Abstract
A family of integral membrane proteins containing a signature DHHC motif has been shown to display protein S-acyltransferase activity, modifying cysteine residues in proteins with fatty acids. The physiological roles of these proteins have largely been unexplored. Here we report that mice homozygous for a hypomorphic allele of a previously uncharacterized member, DHHC5, are born at half the expected rate, and survivors show a marked deficit in contextual fear conditioning, an indicator of defective hippocampal-dependent learning. DHHC5 is highly enriched in a post-synaptic density preparation and co-immunoprecipitates with post-synaptic density protein-95 (PSD-95), an interaction that is mediated through binding of the carboxyl terminus of DHHC5 and the PDZ3 domain of PSD-95. Immunohistochemistry demonstrated that DHHC5 is expressed in the CA3 and dentate gyrus in the hippocampus. These findings point to a previously unsuspected role for DHHC5 in post-synaptic function affecting learning and memory.
Keywords: Gene/Knockout, Membrane Function, Protein Targeting, Protein-Protein Interactions, Synapses
Introduction
Palmitoylation of proteins at intracellular sites is mediated by members of a family of S-fatty acyltransferases that are recognized by a DHHC consensus motif at the active site (1–4). In addition to functioning as protein S-acyltransferases, other functions have been proposed, with certain family members shown to transport divalent cations (5–6). The roles of DHHC proteins in the nervous system have attracted recent interest. Genetic alterations in DHHC9 and DHHC15 are responsible for two different forms of X-linked mental retardation in humans (7–9), whereas DHHC8 is contained within a microdeletion linked to schizophrenia in the Han Chinese population (10), and DHHC17 interacts with huntingtin (Htt) and may play a role in the pathogenesis of Huntington disease (11–13). Another DHHC family member, GODZ (DHHC3), has been shown to affect both excitatory and inhibitory synapse function (14–19), and DHHC23 (neuronal nitric oxide synthetase (nNOS)2-interacting DHHC domain-containing protein with dendritic mRNA (NIDD)) targets neuronal nitric oxide synthetase to synapses via its nNOS PDZ binding domain (20). Given the large number of family members (22 in human and 23 in mouse) and the daunting number of potential substrates (over 250 in the brain, for example) (21), mouse models will become increasingly important for understanding their physiological roles. Of note, the only previously available DHHC gene knock-out mouse model (the DHHC8 model (22)), exhibits behavioral abnormalities and defects in neuronal culture (such as diminished density of dendritic spines) (23).
In this study we show that a previously uncharacterized DHHC family member, DHHC5, is highly enriched in the brain and in synaptic vesicle fractions and that it interacts with the third PDZ binding domain of PSD-95 in vitro and in vivo. Mice homozygous for a hypomorphic allele of DHHC5 show impaired contextual fear conditioning. DHHC5 is shown to contain a hydroxylamine-sensitive acyl group in vivo, but PSD-95 palmitoylation is unchanged in DHHC5 knock-out mice, suggesting that PSD-95 is not a substrate (which is not surprising, as other DHHC family members have been previously shown to perform this function). We speculate that binding to PSD-95 places DHHC5 in proximity to other important synaptic signaling molecules, where it may regulate post-synaptic events at excitatory synapses important for hippocampal learning.
EXPERIMENTAL PROCEDURES
Materials and General Methods
Protein G-agarose beads were obtained from Pierce. FuGENE 6 was obtained from Roche Applied Science. All restriction enzymes were obtained from New England Biolabs (Ipswich, MA). All other reagents and chemicals were from Sigma. Protein concentrations were determined by the DC protein assay (Bio-Rad).
Antibodies
Chicken anti-mouse DHHC5 IgY antibodies were produced (Genetel Laboratories) using a peptide consisting of amino acids 245–261 of mouse DHHC5 (CSSPAPRYLGRPKKEKT) and affinity purified using a peptide column. This antibody was used for immunoblotting following in vitro co-immunoprecipitation assays at a dilution of 10:000. A rabbit anti-mouse DHHC5 polyclonal antibody was obtained from Sigma (catalogue no. HPA014670, distributed in Europe by Atlas Antibodies, Stockholm, Sweden). Immunoblotting with this antibody was performed using a dilution factor of 1:1000. The mouse anti-rat PSD-95 monoclonal antibody (6G6–1C9), the rabbit anti-rat VAMP1 polyclonal antibody, and the rabbit anti-rat calnexin polyclonal antibody were from Abcam (Cambridge, MA). A rabbit anti-rat PSD-95 polyclonal antibody was obtained from Synaptic Systems (Goettingen, Germany). The rabbit anti-human GPRIN1 polyclonal antibody was obtained from Proteintech Group (Chicago, IL). The Alexa Fluor 568 goat anti-mouse IgG secondary antibody was obtained from Invitrogen. Anti-HA2 ascites was obtained from Covance (Emeryville, CA). The anti-Myc mouse monoclonal antibody (9E10), the anti-Myc rabbit polyclonal antibody, the normal mouse IgG control, the anti-rat GAP43 monoclonal antibody, and the anti-bovine COXIV monoclonal antibody were obtained from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmids
A full-length mouse DHHC5 cDNA clone was obtained from Open Biosystems (catalogue number MMM1013-65887); the sequence corresponds to the mouse DHHC5 cDNA in GenBankTM BC_020051. For yeast two-hybrid screening, sequences corresponding to the amino acids 220–715 (the carboxyl-terminal cytosolic domain) were cloned into the pLexN vector (Invitrogen) using EcoRI and BamHI sites. Full-length mouse PSD-95, PICK1, and GRIP1 cDNAs were obtained from Open Biosystems (catalogue numbers EMM1002-11769, MMM1013-9200227, and MMM1013-98478796, respectively, corresponding to GenBankTM BC_014807, GenBankTM BC_048788, and BC_067398). The full-length PSD-95 cDNA and fragments corresponding to amino acids 245–517, 245–447, and 397–721 were cloned into pVP16-3 (Clontech) using NotI and XbaI for use in two-hybrid assays. The mouse PICK1 cDNA (and a fragment corresponding to amino acids 1–200) and GRIP 1 cDNA (and fragment corresponding to amino acids 401–800) were cloned into pVP16-3 at EcoRI and BglII sites. The mouse DHHC5 carboxyl-terminal fragment (amino acids 220–715) was cloned into the pCMV-HA vector (Clontech) using EcoRI and BglII sites to generate the plasmid pHA-CtermDHHC5. Plasmid pHA-CtermDHHC5ΔEISV that deletes the extreme carboxyl terminus of DHHC5 was generated from the parent plasmid by site-directed mutagenesis (QuikChange II XL, Stratagene). The full-length mouse PSD95 was cloned into pcDNA-Myc-His vector (Invitrogen) using NheI and XhoI sites and into pEFGP-C1 (Clontech) using XhoI and SacII sites. The coding regions of all the above plasmids were sequenced to ensure integrity of the constructs. Further details of plasmid construction including primer sequences are available upon request.
Generation of DHHC5-deficient Mice
A mouse embryonic stem cell line (RRD553, strain 129/Ola) with an insertional mutation in DHHC5 was obtained from BayGenomics (24) through the International Gene Trap Consortium (25, 26). The gene-trapping vector, pGT11xf, was designed to introduce an in-frame fusion between the 5′ exons of the trapped gene and a reporter, βgeo (a fusion of β-galactosidase and neomycin phosphotransferase II). To determine the location of the genomic insertion site in the RRD553 stem cell line, genomic DNA was extracted from the embryonic stem cells using the DNeasy blood and tissue kit (Qiagen). PCR was then performed using primers fp1 (within exon 3 of DHHC5, 5′-CCAGGACTAAGCCTGAATGTGTCACC-3′) and rp1 (within the βgeo gene of the gene-trapping vector, 5′-TGCCCAGTCATAGCCGAATA-3′), and the PCR product was sequenced to determine the insertion site. The embryonic stem cells were injected into C57BL/6 blastocysts to create chimeric mice, which were bred with C57BL/6 mice to generate heterozygous DHHC5-deficient mice. The heterozygous mice were then interbred to generate all genotypes of DHHC5-deficient mice. The mice were weaned at 21 days of age and housed in an approved barrier facility with a 12-h light/dark cycle. All animal experiments were performed with the approval of the Institutional Animal Care and Research Advisory Committee at the University of Texas Southwestern Medical Center at Dallas.
