Summary
Ionotropic glutamate receptors (iGluRs) mediate the majority of excitatory neurotransmission in the central nervous system and function by opening a transmembrane ion channel upon binding of glutamate. Despite their crucial role in neurobiology, the architecture and atomic structure of an intact iGluR is unknown. Here we report the crystal structure of the α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA)-sensitive, homotetrameric, rat GluA2 receptor at 3.6 Å resolution in complex with a competitive antagonist. The receptor harbors an overall axis of 2-fold symmetry with the extracellular domains organized as pairs of local dimers and with the ion channel domain exhibiting 4-fold symmetry. A symmetry mismatch between the extracellular and ion channel domains is mediated by two pairs of conformationally distinct subunits, A/C and B/D. Therefore, the stereochemical manner in which the A/C subunits are coupled to the ion channel gate is different from the B/D subunits. Guided by the GluA2 structure and site-directed cysteine mutagenesis we suggest that GluN1/GluN2A NMDA (N-methyl-d-aspartate) receptors have a similar architecture with subunits arranged in a 1-2-1-2 pattern. We exploit the GluA2 structure to develop mechanisms of ion channel activation, desensitization and inhibition by non competitive antagonists and pore blockers.
Introduction
The development and function of the human brain, and its remarkable capacity for experience dependent change, hinges on the organization and dynamics of chemical synapses, communication ‘contact zones’ between neurons. At these specialized sites, chemical transmitters released from presynaptic terminals diffuse across the synaptic cleft and activate receptors localized primarily on the postsynaptic cell1, thereby transmitting the flow of information from one neuron to another. Glutamate is the chemical transmitter of excitatory synapses in the central nervous system2, 3 and receptors for this ubiquitous neurotransmitter are of two classes: metabotropic and ionotropic4. Ionotropic glutamate receptors (iGluRs) are ligand-gated ion channels fundamental to neurotransmission at excitatory synapses and are implicated in nearly all aspects of nervous system development and function5. iGluRs are also involved in chronic neurodegenerative conditions, in psychiatric disorders and in acute injury or trauma6-9.
Comprising the iGluR receptor family are α-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA; GluA1-4), kainate (GluK1-5) and N-methyl-D-aspartate (NMDA; GluN1; GluN2A-D; GluN3A-B) receptors10-14. Whereas NMDA receptors are obligate heterotetramers14, AMPA and kainate subunits form functional homotetramers10-12, 15, although native receptors are almost exclusively heterotetramers16-18. Each subunit has a modular composition19 and includes a large extracellular amino-terminal domain (ATD)20 that participates in subtype-specific receptor assembly, trafficking and modulation, a ligand-binding domain (LBD) central to agonist/competitive antagonist binding and to activation gating21, a transmembrane domain (TMD) that forms the membrane-spanning ion channel22 and a cytoplasmic C-terminal domain involved in receptor localization and regulation23.
AMPA, kainate and NMDA receptors are related in amino acid sequence yet they diverge with respect to function5, 34. Whereas non NMDA receptors exhibit kinetics of activation, deactivation and desensitization on a millisecond time-scale35, NMDA receptors are slower, with corresponding molecular processes occurring on a time-scale of tens to hundreds of milliseconds36. Furthermore, AMPA and kainate receptors only demand glutamate for activation, while NMDA receptors function as coincidence detectors, requiring membrane depolarization to relieve magnesium block37 together with binding of glycine38 and glutamate. AMPA receptors sojourn through multiple sub conductance states contingent upon agonist concentration15, 39, suggesting independent LBDs and a sequential mechanism of activation40. AMPA and kainate receptors undergo profound desensitization, while NMDA receptors desensitize by way of glycine-dependent and glycine-independent mechanisms41.
The pharmacology of iGluR family members is distinct. AMPA receptors, for example, are non competitively antagonized by small molecule binding to the juxta membrane region42. By contrast, the ATDs of NMDA receptors harbor binding sites for polyamines, protons, zinc ions and ifenprodil36, 43. In AMPA receptors, the LBD possesses binding sites for modulators of receptor desensitization and deactivation26, 44. All iGluR subtypes, however, possess binding sites for pore blockers within the transmembrane ion channel45.
Despite divergent functional properties, iGluR family members have a common structural design. Clues to the symmetry and architecture of iGluRs derive from studies of isolated domains, demonstrating that ATDs and LBDs assemble as dimeric entities24-28, from electron microscopy on intact receptors, showing an overall 2-fold symmetry29, 30 and from amino acid sequence analysis and biophysical studies suggesting a ~4-fold symmetric ion channel19, 31. Absent from our understanding, however, is an accurate, atomic resolution description of iGluR architecture and symmetry, a definition of subunit arrangement in homomeric AMPA and heteromeric NMDA receptors32, 33, and proof of the symmetry mismatch between the 2-fold symmetric extracellular domains and the presumably 4-fold symmetric ion channel26. To answer these fundamental questions, we embarked on crystallographic and functional studies of a full length eukaryotic iGluR.
Crystallization and structure determination
We exploited fluorescence-detection size exclusion chromatography (FSEC) 46 to discover that the rat GluA2 receptor10, 11, expressed as the unedited47, ‘flip’ variant48, was a promising candidate for structural studies (Supplementary Fig. 3a). We further harnessed FSEC to find that n-undecyl-β-D-thiomaltoside, the competitive antagonist ZK 20077549 and a modified receptor polypeptide, deemed GluA2cryst, were the optimal detergent, ligand and protein construct for crystallization trials, respectively (Supplementary Figs. 1, 2, 3b-d, 4-6). GluA2cryst binds 3H AMPA with a Kd of 3.5 ± 0.5 nM and yields glutamate-gated currents similar to the wild-type receptor (Supplementary Figs. 7 and 8). Crystals of GluA2cryst belong to the P1 space group, contain one tetrameric receptor in the unit cell and diffract to 3.6 Å resolution (Supplementary Fig. 9; Supplementary Table 1).
We solved the GluA2cryst structure by molecular replacement, using the high resolution structures of the isolated GluA2 ATD27 and the antagonist-bound form of the isolated GluA2 LBD 25 as search probes. Phases were improved by multidomain non crystallographic symmetry (NCS) averaging, solvent flattening and histogram matching. The resulting electron density maps were of sufficient clarity to build the transmembrane helices comprising the ion channel domain and the linking polypeptides connecting the LBD to the ion channel and the ATD to the LBD (Supplementary Figs. 10-12). The weakest density was observed for the S2-M4 linker connecting the LBD and the M4 transmembrane domain. Residues on the cytoplasmic side of the membrane connecting membrane helices M1 to M2, and M2 to M3, were not visible in electron density maps.
We probed the veracity of the polypeptide trace by preparing selenomethionine (SeMet)-labeled receptor of GluA2cryst and of 4 methionine point mutants at Leu542 (M1), Gln586 (Q/R site, M2)47, Ile612 (M3) and Val800 (M4) and by inspecting the corresponding anomalous difference Fourier maps. We formed crystals with a mercury-labeled variant of ZK200775, measured diffraction data at the Hg LIII peak, mapped the antagonist binding site in the LBDs, and found additional mercury binding sites, presumably due to residual methyl mercury chloride (Supplementary Table 1; Supplementary Figs. 13-16). Taken together, the diffraction experiments support the experimental structure, which was refined to good crystallographic statistics and stereochemistry (Supplementary Table 2).
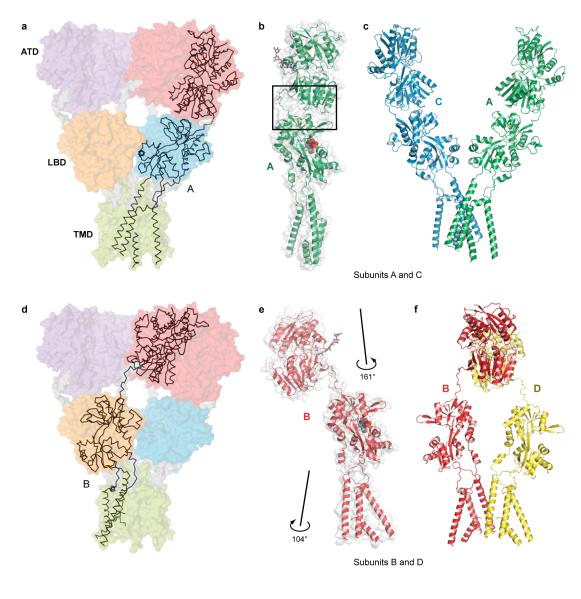
Architecture and symmetry
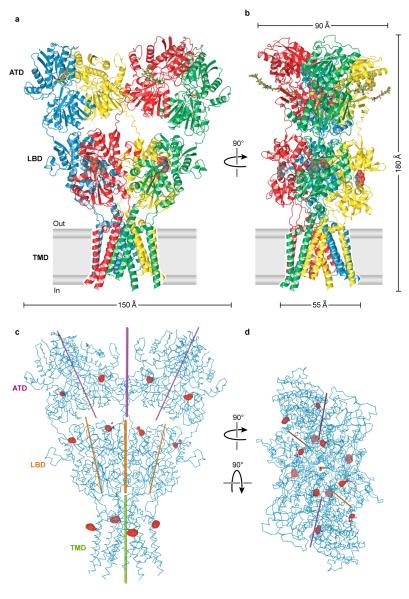
The GluA2 receptor has a shape like the capital letter ‘Y’ where the three major domains are arranged in layers (Fig. 1a, b). The TMDs form the ion channel and define the narrow ’base’, the ATDs are splayed outward, like diverging prongs, at the ’top’ of the ’Y’, and the LBDs, complexed with antagonist molecules, are sandwiched in between the ion channel and ATDs. In this closed, antagonist-bound state of the receptor there are no prominent vestibules or cavities near the ion channel domain and instead the LBD layer rests like a thick slab on top of the ion channel pore (Supplementary Fig. 17).
