve (accumulating fluorescence) crosses the specified horizontal line, or fluorescence threshold.
To determine the number of HIV proviruses within the IS, create a standard curve with the data collected from the HIV copy standard (e.g. 8E5) by plotting HIV copy number against Ct. This curve is used to quantify the number of viral DNA copies in the IS. We assume all HIV DNA copies are integrated in the IS sample since we demonstrated that only integrated DNA was detectable by southern blot.
By using the β-globin assay detailed below (section 2.2.2.1.3), calculate the number of cells assayed in step 8. Use this to calculate proviruses/cell for the IS. This will be used later for making known dilutions of the IS sample for the creation of the IS curve. As mentioned above, we detected ~1 copy of HIV DNA per cell in our IS after sorting for GFP-positive cells. However, when repeating this protocol, the value could be somewhat greater or less than ~1, depending on the multiplicity of infection at the time of inoculation.
2.2.2 Reagents
2.2.2.1 Primers and probes
Below is a list of the primers and probes used in this protocol. We designed HIV-specific primers using the Los Alamos database (34). We chose to use a standard Alu primer that was described in the literature.(45–47). We scanned the entire gag gene, found the most conserved region, and designed our gag primer to this region. We took the same approach to determine the primers for the R and U5 regions by looking at the entire LTR to find the most conserved area, then designing our nested PCR primers and probe to bind these regions. We included two degenerate probes to account for the two most common mutations in the probe site, which were unlinked. The number of sequences analyzed and the percent identity are described in (39). Also listed below are the primers that we use in the β-globin assay for determining the concentration of human genomes in the sample DNA.
2.2.2.1.1 Alu-gag PCR
-
1. Alu (Forward):
5′ GCC TCC CAA AGT GCT GGG ATT ACA G-3′
-
2. HIV gag (Reverse): nucleotides (nt) 1505-1486
5′ GTT CCT GCT ATG TCA CTT CC-3′
2.2.2.1.2 Quantitative real-time (kinetic) RU5 PCR
To date, the sequences in the Los Alamos database (34) for the LTR region of the HIV genome are somewhat limited. However, with the advent of pyrosequencing, we expect that more sequences will soon be available. As more sequences are added to the database, it may be necessary to further refine the primers and probes for this step.
-
3. RU5 (R Forward): nt 518-539
5′-TTA AGC CTC AAT AAA GCT TGC C-3′
-
4. RU5 (U5 Reverse): nt 647-628
5′-GTT CGG GCG CCA CTG CTA GA-3′
-
5. RU5wildtype Probe: nt 584-559
5′-CCA GAG TCA CAC AAC AGA CGG GCA CA-3′
-
6. RU5degenerate1 Probe: nt 584-559
5′-CCA GAG TCA CAT AAC AGA CGG GCA CA-3′
-
7. RU5degenerate2 Probe: nt 584-559
5′-CCA GAG TCA CAC AAC AGA TGG GCA CA-3′
2.2.2.1.3 β-globin kinetic PCR
-
8. β-Globin (Forward)
5′-CCC TTG GAC CCA GAG GTT CT-3′
-
9. β-Globin (Reverse)
5′-CGA GCA CTT TCT TGC CAT GA-3′
-
10. β-Globin Probe
5′-GCG AGC ATC TGT CCA CTC CTG ATG CTG TTA TGG GCG CTC GC-3′
2.2.2.2 Master mixtures
We prepare a large volume of PCR master mixture once a year to minimize systematic variation. The master mixtures contain all the reagents required for PCR except the Platinum Taq enzyme. The master mixture should be aliquoted and stored at −20°C for up to 12 months. At the time of PCR, the master mixture is mixed with the DNA sample and the Platinum Taq enzyme and the PCR protocol is run with this mixture. Before using the new master mixture, we always compare it to an old master mixture at several dilutions of our IS, to check for consistent amplification efficiency between master mixtures. We add the Platinum Taq enzyme immediately before performing PCR in order to optimally preserve enzyme activity and store it separately at −20°C until use. Specifically, we add 1 part Taq Polymerase (5 U/μL) for the β-globin assay, and 2 parts Taq Polymerase for the others (Alu-gag, gag only, and RU5).
