Abstract
Immune responses during infection with pandemic H1N1 2009 influenza A virus (2009-H1N1) are still poorly understood. Using an experimental infection model in ferrets, we examined the pathological features and characterized the host immune responses by using microarray analysis, during infection with 2009-H1N1 A/California/07/2009 and seasonal A/Brisbane/59/2007. Chemokines CCL2, CCL8, CXCL7 and CXCL10 along with the majority of interferon-stimulated genes were expressed early, correlated to lung pathology, and abruptly decreased expression on day 7 following infection of A/California/07/2009. Interestingly, the drop in innate immune gene expression was replaced by a significant increase of the adaptive immune genes for granzymes and immunoglobulins. Serum anti-influenza antibodies were first observed on day 7, commensurate with the viral clearance. We propose that lung pathology in humans occurs during the innate phase of host immunity and a delay or failure to switch to the adaptive phase may contribute to morbidity and mortality during severe 2009-H1N1 infections.
Keywords: influenza, 2009 H1N1, ferrets, immune response, microarrays, cytokines
In the spring of 2009, human transmission of a new influenza A strain was reported in the United States and Mexico (2009a; 2009b; 2009c; 2009d; Cameron, Rowe, and Kelvin, 2009; Chowell et al., 2009; Dawood et al., 2009; Perez-Padilla et al., 2009; Shinde et al., 2009), within a few weeks it spread to other regions of the globe and the World Health Organization raised the pandemic alert phase to “level 6, of moderate severity” (http://www.who.it/influenza/AH1N1/20090611_11). By the end of November, worldwide more than 207 countries and overseas territories had reported laboratory confirmed cases of pandemic H1N1 2009 influenza A virus (2009-H1N1), including over 7820 deaths (http://www.who.int/csr/don/2009_11_27a/en/index.html). This influenza virus is a triple-reassortment of multiple strains of influenza A viruses circulating in the domestic swine population (Smith et al., 2009). While the vast majority of human cases appear to be self limiting and resolve within two weeks of onset of symptoms, a small number of infections lead to severe respiratory illness. Symptoms include fever and cough and possible sore throat, diarrhea and vomiting. In a minority of cases, 6-10 days following onset of symptoms, severe lower respiratory complications arise with radiographic abnormalities. Hospitalization and ventilation in these cases may be required. Fatal cases, while extremely rare, are most often associated with underlying medical conditions that contribute to illness severity. Interestingly, histological examination of a lung specimen from a severe case of 2009-H1N1 showed necrotizing bronchiolitis with alveolar damage and hyaline formation (Perez-Padilla et al., 2009). This finding is similar to histopathological findings in human influenza infected lung specimens from previous pandemics (Taubenberger and Morens, 2008).
The antigenic nature of 2009-H1N1 is distinct from seasonal human A (H1N1) viruses such as A/Brisbane/59/2007, but is similar to circulating North American swine A (H1N1) viruses (Garten et al., 2009). The antigenic dissimilarity between 2009-H1N1 and seasonal H1N1 viruses suggests that the general population has little or no preexisting immunity to 2009-H1N1. This notion is reinforced since some seasonally vaccinated individuals develop classic influenza symptoms and illness caused by 2009-H1N1 infection. In the absence of preexisting immunity in the general population, the modest mortality and morbidity associated with the majority of human 2009-H1N1 infections are puzzling. The paucity of information regarding host immune responses to 2009-H1N1 and the role they play in controlling the viral infection led us to investigate the evolution of innate and adaptive host immune responses following 2009-H1N1 exposure.
To better understand the natural history of 2009-H1N1 infection and underlying immunological, clinical, pathological and molecular features of disease course, we infected twenty-nine ferrets with 106 50% egg infectious doses of the 2009-H1N1 vaccine strain, A/California/07/2009, and observed for clinical signs, changes in body temperature and weight on a daily basis. Additionally we looked for viral replication in respiratory tissue, pathology, host innate and adaptive immune responses and changes in gene expression. The study was completed with data obtained from ferrets infected with seasonal H1N1 influenza A/Brisbane/59/2007, which provided additional insight in a disease that displays different clinical presentations.