Figure 1. Architecture of homomeric rat GluA2 receptor.
a, View of the ‘broad’ face of the receptor, perpendicular to the overall 2-fold axis of molecular symmetry. Each subunit is in different color. b, View of the ‘narrow’ face of the receptor. c-d, Axes of symmetry viewed parallel to the membrane (c) or from the extracellular ‘top’ of the receptor, along the overall 2-fold axis of symmetry (d). Axes of local symmetry for the domains ATD, LBD and TMD are shown in purple, orange and green, respectively. For ATD and LBD, thin lines represent axes of intradimer two-fold symmetry and thick lines represent axes of interdimer two-fold symmetry. For TMD, the thick green line represents the local axis of four-fold symmetry. Red mesh peaks (c,d) define mercury sites derived from an anomalous difference Fourier map of a GluA2cryst mercury derivative. The contour level is 5.0 σ.
The symmetry and subunit arrangement of the tetrameric GluA2 receptor is without precedent. The receptor has an overall yet approximate 2-fold axis of molecular symmetry oriented perpendicular to the membrane plane (Fig. 1c, d; Supplementary Fig. 16). This 2-fold axis of symmetry relates one ATD dimer27 to another, one LBD dimer26 to the second, and half of the pore-forming TMDs to the other half. The ion channel domain has an approximate 4-fold axis of rotational symmetry (Fig. 1c, d).
Each prong of the receptor ‘Y’ is defined by an ATD dimer in which the ‘local’ dimer 2-fold axis is oriented ~24° off of the overall 2-fold axis (Fig. 1c, d). ‘Below’ the ATDs are the LBDs, also organized as a pair of dimers in which the local 2-fold axes within each dimer are also tipped off the overall 2-fold axis of symmetry by ~19° and are thus not aligned with the ‘local’ ATD 2-folds. “Below” the LBDs the 4-fold axis of the ion channel is approximately aligned with the overall 2-fold axis of symmetry. The multiple, non aligned axes of local symmetry, together with the 2-fold symmetry of the LBDs and the 4-fold symmetry of the ion channel, result in symmetry mismatches between the ATDs, LBDs and TMDs.
Extracellular domains
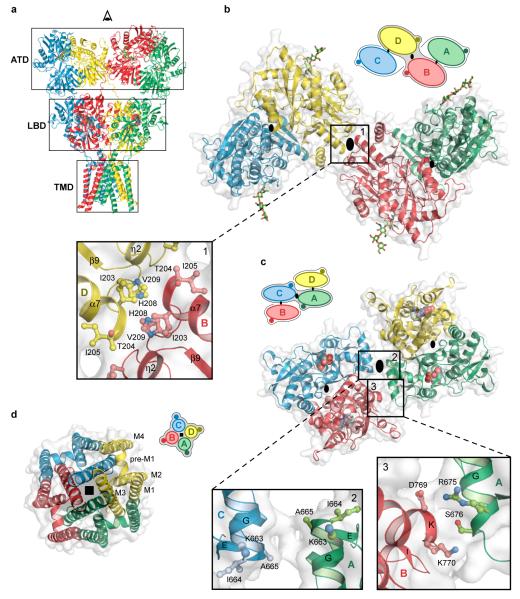
The ATD, implicated in receptor assembly, trafficking and localization, forms two distinct types of subunit-subunit contacts (Fig. 2). On the one hand, within each ATD ‘dimer’, there are extensive subunit - subunit contacts (A-B or C-D) that are indistinguishable from the contacts seen in the high resolution crystal structures of the isolated GluA2 ATDs27 (Supplementary Fig. 18a). On the other hand there is an interface between ATD dimers (~330 Å2), located on the overall axis of 2-fold symmetry, that is mediated by residues on the L2 lobes of the B and D subunits (Fig. 2b). Within the context of the ATD ‘layer’, we define the B/D and A/C subunits as ‘proximal’ and ‘distal’ to the overall 2-fold axis of symmetry. The B-D, dimer-dimer interface, while small, is observed in the packing of GluA2 ATD dimers in crystals of the isolated ATD27 (Supplementary Fig. 18b). We suggest that the ATD B-D interface, together with the subunit ‘crossover’ between the ATD and LBD layers, provides a molecular explanation for the role that the ATD plays in the assembly and stability of the tetrameric receptor.
Figure 2. Domain symmetry and architecture.
a, GluA2cryst structure viewed perpendicular to the overall 2-fold axis. b-d, Domain layers viewed from the ‘top’ of the receptor, parallel to overall 2-fold axis. The simple schematics depict the symmetry and arrangement of domains within each layer. b, The ATD layer. Boxed region highlights dimer-dimer contacts, with overall 2-fold axis (large black oval) in the center. The local, intradimer 2-fold axes of symmetry are shown as smaller black ovals. Subunits B and D are proximal and subunits A and C are distal to the overall 2-fold axis, respectively. c, The LBD layer with the dimer-dimer contacts on and off the overall 2-fold axis shown in panels 2 and 3, respectively. In this layer, subunits A and C are proximal to the overall 2-fold axis. d, The TMD layer and its 4-fold rotational symmetry (black square).
At the LBD layer each agonist binding domain is also a partner in readily identifiable ‘dimers’ and these dimers, in turn, interact across the overall 2-fold axis (Fig. 2c). As a consequence of the subunit crossover between the ATD and LBD layers, the local LBD dimers are formed by the A-D and B-C subunits, with the A and C subunits and the B and D subunits proximal and distal to the overall 2-fold axis, respectively. Within a LBD dimer there are multiple contacts between domain 1 of each subunit, recapitulating the interactions seen in the high resolution crystal structures of the isolated, water-soluble GluA2 LBD25. In the GluA2cryst structure, which corresponds to an antagonist-bound, non desensitized state, the domain 1 - domain 1 interface is ‘intact’, as visualized in the wild-type25, the Leu483 to Tyr mutant or the cyclothiazide26 and aniracetam-bound 44 structures of the isolated LBD. Domain 2, by contrast, does not participate in significant intersubunit interactions within a LBD dimer, a finding that is also in harmony with high resolution studies of the isolated LBD50. Lodged in the ‘clamshell’ of each LBD is a bound antagonist, thus proving that the agonist/competitive antagonist binding site is located within and not between subunits.
There is a small (~224 Å2) interface between LBD dimers, an area consistent with weak dimer - dimer interactions26, 51. Like the ATD dimer - dimer interface, the LBD dimer interface is located on the overall 2-fold axis of symmetry and is composed primarily of residues at the ‘bend’ between helices F and G, with the α-carbon atoms of residues Lys 663, Ile 664 and Ala 665 of subunits A and C 8-13 Å apart. The relationship between LBD dimers is not perfectly 2-fold symmetric, however, and whereas residues at the end of helices G and K of the A and B subunits (Ser 676 and Lys 770) are in van der Waals contact (Cα-Cα distance of 6.1 Å), the equivalent residues in subunits C and D are not in contact (Cα-Cα distance of 9.7 Å).