Genotyping by Southern Blot
Genomic DNA was extracted from mouse tail tips using the Puregene Core Kit A (Qiagen). Approximately 10–20 μg of genomic DNA was digested with EcoRV and PvuII and analyzed by Southern blot. A probe (163 bp) was generated by polymerase chain amplification from mouse genomic DNA using primers 5′-CAGGTGTCCAGGACTAAGCC-3′ and 5′-CAACAGGGAGCTTACATGAGA-3′ derived from sequences within DHHC5 exon 3 (NCBI Entrez Gene ID 228136). The wild-type and mutant alleles are detected as 6.0- and 4.7-kb bands, respectively.
Quantitative Reverse Transcription-PCR
Total RNA was extracted from whole mouse brains using an RNeasy Lipid Tissue Midi kit (Qiagen) following the manufacturer's protocol. First-strand synthesis was performed using RNA (1 μg) and random primers using the iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR was performed using iTaq Supermax with ROX (Bio-Rad). The TaqMan primers, corresponding to sequences in exons 3 and 4 of DHHC5, were obtained from Applied Biosystems (assay ID Mm00523158_m1). Glyceraldehyde-3-phosphate dehydrogenase sequences were used for normalization (assay ID Mm03302249_g1). The thermal cycling conditions were 3 min at 95 °C followed by 50 cycles of denaturation for 15 s at 95 °C and annealing for 45 s at 60 °C. Normalized data were used to compare relative levels of DHHC5 in +/+, +/gt, and gt/gt samples using ΔΔCt analysis (27).
Whole Membrane Lysates
Mice were euthanized by carbon dioxide narcosis, and tissues were quickly extracted on ice. Each tissue sample was homogenized in four volumes of 20 mm HEPES, pH 7.4, 150 mm NaCl, 1 mm EDTA with 1× complete protease inhibitors (Roche Applied Science) using a Brinkman Polytron homogenizer. Unbroken tissue and cells were removed by centrifugation at 1000 × g for 15 min at 4 °C. The supernatant was further centrifuged at 100,000 × g for 1 h. The whole membrane lysate was extracted from the pellet using radioimmune precipitation assay buffer (50 mm Tris, pH 7.2, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) containing 1× complete protease inhibitors at 4 °C for 1 h. The protein sample was then centrifuged at 16,000 × g for 10 min at 4 °C. The cleared supernatant was transferred to a clean Eppendorf tube. The proteins were separated by SDS-PAGE and analyzed by immunoblotting.
Synaptic Plasma Membrane Fractionation
Synaptic plasma membrane fractionation was performed according to a previously published method with some modifications (28). Procedures were carried out at 4 °C, and all buffers contained complete protease inhibitors (Roche Applied Science). Briefly, whole brain tissues were pooled from six adult mice and homogenized in 15 volumes of Tris acetate (TA) buffer (50 mm Tris acetate, pH 7.4, 2 mm CaCl2, 10% (w/v) sucrose) using a glass-Teflon homogenizer. Homogenates were centrifuged at 1000 × g for 15 min to obtain a crude nuclear pellet (P1) and supernatant (S1). The S1 fraction was then centrifuged at 16,000 × g for 30 min, and the resulting mitochondrial pellet (P2) was resuspended in 20 mm HEPES, pH 7.4, and 2 mm CaCl2 and centrifuged at 16,000 × g for 20 min. The resulting pellet (P3) was resuspended in 5 volumes of hypotonic TA buffer (5 mm Tris acetate, pH 8.1, 2 mm CaCl2) and incubated on ice for 45 min. Solid sucrose was added to this fraction to 35% (w/v) and then layered on the bottom of a discontinuous sucrose density gradient. TA buffer containing 20 and 10% (w/v) sucrose constituted the middle and upper layers, respectively. The gradient was centrifuged at 60,000 × g for 2 h in a swinging bucket rotor, and the synaptosomal fraction was collected from the 35–20% sucrose interface. The synaptosomal fraction was diluted with 3 volumes of TA buffer and centrifuged at 30,000 × g for 30 min. The resulting synaptosomal pellet was resuspended in TA buffer containing 1% Triton X-100 and centrifuged at maximum speed in a microcentrifuge for 30 min. The supernatant containing the presynaptic membrane fraction was recovered, and the pellet containing the postsynaptic membrane fraction was resuspended in radioimmune precipitation assay buffer (50 mm Tris, pH 7.2, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS). Equal amounts of protein (20 μg) were loaded onto SDS-PAGE gels, transferred to Immobilon membranes, and subjected to immunoblotting.
Yeast Two-hybrid Assays
A rat brain cDNA library (postnatal day 8) cloned into the prey vector pVP16-3 was a generous gift of Dr. Thomas Sudhof (29). L40 yeast strains harboring the bait vector (pLexN) expressing the carboxyl-terminal fragment of mouse DHHC5 protein (amino acids 220–715 cloned into EcoRI and BamHI sites) and prey vector (pVP16-3) containing the rat brain cDNA library were grown on CSM-WLH (−Trp/−Leu/−His) plates (Clontech). In this protocol, only those clones that contain DHHC5-interacting partners will induce the transcription of the HIS3 gene and survive selection in the absence of histidine. A total of 100 clones were randomly selected for sequencing. The yeast DNA was extracted and subjected to PCR using primers within the vector sequence; V1 (5′-GTTTACCGATGCCCTTGG-3′) and V2 (5′-CGTTGTAAAACGACGGCC-3′). The PCR products were sequenced using V1 primer. Two rounds of selection and sequencing were performed.
To confirm the interactions between DHHC5 and PDZ domain-containing proteins, similarly, L40 yeast strains harboring the bait vector (pLexN) expressing amino acids 220–715 of the mouse DHHC5 protein and prey vectors (pVP16-3) expressing fragments of the PDZ domain-containing proteins were grown on CSM-WLH (−Trp/−Leu/−His) plates. Interactions in yeast were confirmed using a semi-quantitative β-galactosidase assay (29).
Co-immunoprecipitation
For co-immunoprecipitation of DHHC5 and PSD-95 in transfected cells, HEK 293 cells were transfected either with PSD-95-Myc or HA-CtermDHHC5 or HA-CtermDHHC5ΔEISV as indicated. After 48 h, cells were washed twice with cold phosphate-buffered saline and lysed with four volumes of lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA) containing a complete protease inhibitor mixture (Roche Applied Science). For the co-immunoprecipitation assay, 500 μg of cell lysate was diluted to 500 μl using Buffer A (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm EDTA, 0.25% gelatin, 0,02% sodium azide) and pre-cleared with 25 μl of Protein G-agarose beads (Pierce) at 4 °C for 1 h. The pre-cleared solution was incubated with either anti-Myc mouse monoclonal antibody (9E10) (3 μg, Santa Cruz) or anti-HA ascites (6 μg, Covance) at 4 °C for 1 h followed by incubation with 25 μl of Protein G-agarose beads (Pierce) at 4 °C overnight. After incubation, the beads were washed three times with Buffer A and then eluted with 30 μl of SDS-PAGE sample buffer (62.5 mm Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, and 0.001% bromphenol blue) containing 5% β-mercaptoethanol. Eluted samples were analyzed by immunoblotting.