Materials and Methods
Infection and monitoring of Ferrets
Animal experiments with virus A/California/07/2009 or with A/Brisbane/59/2007 were performed in the AAALAC-accredited ABSL-3 facility, University of Pittsburgh or in the ABSL-2 enhanced facility Animal Resource Centre, University Health Network, Toronto, respectively. All procedures were conducted in accordance with the NRC Guide for the Care and Use of Laboratory Animals, the Animal Welfare Act, and the CDC-NIH Biosafety in Microbiological and Biomedical laboratories and approved by the Institutional Animal Care and Use Committee. All ferrets were proven seronegative by HI assay against A/California/04/2009, A/Brisbane/59/2007 and B/Florida/04/2006. Castrated, descented male fitch ferrets (Mustela putorius furo), 5 – 9 months of age and weighing between 800 to 1500 grams (Triple F Farms Inc., Sayre, PA, USA) were pair-housed in individually HEPA-Filtered biocontainment caging (Allentown Inc., Allentown, NJ, USA). Ferrets were given food and water ad libitum. Animals were infected and monitored as previously described (Zitzow et al., 2002), except using 5% isofluorane anesthesia. Briefly, 29 male fitch ferrets were anesthetized with isofluorane and infected intranasally with 1×106 50% egg infectious doses (EID50) (~1×105.75 TCID50/ml) of A/California/07/2009 in a volume of one milliliter. From these, four animals were sacrificed on days 1, 3, 5, 7, and 14 days post infection (PI). Nasal washes, lung tissue and blood samples were collected from each ferret in order to determine viral load, gross and histopathological findings, innate and adaptive host immune responses, and molecular changes associated with disease course. The remaining nine animals were allowed to go to endpoint. A/California/07/2009 is the World Health Organization recommended isolate for generation of 2009-H1N1 vaccines (http://www.who.int/entity/csr/resources/publications/swineflu/H1N1Vaccinevirusrecommendation26May2009.pdf) and was thus the virus used for ferret studies. An additional 4 ferrets were used as Day 0 uninfected controls. Temperatures were measured through use of an implantable temperature transponder (BMDS, Sayre, PA) and were recorded daily at approximately the same time each day. Pre-infection values were averaged to obtain a baseline temperature for each ferret. The change in temperature was calculated daily for each animal. Weight loss and clinical signs of sneezing and nasal discharge, other respiratory distress, and level of activity were assessed daily. A scoring system was used to assess activity level where 0=alert and playful; 1=alert but playful only when stimulated; 2=alert but not playful when stimulated; 3=neither alert nor playful when stimulated. Based on the daily scores for each animal in a group, a relative inactivity index was calculated (Zitzow et al., 2002).
An additional 15 ferrets were infected with 1×106 EID50 of A/Brisbane/59/2007 and monitored for clinical signs. On days 1, 2, 5, 7 and 14 post infection, 3 ferrets were euthanized and organs collected for pathological examination, virus titration and microarray analysis. A group of 3 uninfected controls was also included.
Histopathology
Lung tissue excised from the middle lobes was formalin-fixed and paraffin embedded. Tissue slides were stained with hematoxilyn/eosin for histopathologic assessment. Immunostain was performed by using a polyclonal goat anti-influenza A 1:500 (#PAB7123P, Maine Biotechnology Services, Portland, ME, USA) followed by incubation with a donkey anti-goat b/c antibody 1:200 (#805-7602, Rockland immunochemical, Gilberstville, PA, USA).
Determination of viral load
MDCK cells plated in 6-well tissue culture plates were inoculated with 0.1 ml of virus serially diluted in Dubecco’s modified Eagle’s medium (DMEM). Virus was adsorbed to cells for 1 h, with shaking every 15 min. Wells were overlaid with 1.6% w/v Bacto agar (DIFCO, BD Diagnostic Systems, Palo Alto, CA, USA) mixed 1:1 with L-15 media (Cambrex, East Rutherford, NJ, USA) containing antibiotics and 0.6μg/ml TPCK-trypsin (Sigma, St. Louis, MO, USA). Plates were inverted and incubated for 4 days. Cells were fixed for 10 minutes using 70% v/v Ethanol and then overlaid with 1% w/v crystal violet. Cells were then washed with deionized water to visualize plaques. Plaques were counted and compared to uninfected cells. Additionally, viral load was assessed by HA activity in tissues which were homogenized 10% w/v in DMEM and serially diluted (1/2 log) from 10-0.5 to 10-6.5 in quadruplicate on MDCK cells. After 2 hours incubation at 37°C, 5% CO2 samples were removed and replaced with DMEM containing 1% BSA, 1ug/ml TPCK-Trypsin and antibiotics and incubated. On day 6 post infection, fifty microliters of supernatant were tested for presence of virus by HA activity against 0.5% turkey erythrocytes. The viral loads were determined as the reciprocal of the dilution resulting in 50% HA positivity. Titers are given as TCID50 per gram of tissue or TCID50/ml for nasal washes.
Hemagglutinin Inhibition (HI) assay
RDE-treated sera were serially diluted in v-bottom 96-well microtiter plates followed by the addition of 4 Hemagglutination units (HAU) of influenza virus. Following an incubation of approximately 15 minutes, 0.5% turkey erythrocytes (TRBC) were added and mixed by agitation. The TRBC were allowed to settle for 30 minutes at room temperature and HI titers were determined by the reciprocal value of the last dilution of sera which inhibited hemagglutination of TRBC by the virus. A negative titer was calculated as 10.