Transmembrane domain
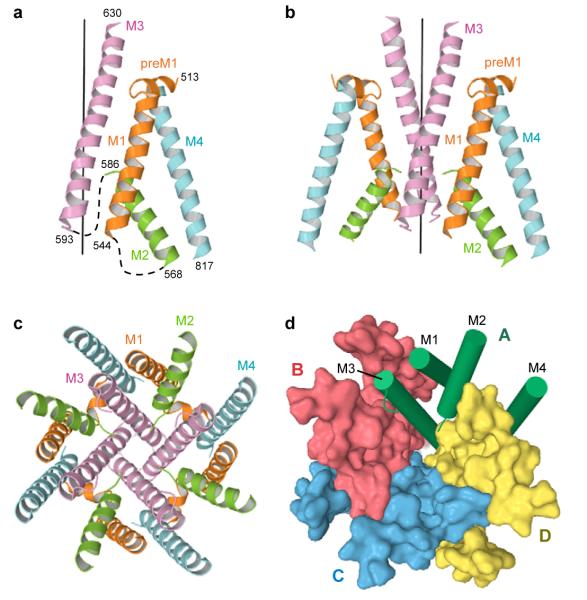
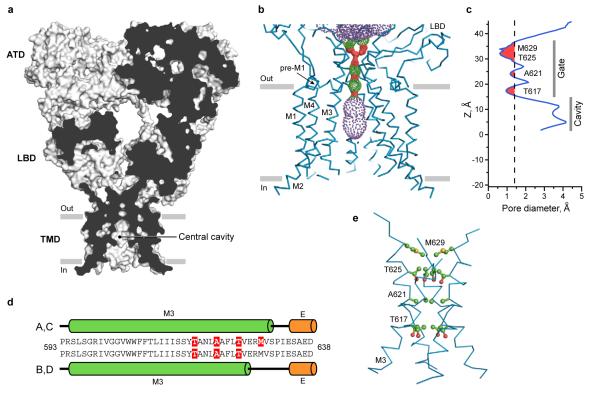
Viewed from the extracellular side of the membrane down the overall 2-fold axis of symmetry, four GluA2cryst subunits arrange their transmembrane domains around an axis of ~4-fold rotational symmetry (Fig. 2d). In harmony with topology studies52, 53, each subunit has 3 transmembrane helices (M1, M3 and M4), a central pore-like helix (M2)54 and a polypeptide pore-lining loop that is disordered in our electron density maps (Fig. 3a, b). Leading from the LBD, the S1-M1 polypeptide segment adopts an extended conformation until reaching the TMD, at which point the polypeptide forms a ~90° turn and initiates a short helix (pre-M1) oriented nearly parallel to the membrane. The pre-M1 helix acts like a cuff around the ‘top’ of the ion channel domain, making contacts with carboxyl and amino terminal ends of helices M3 and M4, respectively. M1 is the first bona fide transmembrane segment and it resides on the exterior of the ion channel domain (Fig. 3c). Within the pore lies the M2 helix, positioned largely on the basis of tube-shaped electron density and the anomalous difference density peak from the SeMet-labeled Gln 586 to Met (Q/R site) mutant. The M3 helices line the inside of the ion channel domain, are ~52 Å in length and, in the present, antagonist bound structure, cross at the level of the pre-M1 ‘cuff’ helices, near the membrane - aqueous solution boundary, forming a ~12 Å occlusion of the putative ion permeation pathway. Residing on the exterior of ion channel domain is the M4 helix, connected to the S2 segment of the LBD by two turns of helix and a short extended region of polypeptide. There are extensive subunit-subunit interactions between the transmembrane segments with the M4 segment of one subunit making interactions primarily with the transmembrane domains of an adjacent subunit (Fig. 3d). These interactions provide a molecular basis for the crucial role of the M4 helix in receptor assembly and function22.
Figure 3. Transmembrane domain architecture.
Fold of transmembrane domain for subunit A (a) and for subunits A and B (b) viewed parallel to the membrane. Transmembrane segments M1 to M4 are depicted in different colors. The ‘vertical’ black line defines the 4-fold symmetry axis. Dashed lines indicate disordered regions. c-d, TMD viewed parallel to the 4-fold axis. c, TM segments are colored as in panels (a,b). d, Surface representation of subunits B to D. For subunit A, the segments M1-M4 are shown as green cylinders.
Probing subunit interfaces
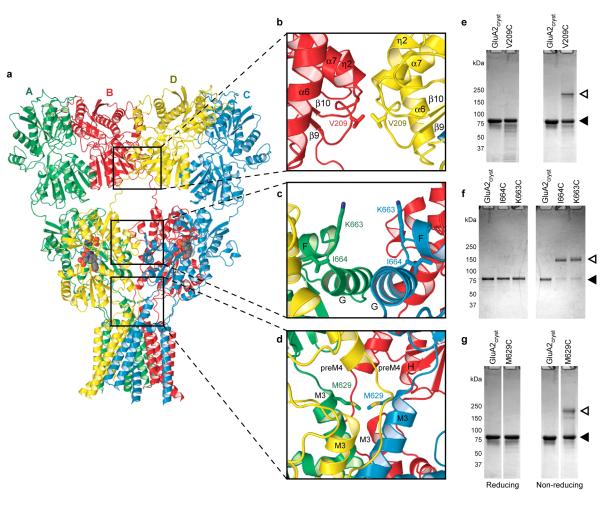
Cognizant that the GluA2cryst structure (Fig. 4a) possesses an unorthodox subunit arrangement and molecular symmetry, we tested whether the subunit arrangement and domain-domain contacts in the crystal structure reflect interactions adopted by the receptor in a non crystalline environment. To accomplish this, we introduced cysteine residues into the three unexplored interfaces at sites that should result in spontaneous disulfide bond formation (Fig. 4b-d). We purified the mutant, tetrameric receptors to homogeneity and probed the extent of spontaneous subunit crosslinking by gel electrophoresis under reducing and non reducing conditions (Fig. 4e-g; Supplementary Fig. 19).
Figure 4. Probing intersubunit interfaces in GluA2 AMPA receptors.
a, Ribbon diagram of the GluA2cryst structure with each subunit in a different color. b-d, Close ups of intersubunit interfaces between two ATD dimers (b), two LBD dimers (c) and at the top of the ion channel (d). e-g, SDS PAGE analysis of spontaneous crosslinking of cysteines introduced at intersubunit interfaces. Left and right panels illustrate experiments carried out in reducing and non-reducing conditions, respectively. Filled and open triangles indicate positions of monomeric and dimeric bands, respectively.
At the ATD, dimer-dimer, B to D subunit interface, we introduced a cysteine at Val 209 (Fig. 4b). For the wild-type-like receptor, in the presence or absence of reducing agent, the GluA2 subunit migrates at a position consistent with its calculated molecular mass. By contrast, for the Val209 to Cys mutant, we observe reducing agent dependent dimer formation, thus supporting the presence of this dimer-dimer interface in the intact receptor under native conditions (Fig. 4e). In the LBD layer, there are two distinct subunit - subunit interfaces. One interface is within a LBD dimer and is formed by extensive contacts between domain 1 of 2-fold related subunits, faithfully mirroring the thoroughly documented dimer interface observed in the isolated LBDs of AMPA 25, 26, 51, kainate55 and NMDA56, 57 receptors. The second interface, between subunits (A and C) proximal to the overall 2-fold axis, is composed of only a handful of intersubunit contacts (Fig. 4c). We therefore tested whether residues in this interface could form inter-dimer disulfide crosslinks. At both Lys 663 and Ile 664 cysteine mutants formed redox dependent dimers (Fig. 4f), supporting the presence of this interface in the intact receptor. This LBD dimer-dimer interface is also important for agonist dependent gating because steady state currents of the Ile 664 to Cys mutant are potentiated ~5-fold following receptor reduction58 (Supplementary Fig. 20).
The apex of the ion channel domain, defined by the C-terminal ends of M3, provides an important test of the GluA2cryst structure, not only because residues at the end of M3 define the gate of the ion channel in this antagonist bound state, but also because the end of M3 and the M3-S2 linker span the transition between the 4-fold symmetry of the ion channel and the 2-fold symmetry of the extracellular domains. In two subunits (A/C), the Met 629 side chains point toward each other and the α-carbons are separated by ~12 Å whereas the corresponding α-carbon atoms in subunits B/D are 30 Å apart (Fig. 4d). In satisfying agreement with the GluA2cryst structure, we find that cysteines introduced at position 629 form redox dependent crosslinks (Fig. 4g), consistent with the proximity of Met 629 residues and the structure of the ion channel domain. We may not see complete crosslinking to a dimer position because of the overall 2-fold symmetry of the extracellular domains or because some of the cysteine residues may have suffered chemical modification during expression or purification.
We propose that the architecture of kainate receptors is similar to that of the GluA2cryst AMPA receptor based on the remarkable observation that the isolated GluR6 ATD dimer forms a similar dimer-of-dimers arrangement in the crystal lattice, yielding a ‘tetrameric’ complex28 like that in the GluA2cryst structure (Supplementary Fig. 21). Superposition of α-carbon atoms for 292 residues per subunit in the structurally conserved regions of GluA2cryst and isolated GluR6 ATD dimer-of-dimers yields a r.m.s. deviation of 3.2 Å. This observation, in combination with the fact that LBDs of AMPA and kainate receptors form similar local dimers55, demonstrates that principles of architecture and symmetry are conserved between AMPA and kainate receptors.
Architecture of NMDA receptors
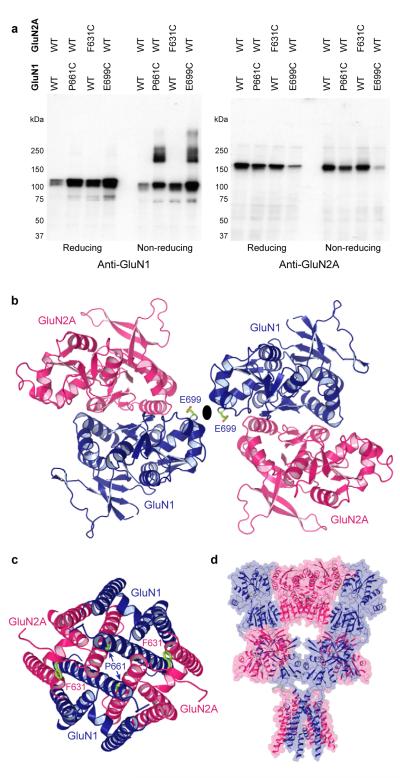
NMDA receptors are the most complex subfamily of ionotropic glutamate receptors, not only because they are obligate heterotetramers, requiring a glycine-binding GluN1 or GluN3 subunit together with a glutamate binding GluN2 subunit, but also because their ATDs bind ions and molecules that modulate receptor activity36. Even though the structure of the GluN1/GluN2A LBD heterodimer is known56, there is no conclusive experimental knowledge of how subunits are arranged in heterotetrameric NMDA receptors. To determine whether the GluA2cryst structure provides a paradigm for understanding NMDA receptor architecture, we carried out cysteine-directed crosslinking experiments on the rat GluN1-GluN2A NMDA receptor.