The following recipes make 2x master mixtures. When we set up the reaction, we add equal amounts of sample and master mixture to the wells, creating a 1x master mixture during PCR. Specifically, for RU5 we add 10 μL sample and 10 μL master mixture to each well (total reaction volume = 20 μL). For the other reactions (β-globin, Alu-gag, and gag only) we add 25 μL sample and 25 μL master mixture to each well (total reaction volume = 50 μL).
2.2.2.2.1 2x β-globin master mixture
Add together the following:
58 parts H2O
21 parts 10X PCR buffer
14 parts 50 mM MgCl2
0.4 parts 100 mM dATP
0.4 parts 100 mM dTTP
0.4 parts 100 mM dGTP
0.4 parts 100 mM dCTP
2 parts 100 μM β-globin Forward Primer
2 parts 100 μM β-globin Reverse Primer
0.4 parts 100 μM β-globin Probe
2.2.2.2.2 2x Alu-gag master mixture
Add together the following:
56.32 parts H2O
20 parts 10X PCR buffer
6 parts 50 mM MgCl2
0.42 parts 100 mM dATP
0.42 parts 100 mM dTTP
0.42 parts 100 mM dGTP
0.42 parts 100 mM dCTP
2 parts 10 μM Alu-forward primer
12 parts 10 μM gag-reverse primer
2.2.2.2.3 2x gag-only master mixture
Add together the following:
58.32 parts H2O
20 parts 10X PCR buffer
6 parts 50 mM MgCl2
0.42 parts 100 mM dATP
0.42 parts 100 mM dTTP
0.42 parts 100 mM dGTP
0.42 parts 100 mM dCTP
12 parts 10 μM gag-reverse primer
2.2.2.2.4 2x RU5 master mixture
Add together the following:
55.36 parts H2O
20 parts 10X PCR buffer
14 parts 50 mM MgCl2
0.6 parts 100 mM dATP
0.6 parts 100 mM dTTP
0.6 parts 100 mM dGTP
0.6 parts 100 mM dCTP
0.52 parts 100 μM RU5 Forward primer
0.52 parts 100 μM RU5 Reverse primer
0.4 parts 100 μM RU5wildtype Probe
0.4 parts 100 μM RU5degenerate1 Probe
0.4 parts 100 μM RU5degenerate2 Probe
4 parts 25 μM carboxy-X-rhodamine (ROX Reference Dye; Invitrogen)
2.2.2.3 Enzyme
We use Platinum Taq DNA Polymerase (Invitrogen Life Technologies). Since different enzyme lots can affect the efficiency of PCR, we prescreen small samples of several lots. We determine the lot with the best activity and then buy sufficient quantity for one year of reactions.
2.2.3 DNA isolation from infected cells
We use the QIAamp DNA Micro Kit as described in the QIAamp DNA Micro Handbook (“Isolation of Genomic DNA from Small Volumes of Blood”), except that we dilute our final sample in 10 mM Tris-HCl pH 8.0 rather than Buffer AE.
2.2.4 β-globin PCR assay
To estimate the number of proviruses per cell, first determine the concentration of human genomes in the isolated sample DNA. For this, our lab uses a β-globin-based PCR assay, described below. Alternatively, OD260/280 can be used.
-
Make the β-globin standard using uninfected DNA isolated from peripheral blood mononuclear cells (PBMCs).
Starting at 24 μg/mL PBMC DNA in 10 mM Tris-HCl pH 8.0, serially dilute at steps of 1:10 to obtain five standard concentrations (24μg/mL – 24/104 μg/mL) in 10 mM Tris-HCl pH 8.0. The top dilution contains 180,000 diploid genomes/25 μL.
Dilute test DNA samples to the desired concentration in 10 mM Tris-HCl pH 8.0.
-
Prepare β-globin master mixture + Platinum Taq enzyme.
Estimate 25 μL for each well (including standard and a no template control) of β-globin master mixture.
Obtain the correct amount of β-globin master mixture, and add 1 part Platinum Taq polymerase enzyme per 99 parts master mixture.
Add 25 μL β-globin master mixture + Platinum Taq to each well of a 96-well PCR plate.
Add the standard (PBMC DNA dilutions) to the appropriate wells, and include a no template control (10 mM Tris-HCl buffer).
Add 25 μL of DNA sample dilutions.
-
Run the following kinetic PCR program. We perform this assay on the MJ research Opticon, which was subsequently purchased by Biorad. The MyiQ™ Single-Color Real-Time PCR Detection System sold by Biorad is the equivalent instrument.