RNA Extraction and Microarray Analysis
Performed as previously published (Cameron et al., 2008) except for the gene classification methods. Briefly, lung sections (~1 g) were obtained during necropsy and immediately put in RNAlater (Ambion, Austin, TX, USA) then homogenized in TriPure reagent, and total RNA was isolated according to the manufacturer’s recommended protocol (Roche, Indianapolis, IN, USA). Total RNA was amplified using MessageAMP kits (Ambion). cRNA (10 g) was labeled and hybridized to Affymetrix GeneChip Canine Genome 2.0 Array (Affymetrix, Santa Clara, CA, USA). Probe-level analysis was performed using the Probe Logarithmic Intensity Error Estimate (PLIER) method in Expression Console Version 1.1 (Affymetrix). The raw intensity values for each target on the Affymetrix arrays were preprocessed with variance stabilization, log2 transformation, and normalization to the control group (Day 0 samples) excluding elements with median signal intensities of <6 log2 units across samples. Time series analysis was performed using EDGE version 1.1.291 (Storey et al., 2005) with Q ≤ 0.005. DAVID bioinformatic tool (Huang da, Sherman, and Lempicki, 2009) was used to perform functional classification in differentially expressed genes, using a biological significance filter of ≥1.5-fold, additional custom enrichment of functional categories (ISGs, cytokine/chemokines and leukocyte markers) was performed. Gene hierarchical clustering and heat map generation was done using the Multi Experiment Viewer 4.4 software (Saeed et al., 2006). Ingenuity Pathway Analysis 7.6 (IPA) (Ingenuity Systems, Redwood City, CA, USA) was used to select, annotate, and visualize genes by function and pathway. Datasets are publicly available at the NCBI’s Gene Expression Omnibus. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=PENDING).
qRT-PCR
Quantitative real-time PCR (qRT-PCR) was performed in triplicate using an ABI-Prism 7900HT (Applied Biosystems, Foster City, CA, USA). PCR reactions were performed in a 10μl reaction volume using SYBR green master mix (Applied Biosystems), 40 amplification cycles and the annealing temperature was 60 degrees. Primer sequences were as follows: for CCL2, 5′-GCTCCCTATTCACTTGCTGTTTC-3′ (forward) and 5′-GATTCGATAGCCCTCCAGCTT-3′ (reverse); for CXCL10, 5′-CTTTGAACCAAAGTGCTGTTCTTATC-3′ (forward) and 5′-AGCGTGTAGTTCTAGAGAGAGGTACTC-3′ (reverse); for IL-1β, 5′-GGACTGCAAATTCCAGGACATAA-3′ and 5′-TTGGTTCACACTAGTTCCGTTGA-3′ (reverse); for TNFα, 5′-CCAGATGGCCTCCAACTAATCA-3′ (forward) and 5′-GGCTTGTCACTTGGAGTTCGA-3′ (reverse). Expression levels were normalized using the house-keeping gene β-actin, results were expressed as arbitrary units. The statistical analysis was performed using the SPSS 16.0 software (SPSS Inc, Chicago, IL, USA) and the Student’s t test was used to assess the statistical differences in the expression levels respect to the control groups.
Results
Pathological features of the infection with A/California/07/2009 in ferrets
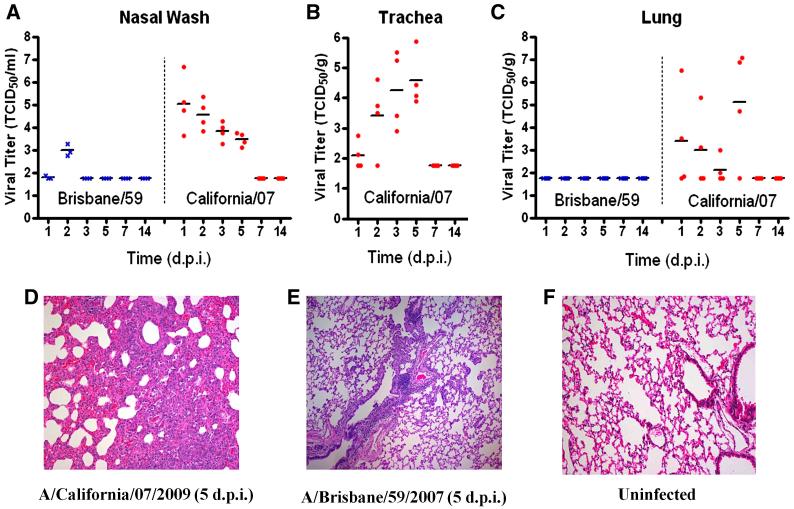
Following challenge with California/07, ferrets exhibited classical clinical signs of influenza infection including elevated temperature, weight loss, nasal discharge, sneeze (table 1 and supplementary Fig. 1) and crusty nose (22% of ferrets on day 6 PI). Animals lost weight throughout the first week post-challenge and slowly recovered by day 14 PI. 3 ferrets on day 6 PI and 2 ferrets on day 7 PI, lost more than 20% of their initial weight and they were euthanized. The resulting mortality rate was 27%. Viral kinetics demonstrated early replication in the upper tract at day 1 PI, which decreased over the following four days (Fig. 1A). Viral load was low on day 1 in the trachea and progressively increased until day 5 PI (Fig. 1B). Viral titers in lung tissue showed great variability, it was initially detected on day 1 and peaked on day 5 PI (Fig. 1C). No virus was detected in any airway tissues on day 7 or later (Fig. 1).
Table 1.