Guided by superpositions of the GluN1-GluN2A LBD heterodimer56 onto the GluA2cryst structure, we designed cysteine substitutions at putative interdimer interfaces. There are three possible arrangements of the GluN1-GluN2A LBD heterodimers within a heterotetrameric receptor and we can distinguish between these models depending on whether redox dependent crosslinking requires cysteine substitutions in only GluN1, in only GluN2A or in both GluN1 and GluN2A subunits (Supplementary Figs. 22 and 23). Remarkably, only a single cysteine substitution in GluN1 at Glu 699 is required to crosslink NMDA receptor subunits at the LBD level (Fig. 5a). Indeed, none of the 6 cysteine substitutions in GluN2A promoted crosslinking, while simultaneous substitutions in both GluN1 and GluN2A resulted in crosslinking similar to substitutions in GluN1 alone. Within the LBD layer of the NMDA receptor, therefore, we propose that GluN1 and GluN2A subunits form diagonal pairs with the GluN1 subunit proximal to the overall 2-fold axis of symmetry (Fig. 5b).
Figure 5. Subunit arrangement in NMDA receptors.
a, Western blot analysis of crosslinking of wild type GluN1-GluN2A (WT) and cysteine-substituted NMDA receptors probed with anti-GluN1 (left) and anti-GluN2A (right) antibodies. b, Model of LBD dimerof-dimers built by superposing two GluN1-GluN2A dimers56 onto the GluA2cryst structure, viewed along the axis of overall two-fold symmetry. Residues substituted with cysteines are shown in green. c, Model of the NMDA receptor ion channel based the GluA2cryst structure and viewed along the axis of pseudo four-fold symmetry. d, Simple model of NMDA receptor architecture based on GluA2cryst structure and using the GluN1-GluN2A LBD heterodimer structure together with the GluA2cryst ATDs and TMDs.
What is the arrangement of NMDA receptors within their TMD? Here we exploit the GluN1 and GluN2A equivalents of GluA2 residue Met 629. Because the GluN1 subunit is proximal to the overall 2-fold axis of symmetry at the LBD layer, we predict that the GluN1 residue, Pro 661, will be proximal to the overall 2-fold axis at the TMD layer and that the GluN2A residue, Phe 631, will be too far away to form a crosslink (Fig. 5c). In concordance with this prediction, GluN1 (Pro 661 to Cys)-GluN2A (wild-type) but not GluN1 (wild-type) - GluN2A (Phe 631 to Cys) receptors demonstrate redox dependent crosslinking (Fig. 5a). Even though we have not explored crosslinking of NMDA receptors ATDs, based on our observation that subunits switch proximity to the overall 2-fold axis between the ATD and LBD layers, we predict that the ATDs are assembled as ‘local’ heterodimers and that NR2 ATDs mediate the dimer-dimer contacts, proximal to an overall 2-fold axis of symmetry (Fig. 5d). This arrangement would appropriately position the L2 lobe of the GluN2A ATD to alter receptor structure and function by either modulating ATD dimer-dimer contacts or ATD-LBD interactions, ultimately influencing ion channel gating via perturbation of the LBD36, 57.
Subunit non equivalence and domain swapping
There is a remarkable swapping of domains, involved in ‘local’ dimers between subunits, that is illustrated by tracing neighboring polypeptide chains through the receptor. Within the ATD layer, subunits A and B interact with each other to form a local ATD ‘dimer’ (red dimer, Fig. 6a, d). When the polypeptide chains of subunits A and B pass to the LBD layer, however, subunit A forms a LBD dimer with the corresponding domain of subunit D (blue dimer, Fig. 6a), while subunit B associates with subunit C (orange dimer, Fig. 6d). Within the TMD, the transmembrane helices of subunits A and B form extensive contacts with each other as well as with subunits C and D (Fig. 3c, d).
Figure 6. Subunit non equivalence and ‘domain swapping’.
a, α-Carbon trace of subunit A and partially transparent solvent accessible surface of the entire receptor. b, Conformation of subunit A. c, Position and conformation of subunits A and C in the intact receptor. d, α-Carbon trace of subunit B showing ‘domain swapping’ in going from the ATD to the LBD layers. e, Conformation of subunit B. f, Position and conformation of subunits B and D in the intact receptor. Note the large differences of the ATD-LBD linkers and interfaces between the A/C and B/D subunit pairs.
The swapping of extensive local dimer interactions between subunits and the symmetry mismatch between the LBDs and TMDs mean that within this homotetrameric receptor, where each subunit is chemically identical, there are two conformationally different subunit pairs related by the overall axis of 2-fold molecular symmetry: subunit A is equivalent to C and subunit B is equivalent to D yet the A/C pair is distinct, in conformation, from the B/D couple (Fig. 6c, f). These differences in conformation can be illustrated by superimposing the LBDs of subunits A and B (Fig. 6b, e), showing the large differences in orientation of the flanking ATDs and TMDs. In subunit A or C, for example, there is a substantial interface between the ATD and LBD (~315 Å2) whereas in subunit B or D there are no similar contacts (Fig. 6b, e). Another fundamental difference is that the LBD of the B/D subunits is ~6 Å closer to the putative membrane plane than the A/C subunit pair.
The conformational difference between the two types of subunits is defined by polypeptides linking the ATDs, LBDs and TMDs that have the same amino acid sequence but adopt different conformations. These polypeptides also mediate symmetry transitions between domain layers. The symmetry mismatch between the ATDs and the LBDs, characterized by different orientations of their local two-fold axes of symmetry (Fig. 1c,d) and domain swapping (Fig. 6a,d), is mediated by the ATD-S1 linkers (Thr 377 - Lys 393). On the one hand, the ATDs of the A and C subunits interact with their LBDs via an ATD-LBD interface (Fig. 6a-c) and the ATD-S1 linkers adopt a more compact conformation. On the other hand, the ATDs of the B and D subunits are suspended between LBD dimers and the ATD-S1 linker nearly spans a LBD dimer, taking on an extended conformation (Fig. 6d-f).
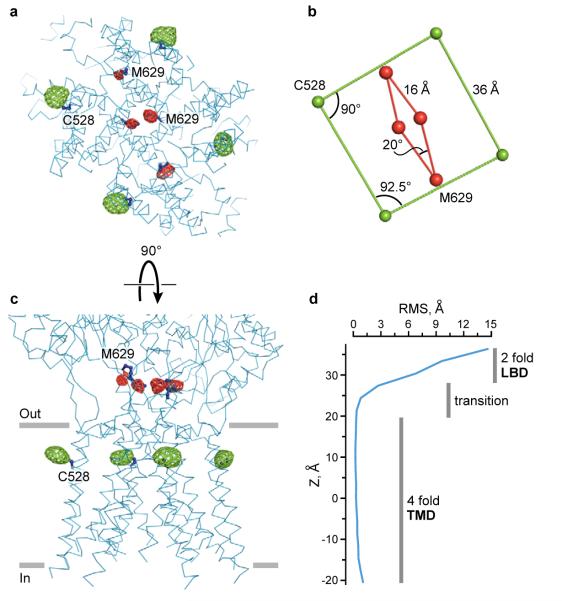
2-Fold to 4-fold symmetry transition
On the extracellular side of the membrane, both the ATD dimers and the LBD dimers are arranged with an overall 2-fold symmetry. By contrast, the GluA2cryst TMD has 4-fold rotational symmetry (Fig. 3c). Where is the 2-fold to 4-fold symmetry mismatch structurally reconciled? We can answer this question by examining selenium and mercury positions derived from SeMet-labeled receptor and from crystals grown with the mercury-containing antagonist. Selenium sites at Met 629, a residue within the highly conserved ‘GluArgMetVal’ sequence at the end of M3 immediately preceding the S2 segment, clearly show 2-fold symmetry (Fig. 7a,b). Mercury sites at Cys 528, near the extracellular end of M1, however, demonstrate unambiguous 4-fold symmetry (Fig. 7a,b). By viewing the receptor perpendicular to the overall 2-fold axis, and defining the approximate membrane boundary based on residue polarity, we see that Met 629 is just ‘outside’ of the membrane-spanning region whereas Cys 528 resides within the membrane bilayer. The 2-fold to 4-fold symmetry transition occurs within this region, a ~10 Å thick slab between the LBDs and TMDs (Fig. 7c).
Figure 7. 2-fold to 4-fold symmetry transition.
a, Ion channel viewed from the cytoplasm, parallel to the local 4-fold axis of symmetry. Mercury (green mesh) and selenium (red mesh) sites, defined by anomalous difference electron density, contoured at 5.0 σ and 2.6 σ, respectively. b, Geometry of the mercury and selenium sites from panel (a). c, View of TMD perpendicular to its local 4-fold axis of molecular symmetry. d, Graph showing the r.m.s. deviations of α-carbon positions following transformation of the A subunit on the B subunit by applying the local 4-fold axis of symmetry, emphasizing that the 2-fold to 4-fold symmetry transition occurs between the LBDs and the membrane embedded TMD.