To analyze co-immunoprecipitation of DHHC5 and PSD-95 in vivo, mouse brain tissues were homogenized in four volumes of buffer containing 20 mm HEPES, 150 mm NaCl, 1 mm EDTA, pH 7.4, with 1× complete protease inhibitors (Roche Applied Science) using a motor-driven homogenizer. The unbroken tissue and cells were removed by centrifugation at 1000 × g for 15 min at 4 °C. The supernatant was further centrifuged at 100,000 - g for 1 h. The whole membrane lysate was extracted from the pellet using Buffer B (50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1% sodium deoxycholate) containing 1× complete protease inhibitors at 4 °C for 1 h. The protein sample was then centrifuged at 16,000 × g for 10 min at 4 °C. The cleared supernatant was transferred to a clean Eppendorf tube. For co-immunoprecipitation assay, 600 μg of brain membrane lysate was pre-cleared with 25 μl of Protein G-agarose beads (Pierce) at 4 °C for 1 h. The pre-cleared solution was incubated with anti-PSD 95 monoclonal antibody (1:40 dilution, 6G6–1C9, Abcam) at 4 °C for 1 h followed by an incubation with 25 μl of Protein G-agarose beads (Pierce) at 4 °C overnight. Normal mouse IgG (Santa Cruz sc-2025) at the same concentration was used as a negative control. After incubation, the beads were washed 3 times with Buffer B and then eluted with 30 μl of SDS-PAGE sample buffer (62.5 mm Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, and 0.001% bromphenol blue) containing 5% β-mercaptoethanol. Eluted samples were subjected to immunoblotting using the anti-DHHC5 (Sigma HPA014670) or anti-PSD-95 (Synaptic Systems) rabbit polyclonal antibodies.
Behavioral Testing
All mice were male and female littermates ∼10–12 weeks of age at the time of testing. Mice were group housed on a 12-h light/dark cycle with food and water available ad libitum. Behavioral testing occurred during the light cycle. Tests for locomotor activity, open field behavior, motor coordination (Rotarod), and fear conditioning were performed; all mice received all tests. Locomotor activity was assessed by placing mice individually into a clean, plastic mouse cage (18 × 28 cm) with a small amount of bedding that was located inside a dark Plexiglas box. Movement was monitored by 5 photobeams in one dimension (Photobeam Activity System, San Diego Instruments, San Diego, CA) for 2 h, with the number of beam breaks recorded every 5 min. For open field activity testing, mice were individually placed in the periphery of a novel open field environment (44 × 44 cm, walls 30 cm high) and allowed to explore for 5 min. The animals were monitored from above by a video camera connected to a computer running video tracking software (Ethovision 3.0, Noldus, Leesburg, VA) to determine the time, distance moved, and number of entries into two areas: the periphery (5 cm from the walls) and the center (14 × 14 cm). The open field arenas were wiped with disinfectant and allowed to dry between mice. For motor coordination testing, mice were placed on a rotarod (model 755, IITC Life Science Inc., Woodland Hills, CA). The rod was then accelerated from 5 to 45 rpm over 5 min. The time that each mouse fell from the rod was recorded. If a mouse held onto the rod and rotated completely around two times, this was recorded as a fall from the rod at that time. Each mouse was tested four times (15–30 min inter-trial interval) each day for two consecutive days. Fear conditioning was measured in boxes equipped with a metal grid floor connected to a scrambled shock generator (Med Associates Inc., St. Albans, VT). On the first day (training) mice were individually placed in the testing chamber and after 2 min received 2 tone-shock pairings (30-s white noise, 95-db tone co-terminated with a 2-s, 0.5-mA foot-shock, 1-min intertrial interval) and were removed from the chamber 1 min after the last shock. Twenty-four hours after training, memory of the context was measured by placing the mice into the same chambers, and freezing was manually measured every 5 s for 5 min. Forty-eight hours after training, memory for the white noise cue was measured by placing the mice in a box with altered floors and walls, lighting, and a vanilla smell. Freezing was manually measured every 5 s for 3 min, then the noise cue was turned on for an additional 3 min, and freezing was measured every 5 s. The experimenter rating the freezing was blind to the genotype of the animals. Approximately 1 week after fear conditioning, the animals were assessed for foot-shock sensitivity. The mice were placed into the same chambers and given a foot-shock every 15 s. For this test, the first foot-shock was 0.05 mA and increased by 0.05-mA intervals. The first shock that evoked a flinch response and jump, and vocalization was recorded.
Immunohistochemistry
To investigate the extent of the DHHC5 immunoreactivity within the brains of wild-type and DHHC5 gt/gt mice, a one-in-six series of free-floating 40-μm frozen coronal sections from each animal was stained via a modified version of a previously published immunoperoxidase protocol (30). Sections underwent antigen retrieval via heating in citrate buffer, pH 6.0, for 30 min at 80 °C and were allowed to cool to room temperature. To quench endogenous peroxidase activity, sections were incubated for 30 min in 1% H2O2 and subsequently blocked for 40 min in 15% normal goat serum in Tris-buffered saline, 0.3% Triton X-100 (TBS-T). Sections were incubated overnight at 4 °C in polyclonal rabbit anti-DHHC5 (1:250) diluted in TBS-T containing 10% normal goat serum. Subsequently, sections were rinsed in TBS and incubated for 2 h with secondary antibody (biotinylated goat anti-rabbit IgG, Vector Laboratories, Peterborough, UK, 1:1000) diluted in TBS-T containing 10% normal goat serum. After rinsing in TBS, sections were incubated for 2 h in avidin-biotin-peroxidase complex (Vectastain Elite ABC kit, Vector Laboratories) diluted in TBS. After rinsing in TBS, DHHC5 immunoreactivity was visualized by incubation in 0.05% diaminobenzidine tetrahydrochloride (Sigma) and 0.001% H202 for 25 min, a sufficient time to ensure saturation for this reaction. Sections were rinsed in TBS, mounted onto slides, air-dried, cleared in xylene, and coverslipped in DPX (Sigma). Appropriate negative controls where the primary antibody was omitted were included in each experiment.
RESULTS
Generation of DHHC5 Gene-trapped Mice
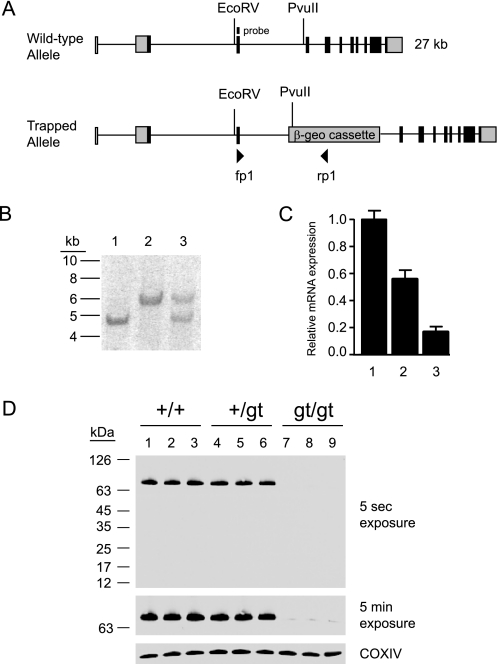
To study the function of the DHHC5 gene in vivo, DHHC5-deficient mice were derived from a mouse embryonic cell line (RRD553) generated by random insertional mutagenesis of 129/Ola embryonic stem cells at BayGenomics and distributed by the International Gene Trap Consortium (25, 26). Genomic DNA obtained from the cell line (and subsequent tail DNA) was found to contain the β-geo cassette insertion in intron 3 on the mouse DHHC5 gene, between nucleotides 5′-cctgtattatcacttatttt-3′ and 5′-tcttttttttttttaattag-3′ (25579683 and 25579684 in GenBankTM accession no. NT_039207.7) (Fig. 1A). The RRD553 cell line was injected into C57BL/6J blastocysts to create chimeric male mice, which were bred with C57BL/6J females to produce an F1 line. A long polypyrimidine tract located immediately 5′ to the insertion site precluded routine PCR as a genotyping method. Therefore, as shown in Fig. 1B, Southern blotting was used for the subsequent genotyping, and wild-type, heterozygous, and homozygous mice were clearly distinguished. DHHC5 heterozygote males and females from the F1 generation were bred to obtain an F2 generation with all genotypes.
FIGURE 1.