Clinical observations and viral load in ferrets infected with influenza A/Brisbane/59/2007 and A/California/07/2009
| Clinical Signs | Peak Viral loade (Log10)TCID50/g or ml |
|||||
|---|---|---|---|---|---|---|
| Lethalitya | Inactivityb | % Weight Lossc (Day) |
Elevated Tempd (Days) |
Tissue | Peak (Day) | |
| Seasonal (H1N1) A/Brisbane/59/07 |
0% | 1.1 | 5% (D7) | 1.9°C (D2) | Nasal Wash | 3.0±0.3 (D2) |
| Trachea | Not Done | |||||
| Lung | <1.8f | |||||
| 2009-H1N1 A/California/07/09 |
27% | 1.4 | 16% (D6) | 1.1°C - 1.3°C (D2 - D5) |
Nasal Wash | 5.0±1.2 (D1) |
| Trachea | 4.6±0.9 (D5) | |||||
| Lung | 5.1±2.5 (D5) | |||||
>20% weight loss from prechallenge (D0) weight
Relative Inactivity Index (D1 - D6) post Challenge
Day post challenge of peak weight loss from prechallenge weight
Average elevated temperatures above baseline °C and (days post challenge elevated)
Average viral load.
Baseline = 1.8 Log10
Fig. 1.
Viral replication of influenza A/California/07/2009 and A/Brisbane/59 in ferrets following intranasal challenge. (A) Nasal wash, (B) trachea and (C) lungs were collected from ferrets at indicated days post-infection and titered on MDCK cells (TCID50). Hematoxylin and eosin stained sections from paraffin embedded lung tissue: (D) A/California/07/2009, (E) A/Brisbane/59/2007 and (F) uninfected.
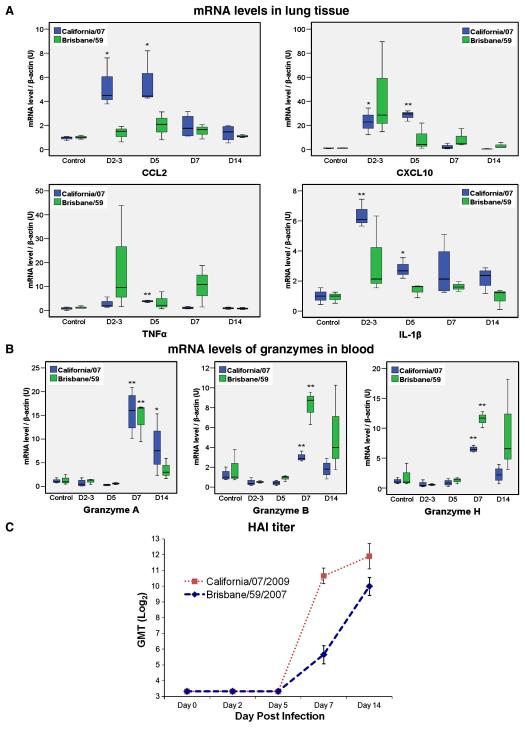
Hemagglutining inhibition assay (HAI) in serum samples revealed the early presence of hemagglutinin inhibiting A/California/07 (HAI) antibodies as soon as day 7 PI (Fig. 5C). This is mostly an IgM response with IgG following at later times (Data not shown).
Fig. 5.
RT-PCR quantitation of differentially expressed genes from ferrets infected with A/California/07/2009 and A/Brisbane/59/2007. A)To validate the expression measurements observed in the H1N1 microarray dataset, we compared the levels of CCL2, CXCL10, IL-1β and TNFα RNA in lung tissue from infected ferrets with uninfected ferret controls during Day 2 (Brisbane/59 only), 3 (California/07 only), 5, 7 and 14 post infection. B) Quantitation of mRNA levels of granzyme A, B and H in blood samples. Data shown is normalized to the amount of endogenous β-actin transcript. *p<0.05, **p<0.01. C) Serum anti-influenza antibodies measured by HI (geometric mean titers).
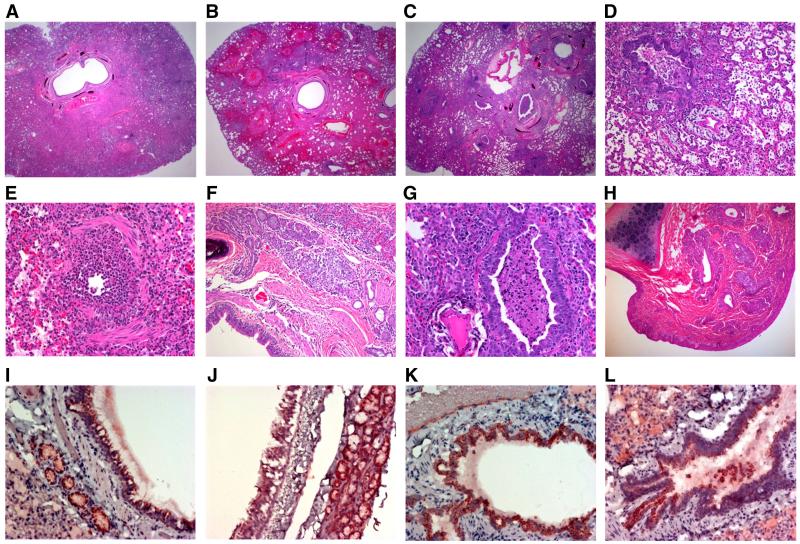
The lungs of California/07 infected ferrets had severe and rapid histopathological changes (Figs. 1D and 2A-L). Early in infection, atelectasis was widespread with intra-alveolar exudate varied from mild to severe (Fig. 2A-C). Bronchial epithelium showed moderate intracellular edema with occasional supra-epithelial exudate containing acute inflammatory cells (Fig. 2E-F). At day 1 post infection, marginating leukocytes invaded the vascular walls and the intense inflammatory infiltration over the ensuing week led to vascular adventitial destruction (Fig. 2K). Early bronchial epithelial necrosis was observed focally on day 1, was widespread by days 2 and 3, peaked between days 3 and 5 and was relatively quiescent by day 7 PI (Fig. 2D-G and I-K). Bronchial submucosal glands also had acute inflammation that was maximal by day 3, but persisted for 1-2 weeks (Data not shown). Two weeks PI, chronic inflammation was mostly restricted to submucosal regions (Data not shown). Intra-alveolar exudate and hemorrhage was pervasive by days 3-5 and were replaced by cellular consolidation with hyperplasia of type-II pneumocytes between days 5-7. At one-week PI, central lung regions were more consolidated while peripheral alveoli contained exudate and inflammatory cells. Gram stain of the tissue did not reveal any organisms in the lung at any time after infection.