To precisely define where the transition occurs, we applied the 4-fold rotational transformation associated with the TMDs to one of the receptor subunits and calculated the root-mean-square (r.m.s.) deviation of α-carbon atom positions between the superimposed subunits. Where the receptor conforms to 4-fold symmetry, the r.m.s. deviations in atom positions are on the order of the coordinate uncertainty. In 2-fold symmetric regions, however, the r.m.s. deviations in atom positions are much greater. Thus, by plotting the r.m.s. deviations in atom positions as a function of distance along the overall 2-fold axis, we can follow the 2-fold to 4-fold symmetry transition (Fig. 7d). This analysis demonstrates that the transition is abrupt and is located at the boundary of the extracellular leaflet of the membrane bilayer, within the region of the ion channel domain encircled by the pre-M1 ‘cuff’ helical segments.
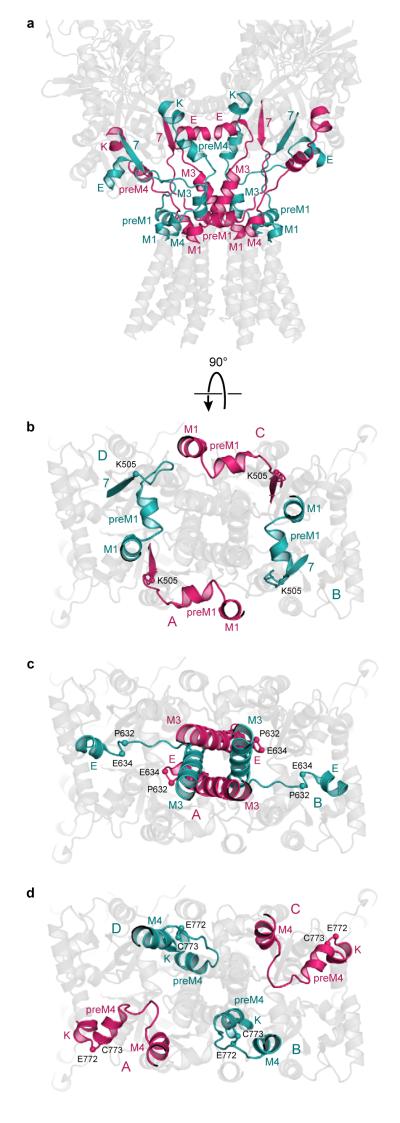
Gating machinery and symmetry mismatch
The symmetry mismatch between the LBDs and the TMDs is resolved by 3 linking peptides - the S1-M1 (Lys 506 - Gly 513), M3-S2 (Val 626 - Glu 634), and S2-M4 (Gly 774 - Ser 788) linkers - making the transition from the 2-fold, parallelogram-like symmetry of the LBDs, to the 4-fold symmetric, square geometry of the TMDs (Fig. 8a). Indeed, these are the central elements of the iGluR gating machinery that transform ligand-induced structural changes in the LBD dimers into the movement of the transmembrane domains that opens and closes the central pore of the 4-fold symmetric ion channel. In bridging 2-fold to 4-fold symmetry transition, the peptide segments linking the LBDs and TMDs can be grouped into two pairs that belong to diagonal subunits A/C or B/D; within each group, the linkers adopt approximately similar conformations whereas between the groups the conformations are clearly distinct.
Figure 8. Gating ‘machinery’ accommodates symmetry mismatch.
a, LBDs and TMDs of GluA2cryst viewed perpendicular to the overall 2-fold axis of molecular symmetry. The elements mediating symmetry mismatch between LBDs and TMDs – the S1-M1, M3-S2 and S2-M4 linkers – are colored pink (subunits A and C) or blue (subunits B or D). b-d, The elements resolving symmetry mismatch between the LBDs and the TMDs viewed from the cytoplasm, parallel to the ion channel 4-fold axis of symmetry. b, The S1-M1 linker, (c) the M3-S2 linker and (d) the S2-M4 linker.
Accommodation of the 2-fold to 4-fold symmetry mismatch is illustrated by the different conformations of the S1-M1 linkers which, when passing from the β7 strand of the LBD to M1 (TMD), either come from inside (A/C) or outside (B/D) of the ‘M1 circle’ (Fig. 8b). The M3-S2 linker provides a particularly striking example of how the conformations of the A/C and B/D linking peptides differ. In the A and C subunits, the M3-S2 linker adopts a helical conformation to Met 629 (Fig. 4d). For the B and D subunits, by contrast, the helical conformation is broken at Val 626 and following this residue, the peptide adopts an extended conformation. The difference in main chain conformation means that the α-carbon atoms of Pro 632 in the A and C subunits are ~27 Å apart whereas the corresponding atoms in the B and D subunits are separated by ~50 Å (Fig. 8c). A third striking illustration is provided by Cys 773, a conserved residue at the end of helix K of the LBD. For Cys 773, in subunits B and D, the α-carbons are 33 Å apart and in the A and C subunits, the span is much larger, 69 Å. To reach the M4 in TMD, which possesses 4-fold symmetry and where the distances between the same atom in the A/C and B/D subunits are necessarily the same, the A/C and B/D S2-M4 linkers take on different conformations and different orientations relative to the overall 2-fold axis (Fig. 8d).
The ion channel
The 4-fold rotationally symmetric GluA2cryst ion channel is shaped like a Mayan temple with a broad cytoplasmic base, ~42 Å on a side, and with a bluntly pointed extracellular ‘top’. In the present, competitive antagonist-bound state, the ion channel unambiguously adopts a closed conformation by the crossing of the M3 helices (Fig. 9). The crossing of the helices occurs near the highly conserved Ser-Tyr-Thr-Ala-Asn-Leu-Ala-Ala-Phe (SYTANLAAF) motif, with Thr 617, Ala 621 and Thr 625 occluding the ion channel permeation pathway22, 59. In the Lurcher mouse60 there is a substitution of Ala 636 by Thr in the GluD2 subunit, leading to spontaneously opening ion channels61. The equivalent residue in GluA2 (Ala 622) participates in close contacts with the M3 helix of a neighboring subunit, suggesting that introduction of bulky residues can directly destabilize the tight helix crossing associated with the resting, closed state of the receptor, leading to constitutively open ion channels. The ion channel permeation pathway is also occluded above the SYTANLAAF motif by a pair of Met 629 residues on the A/C subunits protruding their side chains towards the center of the pore. Remarkably, these methionines are adjacent to the Glu 627-Arg 628 motif that, when mutated, strongly perturbs receptor desensitization62.
Figure 9. Closed conformation of the ion channel pore.
a, Sagittal section of the GluA2cryst receptor illustrates that the occlusion of the putative ion permeation pathway, or the ion channel gate, is formed by the crossing of the M3 helices. b,c, Surface representation of the ion conduction pathway (b) and the pore diameter as a function of distance along the central axis of the channel (c) generated using the program HOLE (red < 1.4 Å < green < 2.8 Å < purple). Residues forming the narrowest portions of the ion conduction pathway are indicated. d, The A/C and B/D M3 segments adopt distinct conformations at their C-termini, proximal to the LBDs. Helical regions are shown as cylinders and colored green for TMD and orange for LBD. Residues forming the narrowest portions of the ion conduction pathway are highlighted in red. e, M3 residues forming the narrowest portion of the ion conduction pathway.
Stimulated by hypotheses19 and experimental data31, 63 proposing a common architecture for the ion channel pores of iGluRs and K+ channels, we superimposed the transmembrane domains of GluA2cryst onto the bacterial K+ channel KcsA54 (Fig. 10, Supplementary Fig. 24). Despite the low pairwise identity of ~20% between the aligned amino acid sequences of the rat GluA2 receptor channel and KcsA (Supplementary Fig. 25), the M1, M2, and M3 segments of GluA2cryst overlap remarkably well with structurally equivalent portions of KcsA (Fig. 10a). In fact, superposition of α-carbon atoms for 64 residues per subunit in GluA2cryst and KcsA yields a r.m.s. deviation of 2.2 Å. Not included in this comparison was transmembrane segment M4, which is absent in KcsA and bacterial glutamate receptors64. In addition to the overall similarity, the structural alignment demonstrates that the occlusion of the ion conductive pathway, i.e. the regions of these channels involved in gating and defined by the carboxyl terminal region of M3 in GluA2cryst and TM2 in KcsA, is remarkably similar (Fig. 10b). By contrast, the region of KcsA that confers ion selectivity - the extended Thr-Val-Gly-Tyr-Gly selectivity filter - is completely different in amino acid sequence in iGluRs and is disordered in the GluA2cryst structure, observations consistent with the fact that K+ channels are highly selective and iGluRs are not.
Figure 10. Closed state conformations of iGluR and K+ channels are similar.
Two diagonal subunits (a) and their bundle crossing (b) in GluA2cryst channel (blue) and KcsA channel54 (grey) viewed parallel to the membrane. Channels were superimposed by aligning main-chain atoms of the M3 and M1 segments in iGluR with the inner and outer helices in KcsA, respectively. Residues forming the narrowest portions of the ion conduction pathway are shown as stick models.
The pre-M1 ‘cuff’ helix is oriented nearly parallel to the membrane, at the interface between the membrane and extracellular solution and is a structural feature that underscores similarities and differences between iGluRs and K+ channels65. Appearing as a ‘collar’ around the TMD bundle, the pre-M1 helices may restrain the mobility of M3 in the closed state and, by virtue of its connection, via the S1-M1 linker, to the LBD, promote opening of the ion channel gate upon agonist binding to the LBD. Whereas some K+ channels have a similar element of structure65, the length of the helical segment as well as its conformation relative to the ion channel diverge from the GluA2cryst pre-M1 helix. In fact, Pro 520 in the ‘elbow’ of the GluA2cryst pre-M1 is highly conserved across iGluR subtypes yet is missing in K+ channels (Supplementary Fig. 25), possibly suggesting a distinct role of the pre-M1 element in iGluR structure and gating.