Insertional mutagenesis of the mouse DHHC5 gene. A, shown is a schematic of the insertional mutation in DHHC5. The β-geo cassette was found to be inserted into intron 3 of the DHHC5 gene. Binding sites for primers fp1 and rp1 are also shown. B, a Southern blot of EcoRv/PvuII digest of genomic mouse tail DNA using an exon 3 probe is shown. The wild-type allele yielded a 6.0-kb band, and the mutant allele yielded a 4.7-kb band. Lane 1, homozygous gt/gt; lane 2, homozygous +/+; lane 3, heterozygous +/gt. C, DHHC5 mRNA levels were determined by quantitative real-time PCR performed on total RNA extracted from DHHC5 +/+, +/gtv and gt/gt mouse brains. Expression levels were normalized to glyceraldehyde-3-phosphate dehydrogenase, n = 5, in each group. D, DHHC5 immunoblotting of membrane extracts (25 μg of protein) prepared from DHHC5 +/+, +/gt, and gt/gt mouse brains is shown. The rabbit polyclonal anti-DHHC5 antibody was used for detection (1:1000, Sigma). Three mice are shown for each genotype. COXIV was immunostained to serve as a loading control.
Genotypes produced by early heterozygous crosses deviated significantly from Hardy-Weinberg equilibrium (+/+, 97 (34%); +/gt, 154 (53%); gt/gt, 37 (13%), n = 288, p < 0.0001), consistent with ∼50% loss of gt/gt homozygotes before birth. The modest decrease in expected heterozygotes did not reach statistical significance (p = 0.08). An increased number of resorption sites was seen in heterozygous matings (data not shown). However, live-born homozygotes appear healthy and reproduce sufficiently to maintain the line in the homozygous state.
The β-geo cassette consists of a splice acceptor site fused in frame to a stop codon, which leads to truncation of the normal protein product. However, inefficient use of the splice acceptor site may in some instances cause a small amount of normal mRNA to be produced (31). Therefore, we determined the DHHC5 mRNA level in all three genotypes using quantitative reverse transcriptase-PCR. Indeed, a residual level of DHHC5 mRNA in the homozygous mice (about 16% compared with wild-type) was detected (Fig. 1C). The amount of DHHC5 protein produced in numerous experiments was determined to be no greater than 7% in brain tissue, as estimated by immunoblotting (Fig. 1D). For this reason, the insertion should be considered a hypomorphic allele and is designated “gt” (for gene-trapped allele).
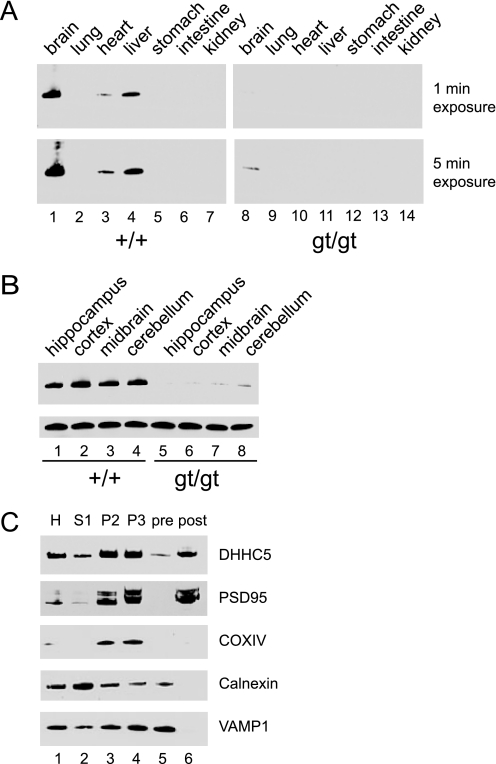
DHHC5 Is Enriched in Brain and Localized to Synaptic Membranes
Extracts prepared from various wild-type and DHHC5 gt/gt tissues were analyzed by immunoblotting using a rabbit polyclonal anti-DHHC5 antibody (Fig. 2A). In wild-type mice, the protein was found to be highly enriched in brain, detectable in liver and heart, and undetectable in a number of other tissues. No detectable DHHC5 was found in gt/gt homozygotes in any tissue examined except the brain. No striking differences were seen upon gross dissection of different brain regions (Fig. 2B).
FIGURE 2.
DHHC5 tissue distribution and subcellular fractionation. A, tissue distribution of DHHC5 in wild-type and DHHC5 gene-targeted mice is shown. Immunoblotting of whole tissue extracts (25 μg of protein/lane) was performed using an anti-DHHC5 polyclonal antibody (1:1000, Sigma). B, regional distribution of DHHC5 is shown. Brain tissue was dissected, and immunoblotting was performed on whole tissue homogenates. COXIV was used as a loading control. C, DHHC5 in pre- and post-synaptic membrane fractions is shown. Twenty micrograms of protein were loaded in each lane. H, homogenate; S1, post-nuclear supernatant; P2, mitochondrial pellet; P3, second mitochondrial pellet; pre, pre-synaptic membrane fractions; post, post-synaptic membrane fraction. Markers shown were used to detect post-synaptic membranes (PSD95), mitochondria (COXIV), endoplasmic reticulum (calnexin), and pre-synaptic membranes (VAMP1). Results shown were from one of two independent experiments giving similar results.
To further localize DHHC5 within brain tissue, we used a synaptic fractionation protocol to prepare pre- and post-synaptic membrane fractions to examine the distribution of DHHC5. As shown in Fig. 2C, a clean separation between pre- and post-synaptic densities was achieved; for example, VAMP1, a presynaptic membrane marker, was only detected in the presynaptic membrane fraction (lane 5), whereas PSD-95, a post-synaptic membrane marker, was only found in the post-synaptic membrane fraction (lane 6). DHHC5 was clearly detected in these synaptic membrane fractions, predominantly in the post-synaptic density.
Effect of DHHC5 Deficiency on Hippocampal-dependent Learning and Memory
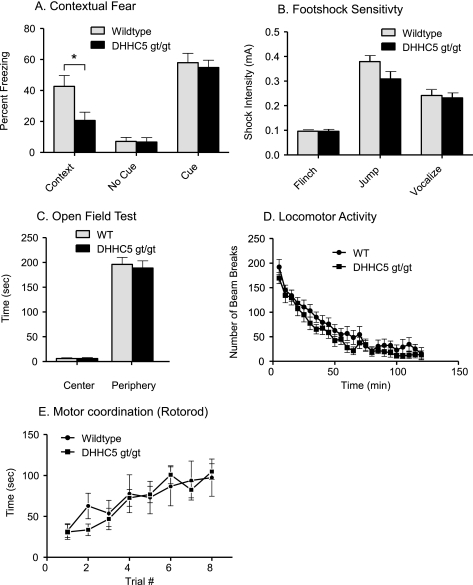
To further delineate the role of DHHC5 in brain function, we subjected the DHHC5 gt/gt and littermate control mice to several standard behavioral tests (Fig. 3). Contextual fear-conditioning is a hippocampus-dependent learning task in which animals associate a specific location and its surroundings with an electric shock. The standard protocol involves a learning (acquisition) phase and a testing phase. Mice tested at 10–12 weeks showed no significant differences in the acquisition phase of testing (data not shown). However, as shown in Fig. 3A, 24 h after training, there was a significant reduction in the amount of freezing for DHHC5 gt/gt mice in the context where they previously received the foot-shocks. There was no difference in the amount of freezing in a context not previously paired to foot-shock, and the freezing to the tone (cued condition) was normal. Foot-shock sensitivity was unimpaired (Fig. 4B), as were a number of other behavioral tests, such as open field activity, locomotion, and motor coordination and learning (Fig. 3, C–E). These results suggest that DHHC5 plays a role in hippocampal learning.
FIGURE 3.