Incubate at 95°C for 2 min
Incubate at 95°C for 15 sec
Incubate at 60°C for 30 sec
Plate read (i.e., the level of fluorescence is recorded)
Incubate at 72°C for 1 min
Return to line 2 for 39 additional iterations (40 total cycles)
End
Collect Ct data as explained above in section 2.2.1.2, step 9. As discussed, the Ct is the cycle number where the fluorescence of each well (PCR amplification curve) crosses the specified threshold.
Calculate the genomic concentrations of samples by making a standard curve relating the number of genomes to Ct using the known dilutions of PBMC DNA. Note that 24 μg/mL PBMC DNA contains 1.8 × 105 genomes in 25 μl. The calculated concentrations will later be used to determine provirus copy number per cell.
2.2.5 Alu-gag (and gag-only) PCR
-
Dilute samples to 2 μg/mL DNA. This equates to 1.5 × 104 cells/well (1 well = 25μL). Perform ~40 repeats each of Alu-gag and gag-only measurement per sample.
Always use 1.5 × 104 cells/well for this step (including standards and controls). This is necessary to keep the concentration of Alu DNA sites constant in each reaction, so that PCR efficiency will not be affected. If necessary, this can be accomplished by the addition of uninfected PBMC DNA. Make sure to take this into account in the calculations.
Allocate 12 wells for standards and interassay variation controls. Use three of these in the first PCR round (the rest will be used in the second round). In each of these three wells, assay 1,000 copies of the IS (in 1.5 × 104 uninfected PBMCs).
-
Prepare the Alu-gag and gag-only master mixtures.
Make enough for 25 μL per well.
Add 2 parts Platinum Taq per 98 parts master mixture (1/50 volume) to both master mixtures.
Add 25 μL of each master mixture and sample/control to appropriate wells.
-
Run the following PCR program:
Incubate at 95°C for 2 min
Incubate at 95°C for 15 sec
Incubate at 50°C for 15 sec
Incubate at 72°C for 3.5 min
Return to line 2 for 39 repeats
End
2.2.6 Detection (RU5) kinetic PCR
-
Prepare the RU5 master mixture.
Make enough for 10 μL per well.
Add 1/50 volume of enzyme to master mixture.
Add 10 μL master mixture (with enzyme) to wells to be used in a 96-well PCR plate.
-
Add 10 μL of each well from the first PCR product to its appropriate well in the new plate.
-
Here, add the following additional controls:
3 repeats of 1,000 copies of IS/well, that was not pre-amplified in the first PCR.
Three dilutions of a copy standard, such as 8E5 DNA. For example, 3, 0.3, and 0.03 μg/mL (2,000, 200, and 20 copies/well, respectively).
Three repeats of sample, that was not pre-amplified.
-
-
Run the following kinetic PCR protocol:
Incubate at 95°C for 20 sec
Incubate at 95°C for 3 sec
Incubate at 60°C for 30 sec
Go to Line 2 for 49 times more
End
Collect Ct data in an excel spreadsheet as described above in section 2.2.1.2, step 9.
Proceed with the analysis outlined below.
2.2.7 Quantitation
2.2.7.1 Making a standard curve
Make a standard curve by running the nested PCR protocol on several dilutions of the IS DNA sample. We use 40 replicates of both Alu-gag and gag-only per dilution, for 8 different dilutions ranging from 1.25 provirus/well to 160 provirus/well (dilute in uninfected PBMC DNA). To make a standard curve, proceed with the following steps:
Determine the natural log (Ln) of the Alu-gag Cts for each well.
For each dilution, average these values, and plot them versus the natural log of provirus/well. This should be a linear correlation.
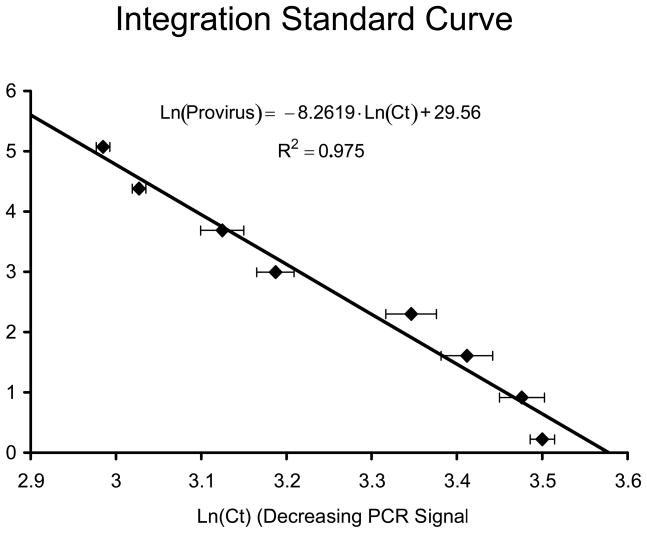
Determine the linear regression of the data. This provides a standard curve similar to that seen in Figure 2, of the form: Ln(Provirus/well) = A·Average(Ln(Ct))+ B, where A and B are constants defined by the linear regression.