Immediately ‘above’ the Q/R site, clearly within the transmembrane portion of the ion channel domain, is a central cavity (Fig. 9a) similar to that observed in potassium channels54. In the GluA2cryst structure there are conspicuous ‘gaps’ between the transmembrane domains that result in a portal between the central cavity, within the ion channel, and the membrane environment. We speculate that this portal may be occupied by residues projecting from the transmembrane domains of AMPA receptor auxiliary subunits66, 67, thus providing a mechanism by which auxiliary subunits modulate ion channel properties, such as the extent of block by polyamines68.
The amino acid sequence and structural relationships between GluA2cryst and K+ ion channel pores allow us to speculate on the conformation of an iGluR in an open channel state (Supplementary Fig. 25 and 26). On the basis of superimposing the ion channels of GluA2cryst and Shaker65, we suggest that the transmembrane helices of iGluR will bend and splay away from the central axis of the channel, mimicking the iris-like opening of K+ channels. Although roughly similar in overall nature, the TMD movements during activation gating of iGluRs and K+ channels may be different in molecular detail69. In fact, the residues lining the ion permeation pathway or the amino acids that perturb gating via genetic or chemical modification, together with the types of non-competitive antagonists and channel blockers that bind to the TMD, both in AMPA (Supplementary Fig. 27) and NMDA (Supplementary Fig. 28) receptors, are highly specific to iGluRs, thus providing further evidence of distinction with potassium channels.
Mechanism of activation
The GluA2cryst structure allows us to interpret decades of studies in the context of an intact receptor. Most fundamentally, the GluA2cryst structure proves that agonist binding sites are located within individual subunit ‘clamshells’ and not between subunits, as is the case with trimeric P2X70 and pentameric Cys-loop71 receptors. This architectural principle is consistent with the observations that binding of multiple agonist molecules and subsequent ion channel activation are largely stepwise, sequential processes15. Indeed, the independence of LBD function is further supported by the fact that they can be genetically excised and studied as isolated soluble domains72. Together, these properties justify exploiting the wealth of structural and biophysical experiments on soluble LBDs of AMPA25, 58, 73, kainate55, 74 and NMDA receptors56, 75, 76 to illuminate principles of gating in full length iGluRs.
The GluA2cryst structure is a complex with the high-affinity competitive antagonist ZK 200775. Antagonist binding stabilizes the binding domain ‘clamshell’ in an open conformation (Supplementary Fig. 29)25 and, in the context of the LBD dimer, places the transmembrane associated linker regions closest together (Supplementary Fig. 30). The conformation of the LBD trapped by ZK 200775 while similar to the apo, resting state, reported for the isolated LBD25, is nevertheless more open or overextended (Supplementary Fig. 31). Binding of full agonists, such as glutamate, quisqualate or AMPA result in closure of the clamshell by movement of domain 2 closer to domain 1 by a ~25° rotation (Supplementary Figs. 30,31)25, 77. In the context of the water-soluble LBD dimer, closure of both clamshells increases the separation of the regions linking the binding domains to the transmembrane domains by ~20 Å, using the α-carbon of Pro 632 (M3-S2 linker) as a reference (Supplementary Fig. 30). This movement, therefore, ‘pulls apart’ the M3 helices at the bundle crossing, opens the ion channel, and is the fundamental conformational change, within the binding domains, that transmits the energy associated with agonist binding to the work required to open the ion channel.
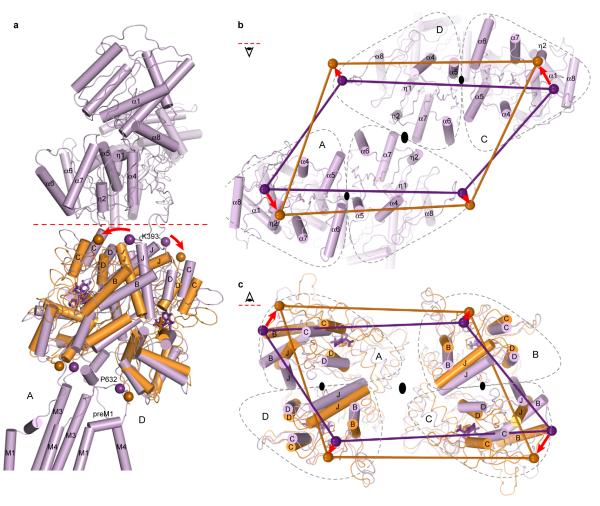
Superposition of two ‘dimer’ structures of the soluble LBD glutamate complex25 onto the LBDs of the GluA2cryst structure allows us to visualize activation-related movements in the context of the tetrameric receptor (Fig. 11, Supplementary Fig. 32). Using α-carbon atoms of residues Lys 506, Pro 632, and Cys 773 as reference points for the S1-M1, M3-S2, and S2-M4 linkers, respectively (Supplementary Fig. 33), the most significant movement involves the M3-S2 linker, an observation in agreement with the critical role of the M3 helix in channel gating. As a consequence of the overall 2-fold symmetry relating one ligand binding domain dimer to the other, the movements of the M3-S2 linkers are ~2-fold symmetric, i.e. there are large conformational changes within dimers and smaller changes between dimers (Fig. 11c, d), consistent with chemical modification studies suggesting a breakdown in the 4-fold symmetry of the ion channel upon receptor activation78. Augmenting the intradimer motions of this simple model, there is almost certainly a perpendicular component of movement, between LBD dimers, to facilitate opening of the ~4-fold symmetric pore. A component of movement between dimers is supported by the observation that reduction of the Ile 664 disulfide bridge potentiates glutamate-induced GluA2 currents58, 79 (Supplementary Fig. 20).
Figure 11. iGluR activation gating.
a, The structure of GluA2cryst with two subunits (A and C) transparent. Red dashed line indicates interface between LBD and TMD. b, Closeup of the LBD-TMD regions of subunits B and D. The structure of the water-soluble GluA2 LBD (S1S2) crystallized in complex with glutamate25 has been superimposed, using the D1 domain, on the corresponding region of GluA2cryst and is shown in green. Helical regions of the ion channel as well as parts of LBD that move upon iGluR activation are shown as cylinders. Purple and green spheres indicate positions of the α-carbons for the residues Lys 393 and Pro 632. Stick models of ZK200775 and glutamate are shown in purple and green, respectively. Red arrows indicate movement during iGluR activation. c-d, Views of the iGluR tetramer from interface between LBD and TMD (red dashed line in a) onto (c) LBD along the overall axis of the two-fold symmetry and (d) ion channel along the axis of four-fold symmetry. Gray dashed lines outline borders of the A, B, C, and D subunits. Purple and green lines connect Pro 632 α-carbon atoms.
The differences in position and conformations of the LBD - TMD ‘linkers’ between the A/C (proximal) and B/D (distal) subunit pairs also mean that the consequences of agonist binding and LBD clamshell closure on the TMD must necessarily be distinct (Fig. 11c, d). We can see that by measuring the distance between a reference residue, such as Pro 632 in the M3-S2 linker, and the overall 2-fold axis of symmetry, an axis which is approximately coaxial with the TMD 4-fold axis (Fig. 11c, d). In doing this, we see that the α-carbon of Pro 632 is ~13 Å and 25 Å distal from the 2-fold axis for the A/C and the B/D subunits, respectively. Upon agonist binding and LBD closure, we predict that the α-carbons of Pro 632 will move away from the 2-fold axis by ~4 Å and 7 Å for the A/C and B/D subunits, respectively. Thus, the extent of conformational movement for the proximal (A/C) and distal (B/D) subunits is substantially different and is greater for the distal (B/D) subunits. We therefore hypothesize that not only will the consequence of agonist binding to ion channel gating be different for the proximal (A/C) and distal (B/D) subunits, but that the agonist-induced conformational changes in the distal subunits may be more important to activation gating, simply because the extent of the predicted conformational change is larger. Conversely, we suggest that agonist-induced conformational changes in the proximal subunits may play a comparatively smaller role in activation gating. This explains, at least in part, why in NMDA receptors, glycine binding to the GluN1 subunit, which we predict occupies the proximal position in the LBD layer and thus transmits a smaller conformational displacement to the TMD in comparison to the distal subunits, does not result in significant ion channel opening in the absence of glutamate.