Fig. 2.
Histopathology of upper and lower respiratory tract of influenza-infected tissues. Low power overview of lungs from day 2 (A), 3 (B) and 5 (C) post-infection showing progressive atelectasis with inflamed bronchi and alveoli filled with acute hemorrhage progressing to intense panlobular pneumonia with bronchial plugs and alveolar consolidation. (D) Day 7: Bronchial lumen with necrotic tissue plugs, adjacent blood vessels with adventitial clearing, surrounding alveoli with inflammatory exudates (E) Day 2: Acutely necrotic bronchus. (F) Day 3: Inflamed bronchial submucosal glands. (G) Day 7: Bronchial lumen with necrotic tissue plugs. (H) Day 7: Nasal turbinates with inflamed submucosal glands. (I) Day 1: Infection of bronchial epithelium and submucosal glands. (J) Day 1: Infection of tracheal epithelium and submucosal glands. (K) Day 1: Infection of bronchial epithelium and submucosal glands. (M) Day 1: Infection of bronchial epithelium with sloughing. Panels A through H-Hematoxylin and eosin stained paraffin embedded sections, panels I through L-anti-influenza immunohistochemistry on paraffin embedded sections.
Beginning at day 1 post infection, immunocytochemistry showed influenza viral antigens predominantly along the apical surfaces of bronchi and bronchial submucosal glandular epithelium (Fig. 2J-L). Immunocytochemical staining is pronounced at day 1, peaked at day 3 and rapidly declined by days 5 and 7 (data not shown). Viral infection was associated with epithelial necrosis and sloughing into the airways. This process was sufficiently rapid and severe that many airways were obstructed by necrotic plugs debris (Fig. 2L). Immunocytochemical staining of gut, lymphoid tissue, pancreas, and brain demonstrated no evidence of viral antigens or inflammatory lesions (data not shown).
Pathological features of the infection with A/Brisbane/59/2007
Ferrets infected with Brisbane/59 displayed mild clinical symptoms such as 5% maximum weight loss, nasal discharge, sneeze and protracted fever, and without lethality (table 1 and Supplementary Fig. 1). The virus was detected in the nasal wash of infected ferrets and the viral load peaked on day 3 post infection. Hemagglutinin inhibiting A/Brisbane/57 antibodies were detected in serum samples on day 7 PI, and higher titers on day 14 PI were found (Fig. 5C). Histological assessment in the nasal turbinates displayed inflammatory infiltrate on days 2 and 5 post infection, with sloughing in some areas (data not shown). In the lung tissue of Brisbane/59 infected ferrets, the viral load was found to be below detection levels (table 1, Fig. 1A and Fig. 1C) and no histological abnormalities were found (Fig. 1E).