Fig. 2.
Integration standard curve. Shown is the curve that we developed for use in estimating proviral levels in samples from infected individuals. The natural log (Ln) of HIV DNA copy number is plotted on the y-axis against the Ln of cycle threshold (Ct) on the x-axis.
2.2.7.2 Using a Standard Curve
The level of HIV integration in the different samples is calculated using the standard curve. First, perform the nested PCR protocol described above and obtain Ct values. Then, follow these steps:
Perform a Student’s t-test to determine that the Cts derived from the Alu-gag PCRs are significantly lower than those from the gag-only reactions. If this is not the case, the level of integration in these samples is below the detection limit of the assay using the correlation method we describe here (see note below). If Alu-gag is statistically lower than gag-only (p < 0.05), proceed with the next step.
Determine the Ln of the Alu-gag Cts, and average these for each sample.
-
Calculate the number of provirus/well by the equation:
where A and B were determined in section 2.2.7.1.
- Calculate 95% confidence intervals by the equation:
The number of provirus/cell is calculated using the number of cells/well determined from the β-globin PCR assay (section 2.2.4).
2.2.8 Notes
The assay described here was specifically designed for use with samples derived from HIV-infected patients. However, it can easily be adapted for experiments performed in basic virology research laboratories, for example, to determine the level of HIV integration in tissue culture cells infected ex vivo. The biggest change is that the PCR primers should be chosen based on the particular virus isolate, rather than by consulting the Los Alamos Database (34) for conserved regions of HIV DNA. With new primers, a new standard curve will need to be generated. However, the rest of the procedure and analysis is the same.
The dynamic range of this assay can be expanded by reducing the number of cycles in the first PCR round from 40 to 20 (38). This will result in less sensitivity, but allows measurement of integration at higher copy numbers. We demonstrated using previously published primers specific for the NL4-3 strain of HIV-1 (38) that the dynamic range of our assay is ~1 to 10,000 proviruses when 20 cycles of Alu-gag PCR are performed in the first reaction.
2.2.8.1 Sensitivity limit of the assay
As the assay is currently described, with 40 Alu-gag repeats, the sensitivity limit with patient samples is ~0.5 proviruses among 10,000 genomes (39). The sensitivity limit is determined by the point when the average Alu-gag signal is no longer statistically different from the gag-only signal. The enhancement in sensitivity is proportional to the number of experimental replicates analyzed in each experiment.
2.2.8.2 Future Directions
It should be possible to enhance the sensitivity of our assay further by simply performing more repeats. In other words, the sensitivity of the assay is only limited by the number of repeats we are willing to perform given that we detect integration approximately 10% of the time (38, 39). However, to detect integration below 0.5 proviruses in 10,000 genomes, it is no longer feasible to correlate the average Alu-gag Ct with proviral copy number because the Alu-gag and gag only signals on average no longer statistically differ. Nonetheless, individual Alu-gag signals are statistically different than the gag only signal.
We are in the process of testing a new mathematical correlation and statistical tools to allow us to detect integration at levels below 0.5 in 10,000 genomes. In this method, we record the percent positive samples and use the binomial distribution to estimate the errors. Positive samples are determined by showing that they differ by greater than ~2 standard deviations from the gag only signal. Using this approach, we have detected as few as 1 provirus in 100,000 cells (unpublished observations). The downside to this approach is the number of samples required. For example, to detect one provirus in 100,000 genomes, we performed 200 Alu-gag PCRs to obtain 5 positive signals.