Impaired contextual fear learning in DHHC5 gt/gt mice. A, contextual and cued fear conditioning is shown. Significant deficits in contextual (p < 0.025, unpaired t test, two-tailed), but not cued freezing, were observed in the DHHC5 gt/gt mice (n = 10), compared with their +/+ littermate controls (n = 12). B, foot-shock sensitivity is shown. The same mice used in A were tested for foot-shock sensitivity. No significant difference was observed between DHHC5 +/+ and gt/gt animals. C, an open field test is shown. The average amount of time spent in the center, non-periphery, and periphery of an open field by DHHC5 +/+ and gt/gt mice was calculated. The means (±S.E.) are presented, and n = 12 for wild-type (WT) and n = 11 for DHHC gt/gt mice. D, locomotor activity is shown. DHHC5 +/+ (n = 12) and DHHC5 gt/gt mice (n = 11) were observed in a clean, plastic mouse cage located inside a dark Plexiglas box. Data are expressed as the average number of beam breaks at each 5-min bin per 2-h test period ± S.E. E, motor coordination (Rotarod test) is shown. The time to fall from a rotating rod that was accelerated from 5 to 45 rpm over 5 min was assessed. Each mouse was tested four times (15–30-min intertrial interval) each day for two consecutive days. n = 12 for wild-type and n = 10 for DHHC5 gt/gt mice.
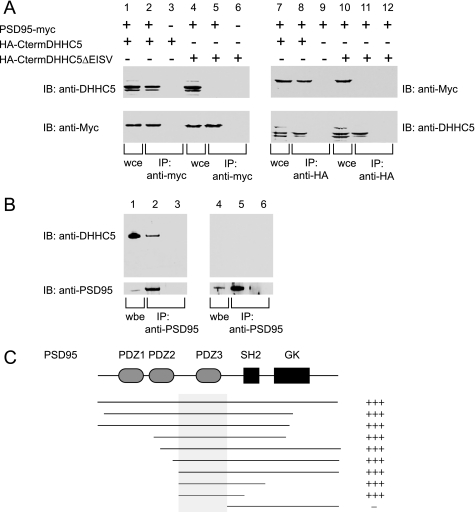
FIGURE 4.
Coimmunoprecipitation of DHHC5 and PSD-95 in transfected cells and brain and determination of the interaction site. A, co-immunoprecipitation of PSD-95 and DHHC5 carboxyl-terminal cytoplasmic domain in transfected cells is shown. HEK-293 cells were transfected with PSD-95-myc and HA-CtermDHHC5, a construct that consisted of an HA epitope tag fused to the carboxyl-terminal cytoplasmic domain of DHHC5. A construct in which the final four amino acids were deleted (ΔEISV) was also tested. In the left panel, immunoprecipitation was performed using an anti-Myc rabbit polyclonal antibody to precipitate PSD-95, and in the right panel, an anti-HA rabbit polyclonal antibody was used to precipitate DHHC5. Whole cell extracts (wce) consisting of 5% of the total mixture (lanes 1, 4, 7, and 10) or entire immunoprecipitate (IP) (other lanes) were loaded onto SDS-PAGE gels and immunoblotted (IB) using an anti-DHHC5 chicken IgY antibody or anti-Myc mouse monoclonal antibody as indicated. B, DHHC5 was detected in an immunoprecipitate of PSD-95 from mouse brain. Mouse brain lysates were prepared (lanes 1 and 4) and incubated with anti-PSD-95 monoclonal antibody (lanes 2 and 5) or control mouse IgG (lanes 4 and 6) and subjected to SDS-PAGE and immunoblotting with the anti-DHHC5 polyclonal antibody (upper panel) or anti-PSD-95 rabbit polyclonal antibody (lower panel). (upper panel, 1% of input; lower panel, 2% of input). Lanes 1–3, wild-type brain, lanes 4–6, DHHC gt/gt brain. The experiment was repeated three times with similar results. C, deletion analysis of PSD-95 clones in a yeast two-hybrid assay with the carboxyl-terminal cytoplasmic domain of DHHC5 is shown. L40 yeast strains harboring the bait vector (pLexN) expressing the amino acids 220–715 of mouse DHHC5 protein and prey vectors (pVP16-3) expressing different fragments of PDZ domain-containing proteins were grown on CSM-WLH (−Trp/−Leu/−His) plates. Only those that contain DHHC5-interacting partners survive the −His selection. GK, guanylate kinase-like domain.
Candidate DHHC5 Interacting Proteins
To further delineate the role of DHHC5 in brain, the 496-amino acid carboxyl terminus of DHHC5 was cloned into a bait vector for yeast two-hybrid screening against a library derived from whole rat brain cDNA. This large domain is predicted to reside in the cytosol based on comparison with homologous proteins (32, 33). In two independent trials, 100 positive clones were recovered from a total of 12 × 106 clones, and 64 of these were sequenced for identification.
As shown in supplemental Table 1, the clones fell into two major classes, PDZ-domain containing proteins and cytoskeletal proteins. The most frequently identified clone was PSD-95 (12/64 hits). Other PDZ domain family members recovered were protein that interacts with C kinase 1 (PICK1) (2 hits), glutamate receptor interacting protein-1 (GRIP1) (1 hit), and GRIP2 (1 hit). These findings suggested that the interactions may be mediated through a PDZ binding motif of DHHC5. Indeed, the last four amino acids of DHHC5, EISV, form a canonical class II PDZ binding motif (XΦXΦCOOH) (34).
Interaction of PSD-95 and DHHC5 in Transfected Cells and in Mouse Brain
As PSD-95 was the most frequently identified DHHC5 interacting partner in the yeast two-hybrid assay, we further tested its interaction with DHHC5 in a mammalian system using a co-immunoprecipitation assay. As we were unable to solubilize full-length DHHC5 when it was overexpressed in heterologous cells, we used the large carboxyl-terminal fragment (amino acids 220–715) with an amino-terminal epitope tag that was similar to that used in the yeast two hybrid screen. A custom made chicken anti-mouse DHHC5 IgY was used in these assays to detect the DHHC5 carboxyl terminus. As shown in Fig. 4A, lane 1, both expressed proteins could be detected in whole cell extracts (wce) when 293 cells were co-transfected with PDS95-myc and the HA-tagged DHHC5 carboxyl terminus (designated HA-CtermDHHC5). The DHHC5 carboxyl terminus appeared as a doublet, which was resistant to alkaline phosphatase or endoglycosidase H or peptide N-glycosidase F digestion (data not shown). Immunoprecipitation using anti-Myc antibodies to pull down PSD-95 were also able to co-immunoprecipitate DHHC5 (lane 2), confirming that the interaction occurs in transfected cells. This interaction was not seen when an empty vector control was used (lanes 3 and 6) or when the final four amino acids of DHHC5 (the PDZ binding domain) were deleted (lane 5). Fig. 4A, lanes 7–12, demonstrate the same interaction when the antibodies are reversed, i.e. when the HA epitope tag is used to pull down the DHHC5 carboxyl terminus and PSD-95 is detected in the immunoprecipitate. These data suggest that PSD-95 and the carboxyl terminus of DHHC5 have the capacity to interact in a mammalian cell and that the interaction is mediated through the DHHC5 carboxyl-terminal PDZ binding domain. Significant colocalization of DHHC5 and PSD-95 could also be demonstrated in transfected CHOK1 cells (supplemental Fig. 1).
To determine whether an endogenous interaction between DHHC5 and PSD-95 could be detected, mouse brain membranes were extracted using a stringent protocol that employs membrane solubilization in 1% sodium deoxycholate. As shown in Fig. 4B, when PSD-95 was immunoprecipitated from brain lysates, DHHC5 was also found in the immunoprecipitates (Fig. 4B, lanes 1–3). The DHHC5 signal was absent from similarly prepared brain lysates obtained from gene-targeted DHHC5 mice (Fig. 4B, lanes 4–6). We were unable to test whether PSD-95 co-immunoprecipitates with endogenous DHHC5 as neither antibody that recognizes DHHC5 on immunoblots would pull down DHHC5 from brain lysates.