Gene expression profiles in lung tissue of infected animals
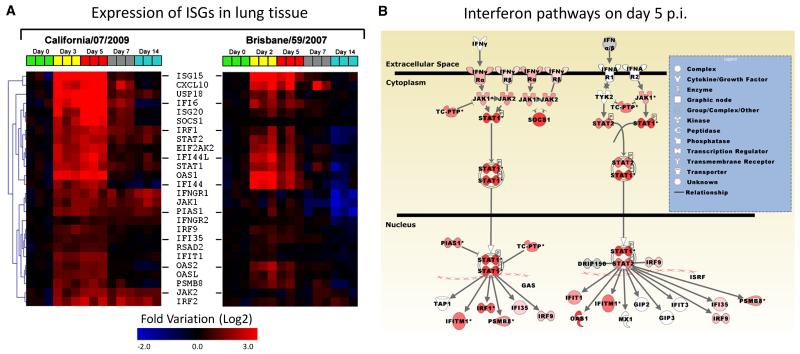
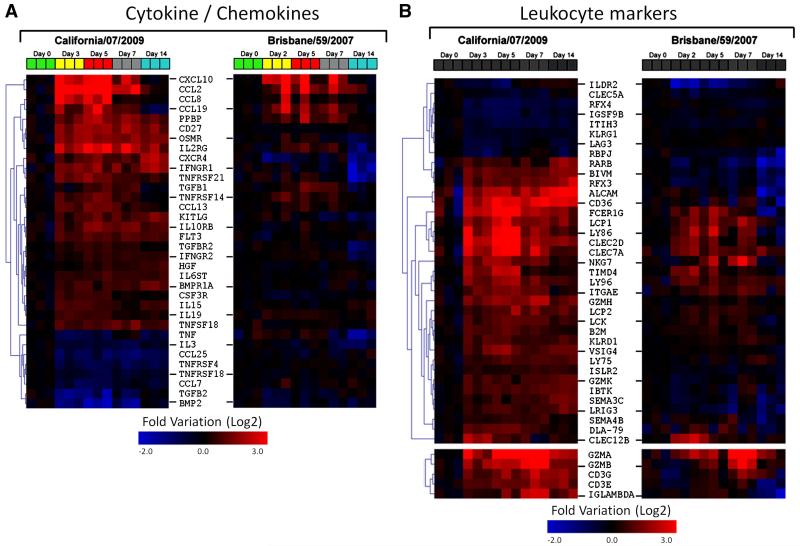
To determine the temporal relationship between host immune responses and the progression and resolution of 2009-H1N1 clinical disease, we conducted a longitudinal study of expression patterns of host immune genes in the lungs of infected animals using high density microarrays (Figs. 3A and 4A-B). On day three post infection, there was a significant increase in interferon stimulated genes (ISG) (Fig. 3A), a functional group that included numerous genes for antiviral proteins (e.g. OAS1&2) and transcriptional regulators (e.g. STAT1) (Malmgaard, 2004). In contrast to the robust expression pattern observed for 2009-H1N1, lungs from animals infected with seasonal H1N1 Brisbane/59 expressed a restricted pattern of ISGs. On day 5, only 12 ISGs were upregulated (mean expression levels were at least 1.5-fold respect to the control group), while lungs from ferrets infected with California/07 displayed 26 upregulated ISGs. Concomitant with early expression of ISGs, California/07 induced the expression of multiple chemokines including CCL2, CCL8, CCL13, CCL19, CXCL7 and CXCL10, and also inflammatory cytokines such as TNFα and IL-1β (Figs. 4A and 5A). These mediators participate in the recruitment of multiple leukocyte subsets including NK cells, monocyte/macrophages, T cells and granulocytes (Charo and Ransohoff, 2006). We observed that their expression patterns coincided with the infiltration of inflammatory mononuclear cells and polymorphonuclear leukocytes in the lungs during infection with California/07 (Fig. 2A-L). Lungs from Brisbane/59-infected animals showed involvement of a limited number of cytokine/chemokines (Fig. 4A) and lower expression levels compared with California/07 (Fig. 5A). The modest lung response during the infection with Brisbane/59, in terms of ISGs, cytokines and chemokines, is likely due to a lack of local host immune activation as the lungs had no detectable viral load (table 1). In the lungs of California/07-infected ferrets, a striking decrease in the expression of ISGs and the chemokines CCL2, CCL8, and CXCL10 takes place on day 7 (Figs. 3A, 4A and 5A). Clinically, on day seven, animals reached the nadir of weight loss and began a slow, but progressive increase, in weight. In contrast to the reduction in ISG and chemokine expression on day seven, expression of genes associated with the effector phase of adaptive immunity (e.g. Ig lambda, CD3, Granzyme A, Granzyme B, and Granzyme H) are increased in 2009-H1N1 infected lungs (Fig. 4B). Similarly, increased expression of granzyme A, B, and H are also observed in peripheral blood on Day 7 (Fig. 5B). The shift from innate to adaptive immune responses is further supported by the appearance of hemagglutinin inhibiting serum antibodies on day 7 (Fig. 5C).
Fig. 3.
Interferon responses in ferret lung tissue. A) Hierarchical gene clustering of interferon-stimulated genes (ISGs) during infection with influenza A/California/07/2009 and A/Brisbane/59/2007. B) Pathway modeling of the interferon responses in the lungs of ferrets infected with influenza A/California/07/2009 using the gene expression data on day 5 post-infection (red, upregulated).
Fig. 4.
Gene expression levels in ferrets during infection with influenza A/California/07/2009 and A/Brisbane/59/2007. Hierarchical gene clustering of different functional categories in lung tissue, A) Cytokine /chemokines, and B) Leukocyte markers.
Discussion
Here, we have used the ferret model of influenza infection as a surrogate for understanding 2009-H1N1 induced pathogenesis and the evolution of host immune responses. Infected ferrets display a number of pathological alterations, such as increased temperature, sneezing, loss of body weight and histological features consistent with virally induced pneumonia, an experimental model that mimics 2009-H1N1 human disease. The results from our study and those from recently published papers, show that the severity can range from a sublethal infection with mild symptoms when using A/California/04/2009 (Itoh et al., 2009; Maines et al., 2009) and A/Netherlands/602/2009 (Munster et al., 2009) influenza strains, to 30% lethality found in our study with California/07, or 50% lethality with A/Mexico/4482/2009 (Maines et al., 2009). All these viral strains infected both the lower and upper respiratory tracts in ferrets. On the contrary, our results and others (Maines et al., 2009), show that seasonal H1N1 Brisbane/59 causes mild illness in ferrets and only affects the upper respiratory tract. While our pathological findings are in keeping with the findings of other authors, our unique immunological data allows for the ability to model host immune responses and pathological changes to 2009-H1N1 over the course of illness.