3. Concluding remarks
We present a method for measuring HIV integration that is more accurate, more sensitive, and more suitable for clinical use than previous assays. HIV integration levels in patients’ PBMCs have only been monitored in a few studies longitudinally (18, 48, 49), and no study has monitored the level of integrated DNA in different CD4+ cell subsets over time. Furthermore, the longitudinal studies that were performed monitoring integration in PBMCs have provided conflicting results regarding the effect of HAART on integrated DNA levels. This may be related to potentially wide and uncertain errors inherent in the assays utilized. With improved methods to measure integration that provide precise confidence intervals, it will be possible to monitor more precisely the level of integration in patients to conclusively determine if it is affected by therapy. The use of the repetitive sampling technique improves the Alu-gag PCR assay by both enhancing sensitivity and permitting calculation of confidence intervals. Thus, this method should provide a means to robustly measure integration and perform rigorous hypothesis testing for the first time within HIV-infected individuals on different therapeutic regimens. Furthermore, these methods may provide new information about the level and the half-life of integrated DNA in CD4+ T and non T cell subsets in the presence of therapy. By combining these measurements with measures of total DNA, important information regarding the potential for ongoing HIV replication within different subsets may be obtained. This in turn should provide a deeper understanding of the differential effect of antiretroviral therapy on different cell subsets.
Acknowledgments
We would like to thank Troy Brady, Bruce Levine, Liz Colston, Wei Yang and Luis Agosto for their assistance in preparation of this manuscript. This study was supported in part by NIH grant R01 AI058862-01 (U.O.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ansari-Lari MA, Donehower LA, Gibbs RA. Virology. 1995;211:332–5. doi: 10.1006/viro.1995.1412. [DOI] [PubMed] [Google Scholar]
- 2.Brown PO. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- 3.Cara A, Guarnaccia F, Reitz MS, Gallo RC, Lori F. Virology. 1995;208:242–8. doi: 10.1006/viro.1995.1148. [DOI] [PubMed] [Google Scholar]
- 4.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. Journal of Virology. 1995;69:2729–36. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Englund G, Theodore TS, Freed EO, Engelman A, Martin MA. Journal of Virology. 1995;69:3216–19. doi: 10.1128/jvi.69.5.3216-3219.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakai H, Kawamura M, Sakuragi J, Sakuragi S, Shibata R, Ishimoto A, Ono N, Ueda S, Adachi A. Journal of Virology. 1993;67:1169–74. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson M, Haggerty S, Lamonica CA, Meier CM, Welch SK, Wasiak AJ. Journal of Virology. 1990;64:2421–5. doi: 10.1128/jvi.64.5.2421-2425.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. European Molecular Biology Organization Journal. 1990;9:1551–60. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiskerchen M, Muesing MA. Journal of Virology. 1995;69:376–86. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vatakis DN, Bristol G, Wilkinson TA, Chow SA, Zack JA. J Virol. 2007;81:3574–82. doi: 10.1128/JVI.02569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swiggard WJ, Baytop C, Yu JJ, Dai J, Li C, Schretzenmair R, Theodosopoulos T, O’Doherty U. J Virol. 2005;79:14179–88. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 13.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Nature. 1997;387:183–88. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 15.Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Nat Rev Microbiol. 2007;5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- 16.Shen L, Siliciano RF. J Allergy Clin Immunol. 2008;122:22–8. doi: 10.1016/j.jaci.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer S, Wiegand AP, Maldarelli F, Bazmi H, Mican JM, Polis M, Dewar RL, Planta A, Liu S, Metcalf JA, Mellors JW, Coffin JM. Journal of Clinical Microbiology. 2003;41:4531–6. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R, Robbins GK, D’Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. J Infect Dis. 2008;197:411–9. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 19.Ostrowski MA, Chun TW, Justement SJ, Motola I, Spinelli MA, Adelsberger J, Ehler LA, Mizell SB, Hallahan CW, Fauci AS. Journal of Virology. 1999;73:6430–35. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellery PJ, Tippett E, Chiu YL, Paukovics G, Cameron PU, Solomon A, Lewin SR, Gorry PR, Jaworowski A, Greene WC, Sonza S, Crowe SM. J Immunol. 2007;178:6581–9. doi: 10.4049/jimmunol.178.10.6581. [DOI] [PubMed] [Google Scholar]