To determine potential sites of interaction between DHHC5 and PSD-95, we performed individual yeast two-hybrid assays using selected regions of PSD-95 and the DHHC5 C carboxyl-terminal bait. As shown in Fig. 4C, all portions of PSD-95 were dispensable for binding to DHHC5 with the exception of the third PSD-95 PDZ domain, which was sufficient for binding. PDZ domains most commonly bind to carboxyl-terminal tails of proteins, with the binding partner tail inserting into a groove within the PDZ domain (35). Deletion of the carboxyl-terminal four amino acids of DHHC5 (EISV) completely abolished its interaction with PSD-95 in this assay as well (data not shown), again suggesting that this interaction with PSD-95 is mediated through DHHC5 carboxyl-terminal PDZ-binding motif, a result we had also obtained in transfected mammalian cells (Fig. 4A).
As DHHC5 is a member of a family of protein palmitoyltransferases based on sequence homology, we wondered whether the S-acylation of PSD-95 or other palmitoylated proteins would be altered in the brains of DHHC5 gt/gt mice. We, therefore, analyzed whole brain lysates using the acyl-biotin exchange method (36), which uses hydroxylamine at neutral pH to remove fatty acids from modified cysteines, exchanging them for HDPD-biotin, which can be captured to a solid-phase support and detected by immunoblotting. No change in the level of S-acylation of PSD-95 or several synaptic vesicles proteins (GAP43, GPRIN1, and VAMP-1) was seen, at least when averaged over the entire brain (supplemental Fig. 2). Of note, DHHC5 was itself shown to be S-acylated in these experiments.
DHHC5 Immunolocalization
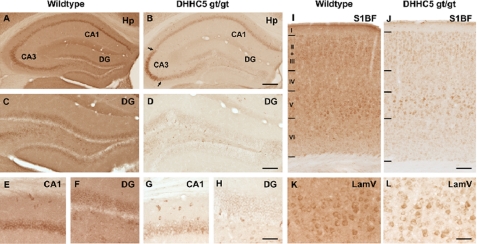
Immunohistochemical staining of brain tissue revealed pronounced changes in the intensity and distribution of staining for DHHC5 in the hippocampus and cortex of DHHC5 gt/gt mice in comparison with wild-type mice (Fig. 5). The staining was virtually absent from the neuropil of these mutant mice, whereas expression was either decreased or retained in regions of the hippocampus, consistent with regional expression from the hypomorphic allele. Decreased DHHC5 immunoreactivity was most pronounced in neurons of the dentate gyrus and CA1 subfields of the hippocampal formation (Fig. 5, B, D, G, and H) but was also evident in the cortex (Fig. 5, J and L). These results are consistent with the notion that DHHC5 acts within neuronal populations of the hippocampus to affect learning.
FIGURE 5.
Altered DHHC5 immunohistochemistry in DHHC5 gt/gt mouse brain. Immunohistochemical staining revealed widespread expression of DHHC5 in the hippocampal formation (A, C, E, F) and cortex (I, K) of wild-type mice, with DHHC5 immunoreactivity present homogeneously in the neuropil, dentate granule neurons, and pyramidal neurons in the CA1–3 of the hippocampal formation (A) and laminae III and V of the cortex (I). In contrast, DHHC5 immunoreactivity was markedly reduced in the neuropil of DHHC5 gt/gt mice, with DHHC5 expression retained in subsets of neurons. The changes in distribution of DHHC5 immunoreactivity were most pronounced in the hippocampal formation, with staining virtually abolished in dentate granule neurons (D and H) and relatively reduced in CA1 (B and G) compared with the retained DHHC5 immunoreactivity evident in the ventrolateral portion of CA3 in these mutant mice (as indicated by arrows in B). Within the primary somatosensory cortex-barrel field (S1BF) of DHHC5 gt/gt mice, staining for DHHC5 was effectively absent from the neuropil (J), and although generally reduced DHHC5 immunoreactivity was present, this was more pronounced in subpopulations of neurons in lamina V (L, LamV). Hp, hippocampus; DG, dentate gyrus; CA1 and CA3, hippocampal subfields. Bars, A and B, 1200 μm; C and D, 500 μm; E–H, 75 μm; I and J, 100 μm, K and L, 50 μm.
DISCUSSION
Synaptic transmission requires precise spatial and temporal control of many proteins within the confines of small synaptic specializations (post-synaptic densities) on dendritic spines. These events are coordinated at glutaminergic synapses by scaffolding proteins (PSD-95, among others) that employ low affinity modular domains (such as PDZ domains) to limit diffusion and assure rapid responses. In the current paper we identify a protein palmitoyltransferase member, DHHC5, that is enriched in a post-synaptic fraction of mouse brain and co-immunoprecipitates with PSD-95 in brain membrane extracts. We further showed that mice homozygous for a gene-targeted insertion that leads to reduced levels of DHHC5 protein in subpopulations of hippocampal neurons are impaired in contextual fear conditioning, suggesting a role for DHHC5 in hippocampal learning. The palmitoylation of PSD-95 was not reduced in the brains of these mice, suggesting that the binding may serve to position DHHC5 for accessing other substrates.
PSD-95 is a major protein of the synaptic density that consists of several modular domains that may function either singly or in tandem (35). These include three PDZ domains, an SH2 domain, and a guanylate kinase-like domain (Fig. 4C). PDZ domains most commonly interact with other proteins through the carboxyl-terminal tails of their binding partners, recognizing the distal 5–7 amino acids (although more promiscuous binding to internal sequences (37–39) and even phosphatidylinositol lipids (40) has been reported). The amino-terminal two PDZ domains (PDZ1 and PDZ2) of PSD-95 appear to function in tandem as a single unit (referred to as a supermodule, PDZ12) to form adjacent parallel high affinity binding sites for the carboxyl-terminal tails of dimerized N-methyl-d-aspartate receptors, potassium channels, NOS, and other molecules (for review, see Ref. 35). In binding to PDZ3, DHHC5 would be positioned to access these or other important signaling molecules. A number of known (and candidate) palmitoylated proteins that bind to PDZ12 were recently identified in a global “palmitoylome” survey, such as glutaminergic N-methyl-d-aspartate receptors 2A and 2B (Grin2a and Grin2b) (21). These may be candidates for palmitoylation by DHHC5; however, we did not find gross differences in palmitoylation of these receptors using the acyl biotin exchange assay applied to whole brain (data not shown). The palmitoylation of N-methyl-d-aspartate receptor subunits is complex (14), involving multiple sites subject to differential regulation, and so more detailed studies will be needed to definitively address this question. It is not surprising that the palmitoylation of PSD-95 itself was not affected in the DHHC5 hypomorphic mice, as the palmitoylation of PSD-95 is known to be performed by other DHHC enzymes (41). A palmitoylation cycle that regulates synaptic localization of PSD-95 has been described that involves a DHHC2 protein palmitoyltransferase whose vesicular movement in dendrites is regulated by synaptic activity (41, 42). Constitutive palmitoylation of PSD-95 is mediated by a second enzyme, DHHC3, that resides in the Golgi. DHHC3 (also known as GODZ) also plays a role at inhibitory synapses, where it palmitoylates the γ2 subunits of γ-aminobutyric acid, type A receptors (17). Interestingly, this earlier paper suggests that DHHC3 is dispensable at glutamate synapses, and the reason for this discrepancy is unclear but may relate to functional redundancies among DHHC enzymes, highlighting the complexity of these interactions. A third DHHC family member (DHHC17, also known as HIP14) has also been shown to palmitoylate PSD-95 in heterologous cell assays (43), and interference with endogenous expression of DHHC17 reduces clustering of PSD-95 in neurons (12). DHHC17 is believed to localize to the Golgi (13), binds to the polyglutamine repeat of huntingtin, and may play a role in Huntington disease (11). In addition to these PSD-95 palmitoylating enzymes, DHHC23 (NIDD) is a brain-specific DHHC family member that interacts with nNOS at the post-synaptic membrane. In an analogous fashion to the interaction of DHHC5 and PSD-95 at PDZ3, DHHC23 binds through its carboxyl terminus to a PDZ domain on nNOS (20), causing an increase in nNOS activity by promoting nNOS membrane localization.