As expected, increased levels of ISGs appeared early in the course of the viral infection, these genes are usually stimulated by the type I interferon signaling pathway (Fig. 3B) (Malmgaard, 2004) and disruption of this pathway in mice leads to uncontrolled viral infection and death (Durbin et al., 2000). It is well established the relationship between high levels of proinflammatory mediators and degree of lung pathology (de Jong et al., 2006), however the contribution of other factors, such as the direct cytopatic effect caused by the virus, still needs to be assessed. The progressive increase in weight and overall health of animals between days 7 to 14, and the elimination of viral load in respiratory tissues by day 7, indicates that adaptive immune mechanisms were responsible for resolution of 2009-H1N1 induced disease. Infection with Brisbane/59 also produced the early production of neutralizing antibodies and granzymes on day 7, however it is possible that the virus had already been cleared by the innate response (Brisbane/59 is below detection levels on day 3) by the time when the adaptive response was triggered in naïve ferrets.
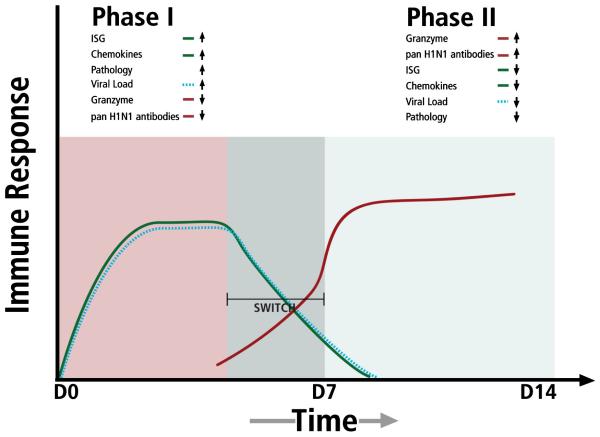
The global analysis of our data support the idea that the host immune responses were biphasic in nature, with an early robust innate ISG and chemokine response that shut down as an adaptive immune phase was initiated (Fig. 6). The timing of the “switch” between these two phases may be important for eliminating viral load and orchestrating the reduction of inflammation associated with early stages of disease. A failure to effectively switch from innate to adaptive responses may be, in part, responsible for severe cases of 2009-H1N1 that result in hospitalization and in some cases death.
Fig. 6.
Stylized representation of bi-phasic host immune response against 2009-H1N1 influenza A. Phase I is characterized by increases in interferon response genes (ISG), chemokines and proinflammatory cytokines (green line) that drive lung pathology. A cellular/molecular switch causes a transition from innate to adaptive immunity, signaling a shut down in ISGs and chemokines and an increase in effector adaptive genes and molecules (red line) and a decrease in viral load (dashed line).
Gene expression of some ISGs, chemokines CCL2, CCL8, and CXCL10, and the inflammatory cytokines TNFα and IL-1β appear to be related to the pathological phase of 2009-H1N1. Moreover, levels of CXCL10 and CCL2 correlate well with respiratory illness severity and they have been associated with human severe cases of SARS and H5N1 infections (Cameron et al., 2007; de Jong et al., 2006), therefore CXCL10 and CCL2 may be useful bio-markers for human disease severity. It will be important to determine if underlying medical conditions associated with severe 2009-H1N1 disease cause aberrant or delayed innate-adaptive immune switching leading to severe influenza H1N1 disease.
Supplementary Material
Acknowledgements
We gratefully appreciate receiving 2009-H1N1 influenza A, A/California/07/2009 from Dr. Steve Lindstrom from the Centers of Disease Control in Atlanta. We thank Guoji Wang and Jonette Werley for technical assistance in preparation of histopathological specimens. We also appreciate the informatics contributions of Fiona Almeida, MacKenzie Howatt, Ben Wang, Joanne Leung and William Li from the Emerging Infectious Disease Training course, Shantou Medical College, summer 2009. We would also like to acknowledge the assistance of Amanda Li and Joey Kelvin in generating the manuscript.
Financial support: This research was supported by National Institute of Health Award U01AI077771 to C.A.W and T.M.R. This research was also supported by NIH and the Li Ka-Shing foundation of Canada to DJK. The Canada-China Training Course in Emerging Infectious Disease, Shantou Medical College was supported by the Li Ka-Shing Foundation Canada.