Mechanism of desensitization
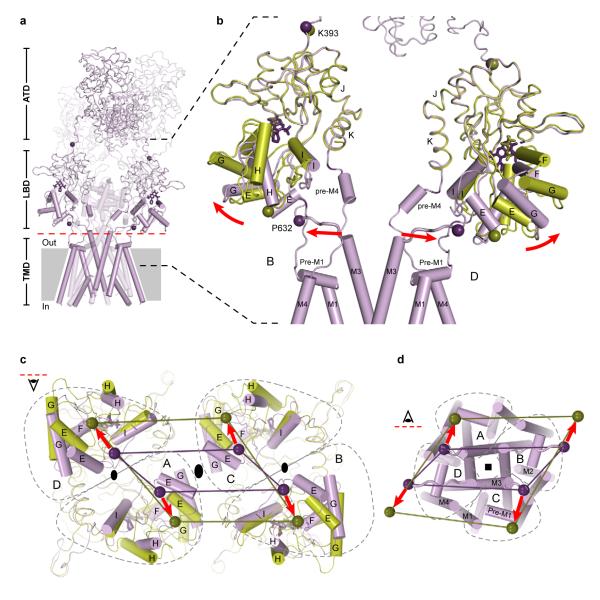
A hallmark of AMPA and kainate receptors is rapid and profound desensitization, or ion channel closure, following receptor activation12, 48. The molecular principles of desensitization and the structural relationships between the resting/closed and desensitized states of the ion channel pore are not yet understood for an intact receptor. To address these questions we can exploit the desensitized-like structure of the Ser 729 to Cys (S729C) mutant of the isolated LBD dimer58. By superimposing the desensitized-like S729C dimer onto the GluA2cryst structure we find that both D2 lobes of the isolated dimer superimpose well on the corresponding lobes of the tetrameric receptor, even though the isolated domains are bound with glutamate (closed clamshell) and in GluA2cryst, the LBDs are ‘open’ and bound with antagonist. This analysis demonstrates that peptide linkers connecting the LBDs to the TMDs can adopt a similar separation in the antagonist-bound GluA2cryst state and in the glutamate-bound S729C desensitized-like dimer form (Fig. 12 and Supplementary Fig. 30). Desensitization therefore simply involves rupture of the LBD D1-D1 interface and rotation of entire binding domain subunits to allow for the D2 domains and the linkers to the ion channel to adopt a closed state-like conformation.
Figure 12. iGluR desensitization.
a, Structure of GluA2cryst subunits A and D (purple) with superposed structure of the S729C dimer LBD (orange)58. Red dashed line indicates interface between ATD and LBD. Purple and orange spheres indicate positions of the α-carbons for the residues Lys 393 and Pro 632. Stick models of ZK200775 and glutamate are shown in purple and orange, respectively. Red arrows indicate movement of Lys 393 during GluR desensitization. b-c, Views on the GluA2cryst tetramer along the overall axis of two-fold symmetry from interface between ATD and LBD (red dashed line in a) looking into the ATD (b) and into the LBD (c). Gray dashed lines define boundaries of the A, B, C, and D subunits.
In the context of the GluA2cryst structure, rearrangement of D1-D1 LBD interface during desensitization (Fig. 12c) demands movement of the ATDs and the ATD-LBD linkers (Fig. 12b). Upon receptor desensitization, we predict changes in both the distances between and within ATD dimers. These observations provide mechanisms, grounded in three-dimensional structure, by which binding of ions and small molecules to the ATDs of NMDA receptors can modulate receptor function80. Ligands alter the conformation of the ATD ‘clamshell’ and propagate these conformational changes throughout the receptor either directly, through the ATD - LBD linkers, or indirectly, via changes in the ATD dimer-dimer contacts across the overall 2-fold axis of symmetry.
Conclusion
The GluA2cryst structure uncovers domain organization and molecular symmetry of iGluRs. Crystallographic and site-directed chemical modification data demonstrate that AMPA, kainate and NMDA receptors have ~2-fold symmetric ‘dimeric’ extracellular domains, a ~4-fold symmetric ion channel, and a symmetry mismatch that renders diagonally related subunit pairs distinct. In heterotetrameric GluN1/GluN2A NMDA receptors, subunits are positioned in a GluN1-GluN2A-GluN1-GluN2A pattern. Activation gating of iGluRs originates within individual LBDs with agonist binding inducing closure of the clamshell, separation of the transmembrane domain linkers, and opening of the ion channel gate. Desensitization (inactivation) results from rupture of the agonist-bound LBD dimer D1-D1 interface, leading to rigid body rotation of individual LBD domains within the dimer and allowing for D2 domains, linkers to the ion channel and the ion channel gate to adopt a closed state-like conformation. Gating can be perturbed at many sites by small molecule inhibitors,and modulators and auxiliary protein subunits (Supplementary Fig. 34). The underlying architecture and symmetry of iGluRs, as revealed by the GluA2cryst structure, has implications for understanding how these molecules perturb receptor function.
Methods summary.
The GluA2cryst – green fluorescent protein (GFP) fusion (see Supplementary Materials) was expressed in baculovirus-infected Sf9 insect cells and was purified using metal-affinity chromatography in buffers supplemented with 1.0 mM n-dodecyl-β-D-maltoside. Following cleavage by thrombin to remove GFP, the receptor was further purified by size-exclusion chromatography in a buffer supplemented with 1 mM n-undecyl-β-D-thiomaltoside and synthetic lipids. Crystallization was performed under paraffin oil, at 4° C, using a precipitating solution composed of 7-11% (w/v) PEG 20,000, 0.1 M 2-(N-morpholino)-ethanesulfonic acid (MES; pH 6.0-6.5) and 300 μM [[3,4-dihydro-7-(4-morpholinyl)-2,3-dioxo-6-(trifluoromethyl)-1(2H)-quinoxalinyl]methyl]phosphonic acid (ZK 200775). Selenomethionine-labeled receptor was prepared and crystallized using similar conditions. Diffraction data sets were indexed, integrated and scaled using HKL2000. Initial phase information was obtained by molecular replacement using the isolated amino terminal domains and ligand-binding domains as search probes. The ion channel and interdomain polypeptides were iteratively built using the computer graphics program COOT and the structure was refined using the computer program Phenix. The ion channel functional activity of the GluA2cryst construct was measured by whole-cell or outside-out patch clamp experiments. Ligand binding activity was evaluated by 3H-AMPA saturation and competition assays. NMDA receptor crosslinking experiments were done by transiently transfecting human embryonic kidney tsA 201 cells with wild type or mutant GluN1 and GluN2A plasmid DNA and analyzing oligomeric behavior of protein on gradient SDS PAGE gels by Western blot. Structure superpositions were done using Superpose (CCP4).
Supplementary Material
Acknowledgements
We thank the personnel at beamlines 5.0.2, 8.2.1, and 8.2.2 of the Advanced Light Source and at beamline 24-ID-E of the Advanced Photon Source. We also thank T. Homrichhausen for help with cloning and FSEC screening, L. Vaskalis for assistance with illustrations, and Gouaux lab members for discussion. M.P.R. was supported by an individual NIH National Research Service Award. This work was supported by the NIH. E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Cowan WM, Sudhof TC, Stevens CF, editors. Synapses. The Johns Hopkins University Press; Baltimore, MD: 2001. [Google Scholar]
- 2.Hayashi T. Effects of sodium glutamate on the nervous system. Keio J. Med. 1954;3:183–192. [Google Scholar]
- 3.Curtis DR, Phillis JW, Watkins JC. Chemical excitation of spinal neurons. Nature. 1959;183:611. doi: 10.1038/183611a0. [DOI] [PubMed] [Google Scholar]
- 4.Sugiyama H, Ito I, Watanabe M. Glutamate receptor subtypes may be classified into two major categories: A study on Xenopus oocytes injected with rat brain mRNA. Neuron. 1989;3:129–132. doi: 10.1016/0896-6273(89)90121-9. [DOI] [PubMed] [Google Scholar]
- 5.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacological Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 6.Jane DE, Lodge D, Collingridge GL. Kainate receptors: Pharmacology, function and therapeutic potential. Neuropharmacology. 2009;56:90–113. doi: 10.1016/j.neuropharm.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 7.Lipton SA. Paradigm shift in neuroprotection by NMDA receptor blockage: memantine and beyond. Nat. Rev. Drug Discov. 2006;5:160–170. doi: 10.1038/nrd1958. [DOI] [PubMed] [Google Scholar]
- 8.Alt A, Nisenbaum ES, Bleakman D, Witkin JM. A role for AMPA receptors in mood disorders. Biochem. Pharmacol. 2006;71:1273–1288. doi: 10.1016/j.bcp.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Labrie V, Roder JC. The involvement of the NMDA receptor d-serine/glycine site in the pathophysiology and treatment of schizophrenia. Neurosci. Biobehav. Rev. 2009 doi: 10.1016/j.neubiorev.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Boulter J, et al. Molecular cloning and functional expression of glutamate receptor subunit genes. Science. 1990;249:1033–1037. doi: 10.1126/science.2168579. [DOI] [PubMed] [Google Scholar]
- 11.Keinänen K, et al. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- 12.Sommer B, et al. A glutamate receptor channel with high affinity for domoate and kainate. EMBO J. 1992;11:1651–1656. doi: 10.1002/j.1460-2075.1992.tb05211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moriyoshi K, et al. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- 14.Monyer H, et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 15.Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- 16.Lu W, et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulle C, et al. Subunit composition of kainate receptors in hippocampal interneurons. Neuron. 2000;28:475–484. doi: 10.1016/s0896-6273(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 18.Christensen JK, Paternain AV, Selak S, Ahring PK, Lerma J. A mosaic of functional kainate receptors in hippocampal interneurons. J. Neurosci. 2004;24:8986–8993. doi: 10.1523/JNEUROSCI.2156-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wo ZG, Oswald RE. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 1995;18:161–168. doi: 10.1016/0166-2236(95)93895-5. [DOI] [PubMed] [Google Scholar]