It is pertinent to note that in addition to PSD-95, DHHC5 was found to potentially interact with several scaffolding molecules that functionally compensate for each other in mouse knock-out or inhibitory RNA experiments (PSD-93 and SAP102) (44, 45). This observation raises the possibility of a more general role for DHHC5 at the synapse that would include interactions with proteins other than PSD-95, further broadening potential DHHC5 substrates. Of note, 2-amino-3-(5-methyl-3-oxo-1,2-oxazol-4-yl)propanoic acid receptor subunits are regulated by palmitoylation (15, 16) and interact with neuronal scaffolding proteins GRIP1 and GRIP2 (46), which were identified here as potential binding partners of DHHC5, and so it would be of interest to examine these as potential substrates of DHHC5. In addition, DHHC5 binding partners identified in the assay include the kinesin motor proteins KIF1A and KIF5A, which are the major kinesins of dendritic spines. Kinesins are motor proteins that drive the transport of cargo, scaffold, or adaptor molecules along microtubules through interactions involving their coiled-coil domains (47). The interaction site between DHHC5 and the kinesins remains to be determined, but the function of the interaction is likely to involve vesicular transport of DHHC5. Clearly, further experiments that follow the trafficking of DHHC5 in neurons are needed.
A number of recent papers have presented proteomic analyses of synaptosomes and post-synaptic density (48–50). DHHC enzymes have not appeared prominently in these reports, which is consistent with a low abundance in cells. Interestingly, DHHC5 was the only DHHC member identified among 499 phosphoproteins in synaptic membranes (51). In this paper stoichiometry of the phosphorylation of DHHC5 could not be assessed. However, a “deep” phosphoproteome survey identified DHHC5 as one of many proteins in which phosphorylation was stimulated by growth factors (albeit in a non-neuronal cell) (52). So it appears that DHHC5 may be in turn regulated by phosphorylation under some circumstances. In addition, regulation of DHHC5 by palmitoylation may also be possible, as DHHC5 palmitoylation (outside of the catalytic site) was recently identified in a global screen for palmitoylated proteins of lipid rafts (53).
Many questions regarding the role of DHHC5 in the brain remain to be answered. Developmental expression and cellular distribution and seem particularly important questions as is the effect of DHHC5 expression on synaptic structure and function. Identifying substrates recognized by DHHC5 is important and may be challenging. It is now possible to identify a large number of S-acylated proteins in bulk tissue (21, 36, 54), but given the varied distribution of DHHC5 in different neuronal populations, refinements to these methods will be needed for comparisons to be meaningful and will require further study. New methods that incorporate stable isotope labeling with amino acids in cell culture (SILAC) and use azide or alkyne-containing palmitate analogs (and mass spectrometry) are on the horizon and may even be used to identify discrete acylation sites (53, 55–57). However, these methods have yet to be applied to cultured neurons. It is anticipated that we will soon learn from an unbiased method the substrates of DHHC5 from comparisons of brain tissue or cells and gain new insight into its role in central nervous system function.
Supplementary Material
Acknowledgments
We thank Jui-Yun Lu and Abigail Soyombo for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant NS36867 (to S. L. H.) and NS41930 (to J. D. C.). This work was also supported by Robert A. Welch Foundation Grant I-1187 (to S. L. H.), Wellcome Trust Grant GR079491MA (to J. D. C.), an Erasmus Studentship (to K. H.), the Batten Disease Support and Research Association (to J. D. C.), the Batten Disease Family Association (to J. D. C.), and the Natalie Fund (to J. D. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1 and 2.
- nNOS
- neuronal nitric oxide synthetase
- gt
- gene-trapped allele
- HA
- hemagglutinin
- PSD-95
- post-synaptic density protein of 95 kDa
- TA
- Tris acetate.
REFERENCES
- 1.Bartels D. J., Mitchell D. A., Dong X., Deschenes R. J. (1999) Mol. Cell Biol. 19, 6775–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lobo S., Greentree W. K., Linder M. E., Deschenes R. J. (2002) J. Biol. Chem. 277, 41268–41273 [DOI] [PubMed] [Google Scholar]
- 3.Roth A. F., Feng Y., Chen L., Davis N. G. (2002) J. Cell Biol. 159, 23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Putilina T., Wong P., Gentleman S. (1999) Mol. Cell Biochem. 195, 219–226 [DOI] [PubMed] [Google Scholar]
- 5.Hines R. M., Kang R., Goytain A., Quamme G. A. (2010) J. Biol. Chem. 285, 4621–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goytain A., Hines R. M., Quamme G. A. (2008) J. Biol. Chem. 283, 33365–33374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymond F. L., Tarpey P. S., Edkins S., Tofts C., O'Meara S., Teague J., Butler A., Stevens C., Barthorpe S., Buck G., Cole J., Dicks E., Gray K., Halliday K., Hills K., Hinton J., Jones D., Menzies A., Perry J., Raine K., Shepherd R., Small A., Varian J., Widaa S., Mallya U., Moon J., Luo Y., Shaw M., Boyle J., Kerr B., Turner G., Quarrell O., Cole T., Easton D. F., Wooster R., Bobrow M., Schwartz C. E., Gecz J., Stratton M. R., Futreal P. A. (2007) Am. J. Hum. Genet. 80, 982–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tarpey P. S., Smith R., Pleasance E., Whibley A., Edkins S., Hardy C., O'Meara S., Latimer C., Dicks E., Menzies A., Stephens P., Blow M., Greenman C., Xue Y., Tyler-Smith C., Thompson D., Gray K., Andrews J., Barthorpe S., Buck G., Cole J., Dunmore R., Jones D., Maddison M., Mironenko T., Turner R., Turrell K., Varian J., West S., Widaa S., Wray P., Teague J., Butler A., Jenkinson A., Jia M., Richardson D., Shepherd R., Wooster R., Tejada M. I., Martinez F., Carvill G., Goliath R., de Brouwer A. P., van Bokhoven H., Van Esch H., Chelly J., Raynaud M., Ropers H. H., Abidi F. E., Srivastava A. K., Cox J., Luo Y., Mallya U., Moon J., Parnau J., Mohammed S., Tolmie J. L., Shoubridge C., Corbett M., Gardner A., Haan E., Rujirabanjerd S., Shaw M., Vandeleur L., Fullston T., Easton D. F., Boyle J., Partington M., Hackett A., Field M., Skinner C., Stevenson R. E., Bobrow M., Turner G., Schwartz C. E., Gecz J., Raymond F. L., Futreal P. A., Stratton M. R. (2009) Nat. Genet. 41, 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansouri M. R., Marklund L., Gustavsson P., Davey E., Carlsson B., Larsson C., White I., Gustavson K. H., Dahl N. (2005) Eur. J. Hum. Genet. 13, 970–977 [DOI] [PubMed] [Google Scholar]
- 10.Chen W. Y., Shi Y. Y., Zheng Y. L., Zhao X. Z., Zhang G. J., Chen S. Q., Yang P. D., He L. (2004) Hum. Mol. Genet. 13, 2991–2995 [DOI] [PubMed] [Google Scholar]