- 21.Coffin JM. Science. 1995;267:483–89. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 22.KewalRamani VN, Coffin JM. Science. 2003;301:923–5. doi: 10.1126/science.1088965. [DOI] [PubMed] [Google Scholar]
- 23.Achaz G, Palmer S, Kearney M, Maldarelli F, Mellors JW, Coffin JM, Wakeley J. Molecular Biology & Evolution. 2004;21:1902–12. doi: 10.1093/molbev/msh196. [DOI] [PubMed] [Google Scholar]
- 24.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT, Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. Journal of Clinical Microbiology. 2005;43:406–13. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Proceedings of the National Academy of Sciences USA. 1997;94:13193–97. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler SL, Hansen MST, Bushman FD. Nature Medicine. 2001;7:631–34. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 27.Brussel A, Sonigo P. J Virol. 2003;77:10119–24. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carr JM, Cheney KM, Coolen C, Davis A, Shaw D, Ferguson W, Chang G, Higgins G, Burrell C, Li P. J Clin Microbiol. 2007;45:1288–97. doi: 10.1128/JCM.01926-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brussel A, Delelis O, Sonigo P. Methods Mol Biol. 2005;304:139–54. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- 30.Lehrman G, Hogue IB, Palmer S, Jennings C, Spina CA, Wiegand A, Landay AL, Coombs RW, Richman DD, Mellors JW, Coffin JM, Bosch RJ, Margolis DM. Lancet. 2005;366:549–55. doi: 10.1016/S0140-6736(05)67098-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. Nature Medicine. 1995;1:1284–90. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 32.Lewis P, Hensel M, Emerman M. European Molecular Biology Organization Journal. 1992;11:3053–58. doi: 10.1002/j.1460-2075.1992.tb05376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandegraaff N, Kumar R, Burrell CJ, Li P. Journal of Virology. 2001;75:11253–60. doi: 10.1128/JVI.75.22.11253-11260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leitner TFB, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B. Published by Theoretical Biology and Biophysics Group,; 2005. http://www.hiv.lanl.gov/ [Google Scholar]
- 35.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Nature. 1997;387:183–88. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 36.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. Nat Med. 1995;1:1284–90. doi: 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 37.O’Doherty U, Swiggard WJ, Jeyakumar D, McGain D, Malim MH. J Virol. 2002;76:10,942–10,50. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agosto LM, Yu JJ, Dai J, Kaletsky R, Monie D, O’Doherty U. Virology. 2007;368:60–72. doi: 10.1016/j.virol.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu JJ, Wu TL, Liszewski MK, Dai J, Swiggard WJ, Baytop C, Frank I, Levine BL, Yang W, Theodosopoulos T, O’Doherty U. Virology. 2008;379:78–86. doi: 10.1016/j.virol.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelinek WR, Schmid CW. Annual Review of Biochemistry. 1982;51:813–44. doi: 10.1146/annurev.bi.51.070182.004121. [DOI] [PubMed] [Google Scholar]
- 41.Mighell AJ, Markham AF, Robinson PA. FEBS Letters. 1997;417:1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 42.Vogt VM. In: Retroviruses. Coffin JM, Hughes SH, Varmus HE, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 27–69. [Google Scholar]
- 43.O’Doherty U, Swiggard WJ, Malim MH. J Virol. 2000;74:10074–80. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Folks TM, Powell D, Lightfoote M, Koenig S, Fauci AS, Benn S, Rabson A, Daugherty D, Gendelman HE, Hoggan MD, Venkatesan S, Martin MA. Journal of Experimental Medicine. 1986;164:280–90. doi: 10.1084/jem.164.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benkirane M, Corbeau P, Housset V, Devaux C. EMBO Journal. 1993;12:4909–21. doi: 10.1002/j.1460-2075.1993.tb06185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roy-Engel AM, Carroll ML, Vogel E, Garber RK, Nguyen SV, Salem AH, Batzer MA, Deininger PL. Genetics. 2001;159:279–90. doi: 10.1093/genetics/159.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batzer MA, Deininger PL. Nature Reviews Genetics. 2002;3:370–9. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 48.Izopet J, Cazabat M, Pasquier C, Sandres-Saune K, Bonnet E, Marchou B, Massip P, Puel J. Virology. 2002;302:393–404. doi: 10.1006/viro.2002.1621. [DOI] [PubMed] [Google Scholar]
- 49.Ibanez A, Puig T, Elias J, Clotet B, Ruiz L, Martinez MA. Aids. 1999;13:1045–9. doi: 10.1097/00002030-199906180-00007. [DOI] [PubMed] [Google Scholar]