- 20.O’Hara PJ, et al. The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron. 1993;11:41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- 21.Stern-Bach Y, et al. Agonist selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid-binding proteins. Neuron. 1994;13:1345–1357. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 22.Wollmuth LP, Sobolevsky AI. Structure and gating of the glutamate receptor ion channel. Trends Neurosci. 2004;27:321–328. doi: 10.1016/j.tins.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Soderling TR, Derkach VA. Postsynaptic protein phosphorylation and LTP. Trends Neurosci. 2000;23:75–80. doi: 10.1016/s0166-2236(99)01490-3. [DOI] [PubMed] [Google Scholar]
- 24.Kuusinen A, Abele R, Madden DR, Keinänen K. Oligomerization and ligand-binding properties of the ectodomain of the a-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunit GluRD. J. Biol. Chem. 1999;41:28937–28943. doi: 10.1074/jbc.274.41.28937. [DOI] [PubMed] [Google Scholar]
- 25.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 27.Jin R, et al. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–23. doi: 10.1038/emboj.2009.140. [PubMed: 19461580] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat. Struct. Mol. Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tichelaar W, Safferling M, Keinänen K, Stark H, Madden DR. The three-dimensional structure of an ionotropic glutamate receptor reveals a dimer-of-dimers assembly. J. Mol. Biol. 2004;344:435–442. doi: 10.1016/j.jmb.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–9. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 31.Chen G-Q, Cui C, Mayer M, Gouaux E. Functional characterization of a potassium-selective prokaryotic glutamate receptor. Nature. 1999;402:817–821. doi: 10.1038/45568. [DOI] [PubMed] [Google Scholar]
- 32.Mansour M, Nagarajan N, Nehring RB, Clements JD, Rosenmund C. Heteromeric AMPA receptors assemble with a preferred subunit stoichiometry and spatial arrangement. Neuron. 2001;32:841–853. doi: 10.1016/s0896-6273(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 33.Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J. Neurosci. 2003;23:1151–1158. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Erreger K, Chen PE, Wyllie DJ, Traynelis SF. Glutamate receptor gating. Crit. Rev. Neurobiol. 2004;16:187–224. doi: 10.1615/critrevneurobiol.v16.i3.10. [DOI] [PubMed] [Google Scholar]
- 35.Hansen KB, Yuan H, Traynelis SF. Structural aspects of AMPA receptor activation, desensitization and deactivation. Curr. Opin. Neurobiol. 2007;17:281–288. doi: 10.1016/j.conb.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 36.Paoletti P, Neyton J. NMDA receptor subunits: function and pharmacology. Curr. Op. Pharmacol. 2007;7:39–47. doi: 10.1016/j.coph.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 37.Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- 38.Johnson JW, Ascher P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 1987;325:529–531. doi: 10.1038/325529a0. [DOI] [PubMed] [Google Scholar]
- 39.Smith TC, Howe JR. Concentration-dependent substate behavior of native AMPA receptors. Nature Neurosci. 2000;3:992–997. doi: 10.1038/79931. [DOI] [PubMed] [Google Scholar]
- 40.Klein RM, Howe JR. Effects of the lurcher mutation on GluR1 desensitization and activation kinetics. J. Neurosci. 2004;24:4941–4951. doi: 10.1523/JNEUROSCI.0660-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vyklicky L, Benveniste M, Mayer ML. Modulation of N-methyl-D-aspartic acid receptor desensitization by glycine in mouse cultured hippocampal neurones. J. Physiol. (Lond.) 1990;428:313–331. doi: 10.1113/jphysiol.1990.sp018214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balannik V, Menniti FS, Paternain AV, Lerma J, Stern-Bach Y. Molecular mechanisms of AMPA receptor noncompetitive antagonism. Neuron. 2005;48:279–288. doi: 10.1016/j.neuron.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 43.Mony L, Kew JN, Gunthorpe MJ, Paoletti P. Allosteric modulators of NR2B-containing NMDA receptors: molecular mechanisms and therapeutic potential. Br J Pharmacol. 2009;157:1301–17. doi: 10.1111/j.1476-5381.2009.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin R, et al. Mechanism of positive allosteric modulators acting on AMPA receptors. J. Neurosci. 2005;25:9027–9036. doi: 10.1523/JNEUROSCI.2567-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobolevsky AI. Channel Block of Glutamate Receptors. In: Pandalai SG, editor. Recent Research Developments in Physiology. Research Signpost; Trivandrum: 2003. pp. 1–38. [Google Scholar]
- 46.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Sommer B, Köhler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 48.Sommer B, et al. Flip and flop: A cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 49.Turski L, et al. ZK200775: a phosphonate quinoxalinedione AMPA antagonist for neuroprotection in stroke and trauma. Proc Natl Acad Sci U S A. 1998;95:10960–5. doi: 10.1073/pnas.95.18.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayer ML, Armstrong N. Structure and function of glutamate receptor ion channels. Annu. Rev. Physiol. 2004;66:161–181. doi: 10.1146/annurev.physiol.66.050802.084104. [DOI] [PubMed] [Google Scholar]
- 51.Horning M, Mayer M. Regulation of AMPA receptor gating by ligand binding core dimers. Neuron. 2004;41:379–388. doi: 10.1016/s0896-6273(04)00018-2. [DOI] [PubMed] [Google Scholar]
- 52.Hollmann M, Maron C, Heinemann S. N-Glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron. 1994;13:1331–1343. doi: 10.1016/0896-6273(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 53.Wo ZG, Oswald RE. Transmembrane topology of two kainate receptor subunits revealed by N-glycosylation. Proc. Natl. Acad. Sci. USA. 1994;91:7154–7158. doi: 10.1073/pnas.91.15.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 55.Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nature Struct. Mol. Biol. 2006;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- 56.Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 57.Gielen M, et al. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 59.Chang H-R, Kuo C-C. The activation gate and gating mechanism of the NMDA receptor. J. Neurosci. 2008;28:1546–1556. doi: 10.1523/JNEUROSCI.3485-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kashiwabuchi N, et al. Impairment of motor coordination, Purkinje cell synapse formation, and cerebellar long-term depression in GluR delta 2 mutant mice. Cell. 1995;81:245–252. doi: 10.1016/0092-8674(95)90334-8. [DOI] [PubMed] [Google Scholar]
- 61.Zuo J, et al. Neurodegeneration in Lurcher mice caused by mutation in d2 glutamate receptor gene. Nature. 1997;388:769–773. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]
- 62.Yelshansky MV, Sobolevsky AI, Jatzke C, Wollmuth LP. Block of AMPA receptor desensitization by a point mutation outside the ligand-binding domain. J Neurosci. 2004;24:4728–36. doi: 10.1523/JNEUROSCI.0757-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Panchenko VA, Glasser CR, Mayer ML. Structural similarities between glutamate receptor channels and K(+) channels examined by scanning mutagenesis. J. Gen. Physiol. 2001;117:345–360. doi: 10.1085/jgp.117.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuner T, Seeburg PH, Guy H. Robert. A common architecture for K(+) channels and ionotropic glutamate receptors? Trends Neurosci. 2003;26:27–32. doi: 10.1016/s0166-2236(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 65.Long SB, Campbell EB, MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 66.Chen L, et al. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 67.Schwenk J, et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- 68.Soto D, Coombs ID, Kelly L, Farrant M, Cull-Candy SG. Stargazin attenuates intracellular polyamine block of calcium-permeable AMPA receptors. Nat. Neurosci. 2007;10:1260–1270. doi: 10.1038/nn1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sobolevsky AI, Yelshansky MV, Wollmuth LP. Different gating mechanisms in glutamate receptor and K+ channels. J. Neurosci. 2003;23:7559–7568. doi: 10.1523/JNEUROSCI.23-20-07559.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marquez-Klaka B, Rettinger J, Bhargava Y, Eisele T, Nicke A. Identification of an intersubunit cross-link between substituted cysteine residues located in the putative ATP binding site of the P2X1 receptor. J. Neurosci. 2007:1456–1466. doi: 10.1523/JNEUROSCI.3105-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pedersen SE, Cohen JB. d-Turbocurarine binding sites are located at a-g and a-d subunit interfaces of the nicotinic acetylcholine receptors. Proc Natl Acad Sci USA. 1990;87:2785–2789. doi: 10.1073/pnas.87.7.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuusinen A, Arvola M, Keinänen K. Molecular dissection of the agonist binding site of an AMPA receptor. EMBO J. 1995;14:6327–6332. doi: 10.1002/j.1460-2075.1995.tb00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Armstrong N, Sun Y, Chen G-Q, Gouaux E. Structure of a glutamate receptor ligand binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 74.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–52. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 75.Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 76.Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J. 2008;27:2158–2170. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin R, Horning M, Mayer ML, Gouaux E. Mechanism of activation and selectivity in a ligand-gated ion channel: Structural and functional studies of GluR2 and quisqualate. Biochemistry. 2002;41:15635–15643. doi: 10.1021/bi020583k. [DOI] [PubMed] [Google Scholar]
- 78.Sobolevsky AI, Yelshansky MV, Wollmuth LP. The outer pore of the glutamate receptor channel has 2-fold rotational symmetry. Neuron. 2004;41:367–378. doi: 10.1016/s0896-6273(04)00008-x. [DOI] [PubMed] [Google Scholar]
- 79.Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 80.Gielen M, Retchless B. Siegler, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.