Footnotes
Potential conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Outbreak of swine-origin influenza A (H1N1) virus infection - Mexico, March-April 2009. MMWR Morb Mortal Wkly Rep. 2009a;58(17):467–70. [PubMed] [Google Scholar]
- Swine-origin influenza A (H1N1) virus infections in a school - New York City, April 2009. MMWR Morb Mortal Wkly Rep. 2009b;58(17):470–2. [PubMed] [Google Scholar]
- Update: infections with a swine-origin influenza A (H1N1) virus--United States and other countries, April 28, 2009. MMWR Morb Mortal Wkly Rep. 2009c;58(16):431–3. [PubMed] [Google Scholar]
- Update: swine influenza A (H1N1) infections--California and Texas, April 2009. MMWR Morb Mortal Wkly Rep. 2009d;58(16):435–7. [PubMed] [Google Scholar]
- Cameron CM, Cameron MJ, Bermejo-Martin JF, Ran L, Xu L, Turner PV, Ran R, Danesh A, Fang Y, Chan PK, Mytle N, Sullivan TJ, Collins TL, Johnson MG, Medina JC, Rowe T, Kelvin DJ. Gene expression analysis of host innate immune responses during Lethal H5N1 infection in ferrets. J Virol. 2008;82(22):11308–17. doi: 10.1128/JVI.00691-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, Muller MP, Gold WL, Richardson SE, Poutanen SM, Willey BM, DeVries ME, Fang Y, Seneviratne C, Bosinger SE, Persad D, Wilkinson P, Greller LD, Somogyi R, Humar A, Keshavjee S, Louie M, Loeb MB, Brunton J, McGeer AJ, Kelvin DJ. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. 2007;81(16):8692–706. doi: 10.1128/JVI.00527-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron MJ, Rowe T, Kelvin DJ. Possible Link between Severe Respiratory Outbreak in Mexico and Swine Flu in Southwestern United States? J Infect Developing Countries. 2009;3(3):157–158. [PubMed] [Google Scholar]
- Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354(6):610–21. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- Chowell G, Bertozzi SM, Colchero MA, Lopez-Gatell H, Alpuche-Aranda C, Hernandez M, Miller MA. Severe Respiratory Disease Concurrent with the Circulation of H1N1 Influenza. N Engl J Med. 2009 doi: 10.1056/NEJMoa0904023. [DOI] [PubMed] [Google Scholar]
- Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360(25):2605–15. doi: 10.1056/NEJMoa0903810. [DOI] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha do Q, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–7. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin JE, Fernandez-Sesma A, Lee CK, Rao TD, Frey AB, Moran TM, Vukmanovic S, Garcia-Sastre A, Levy DE. Type I IFN modulates innate and specific antiviral immunity. J Immunol. 2000;164(8):4220–8. doi: 10.4049/jimmunol.164.8.4220. [DOI] [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, López-Gatell H, Olivera H, López I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. Antigenic and Genetic Characteristics of Swine-Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans. Science. 2009 doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, Sakabe S, Imai M, Hatta Y, Watanabe S, Li C, Yamada S, Fujii K, Murakami S, Imai H, Kakugawa S, Ito M, Takano R, Iwatsuki-Horimoto K, Shimojima M, Horimoto T, Goto H, Takahashi K, Makino A, Ishigaki H, Nakayama M, Okamatsu M, Warshauer D, Shult PA, Saito R, Suzuki H, Furuta Y, Yamashita M, Mitamura K, Nakano K, Nakamura M, Brockman-Schneider R, Mitamura H, Yamazaki M, Sugaya N, Suresh M, Ozawa M, Neumann G, Gern J, Kida H, Ogasawara K, Kawaoka Y. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460(7258):1021–5. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. Transmission and Pathogenesis of Swine-Origin 2009 A(H1N1) Influenza Viruses in Ferrets and Mice. Science. 2009 doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmgaard L. Induction and regulation of IFNs during viral infections. J Interferon Cytokine Res. 2004;24(8):439–54. doi: 10.1089/1079990041689665. [DOI] [PubMed] [Google Scholar]
- Munster VJ, de Wit E, van den Brand JM, Herfst S, Schrauwen EJ, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. Pathogenesis and Transmission of Swine-Origin 2009 A(H1N1) Influenza Virus in Ferrets. Science. 2009 doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Padilla R, de la Rosa-Zamboni D, Ponce de Leon S, Hernandez M, Quinones-Falconi F, Bautista E, Ramirez-Venegas A, Rojas-Serrano J, Ormsby CE, Corrales A, Higuera A, Mondragon E, Cordova-Villalobos JA. Pneumonia and Respiratory Failure from Swine-Origin Influenza A (H1N1) in Mexico. N Engl J Med. 2009 doi: 10.1056/NEJMoa0904252. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, Lindstrom S, Gubareva LV, Deyde V, Garten RJ, Harris M, Gerber S, Vagasky S, Smith F, Pascoe N, Martin K, Dufficy D, Ritger K, Conover C, Quinlisk P, Klimov A, Bresee JS, Finelli L. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N Engl J Med. 2009;360(25):2616–25. doi: 10.1056/NEJMoa0903812. [DOI] [PubMed] [Google Scholar]
- Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005;102(36):12837–42. doi: 10.1073/pnas.0504609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitzow LA, Rowe T, Morken T, Shieh WJ, Zaki S, Katz JM. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J Virol. 2002;76(9):4420–9. doi: 10.1128/JVI.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.