- 11.Yanai A., Huang K., Kang R., Singaraja R. R., Arstikaitis P., Gan L., Orban P. C., Mullard A., Cowan C. M., Raymond L. A., Drisdel R. C., Green W. N., Ravikumar B., Rubinsztein D. C., El-Husseini A., Hayden M. R. (2006) Nat. Neurosci. 9, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang K., Yanai A., Kang R., Arstikaitis P., Singaraja R. R., Metzler M., Mullard A., Haigh B., Gauthier-Campbell C., Gutekunst C. A., Hayden M. R., El-Husseini A. (2004) Neuron 44, 977–986 [DOI] [PubMed] [Google Scholar]
- 13.Singaraja R. R., Hadano S., Metzler M., Givan S., Wellington C. L., Warby S., Yanai A., Gutekunst C. A., Leavitt B. R., Yi H., Fichter K., Gan L., McCutcheon K., Chopra V., Michel J., Hersch S. M., Ikeda J. E., Hayden M. R. (2002) Hum. Mol. Genet. 11, 2815–2828 [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T., Thomas G. M., Huganir R. L. (2009) Neuron 64, 213–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin D. T., Makino Y., Sharma K., Hayashi T., Neve R., Takamiya K., Huganir R. L. (2009) Nat. Neurosci. 12, 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi T., Rumbaugh G., Huganir R. L. (2005) Neuron 47, 709–723 [DOI] [PubMed] [Google Scholar]
- 17.Fang C., Deng L., Keller C. A., Fukata M., Fukata Y., Chen G., Lüscher B. (2006) J. Neurosci. 26, 12758–12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller C. A., Yuan X., Panzanelli P., Martin M. L., Alldred M., Sassoè-Pognetto M., Lüscher B. (2004) J. Neurosci. 24, 5881–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uemura T., Mori H., Mishina M. (2002) Biochem. Biophys. Res. Commun. 296, 492–496 [DOI] [PubMed] [Google Scholar]
- 20.Saitoh F., Tian Q. B., Okano A., Sakagami H., Kondo H., Suzuki T. (2004) J. Biol. Chem. 279, 29461–29468 [DOI] [PubMed] [Google Scholar]
- 21.Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukai J., Liu H., Burt R. A., Swor D. E., Lai W. S., Karayiorgou M., Gogos J. A. (2004) Nat. Genet. 36, 725–731 [DOI] [PubMed] [Google Scholar]
- 23.Mukai J., Dhilla A., Drew L. J., Stark K. L., Cao L., MacDermott A. B., Karayiorgou M., Gogos J. A. (2008) Nat. Neurosci. 11, 1302–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stryke D., Kawamoto M., Huang C. C., Johns S. J., King L. A., Harper C. A., Meng E. C., Lee R. E., Yee A., L'Italien L., Chuang P. T., Young S. G., Skarnes W. C., Babbitt P. C., Ferrin T. E. (2003) Nucleic Acids Res. 31, 278–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skarnes W. C., von Melchner H., Wurst W., Hicks G., Nord A. S., Cox T., Young S. G., Ruiz P., Soriano P., Tessier-Lavigne M., Conklin B. R., Stanford W. L., Rossant J. (2004) Nat. Genet. 36, 543–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nord A. S., Chang P. J., Conklin B. R., Cox A. V., Harper C. A., Hicks G. G., Huang C. C., Johns S. J., Kawamoto M., Liu S., Meng E. C., Morris J. H., Rossant J., Ruiz P., Skarnes W. C., Soriano P., Stanford W. L., Stryke D., von Melchner H., Wurst W., Yamamura K., Young S. G., Babbitt P. C., Ferrin T. E. (2006) Nucleic Acids Res. 34, D642–D648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S. H., Shrivastav A., Kosinski C., Sharma R. K., Chen M. H., Berthiaume L. G., Peters L. L., Chuang P. T., Young S. G., Bergo M. O. (2005) J. Biol. Chem. 280, 18990–18995 [DOI] [PubMed] [Google Scholar]
- 28.Bouvier D., Corera A. T., Tremblay M. E., Riad M., Chagnon M., Murai K. K., Pasquale E. B., Fon E. A., Doucet G. (2008) J. Neurochem. 106, 682–695 [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi T., Dulubova I., Min S. W., Chen X., Rizo J., Südhof T. C. (2002) Dev. Cell 2, 295–305 [DOI] [PubMed] [Google Scholar]
- 30.Bible E., Gupta P., Hofmann S. L., Cooper J. D. (2004) Neurobiol. Dis. 16, 346–359 [DOI] [PubMed] [Google Scholar]
- 31.Yang S. H., Bergo M. O., Farber E., Qiao X., Fong L. G., Young S. G. (2009) Transgenic Res. 18, 483–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Politis E. G., Roth A. F., Davis N. G. (2005) J. Biol. Chem. 280, 10156–10163 [DOI] [PubMed] [Google Scholar]
- 33.Ohno Y., Kihara A., Sano T., Igarashi Y. (2006) Biochim. Biophys. Acta 1761, 474–483 [DOI] [PubMed] [Google Scholar]
- 34.Harris B. Z., Lim W. A. (2001) J. Cell Sci. 114, 3219–3231 [DOI] [PubMed] [Google Scholar]
- 35.Feng W., Zhang M. (2009) Nat. Rev. Neurosci. 10, 87–99 [DOI] [PubMed] [Google Scholar]
- 36.Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hillier B. J., Christopherson K. S., Prehoda K. E., Bredt D. S., Lim W. A. (1999) Science 284, 812–815 [PubMed] [Google Scholar]
- 38.Wong H. C., Bourdelas A., Krauss A., Lee H. J., Shao Y., Wu D., Mlodzik M., Shi D. L., Zheng J. (2003) Mol. Cell 12, 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penkert R. R., DiVittorio H. M., Prehoda K. E. (2004) Nat. Struct. Mol. Biol. 11, 1122–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H., Feng W., Chen J., Chan L. N., Huang S., Zhang M. (2007) Mol. Cell 28, 886–898 [DOI] [PubMed] [Google Scholar]
- 41.Noritake J., Fukata Y., Iwanaga T., Hosomi N., Tsutsumi R., Matsuda N., Tani H., Iwanari H., Mochizuki Y., Kodama T., Matsuura Y., Bredt D. S., Hamakubo T., Fukata M. (2009) J. Cell Biol. 186, 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalva M. B. (2009) J. Cell Biol. 186, 7–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004) Neuron 44, 987–996 [DOI] [PubMed] [Google Scholar]
- 44.Elias G. M., Funke L., Stein V., Grant S. G., Bredt D. S., Nicoll R. A. (2006) Neuron 52, 307–320 [DOI] [PubMed] [Google Scholar]
- 45.Migaud M., Charlesworth P., Dempster M., Webster L. C., Watabe A. M., Makhinson M., He Y., Ramsay M. F., Morris R. G., Morrison J. H., O'Dell T. J., Grant S. G. (1998) Nature 396, 433–439 [DOI] [PubMed] [Google Scholar]
- 46.Hirbec H., Francis J. C., Lauri S. E., Braithwaite S. P., Coussen F., Mulle C., Dev K. K., Coutinho V., Meyer G., Isaac J. T., Collingridge G. L., Henley J. M., Couthino V. (2003) Neuron 37, 625–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirokawa N., Noda Y., Tanaka Y., Niwa S. (2009) Nat. Rev. Mol. Cell Biol. 10, 682–696 [DOI] [PubMed] [Google Scholar]
- 48.Cheng D., Hoogenraad C. C., Rush J., Ramm E., Schlager M. A., Duong D. M., Xu P., Wijayawardana S. R., Hanfelt J., Nakagawa T., Sheng M., Peng J. (2006) Mol. Cell Proteomics 5, 1158–1170 [DOI] [PubMed] [Google Scholar]
- 49.Klemmer P., Smit A. B., Li K. W. (2009) J. Proteomics 72, 82–90 [DOI] [PubMed] [Google Scholar]
- 50.Witzmann F. A., Arnold R. J., Bai F., Hrncirova P., Kimpel M. W., Mechref Y. S., McBride W. J., Novotny M. V., Pedrick N. M., Ringham H. N., Simon J. R. (2005) Proteomics 5, 2177–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munton R. P., Tweedie-Cullen R., Livingstone-Zatchej M., Weinandy F., Waidelich M., Longo D., Gehrig P., Potthast F., Rutishauser D., Gerrits B., Panse C., Schlapbach R., Mansuy I. M. (2007) Mol. Cell Proteomics 6, 283–293 [DOI] [PubMed] [Google Scholar]
- 52.Gnad F., Ren S., Cox J., Olsen J. V., Macek B., Oroshi M., Mann M. (2007) Genome Biol. 8, R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010) Mol. Cell Proteomics 9, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drisdel R. C., Green W. N. (2004) Biotechniques 36, 276–285 [DOI] [PubMed] [Google Scholar]
- 55.Martin B. R., Cravatt B. F. (2009) Nat. Methods 6, 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kostiuk M. A., Corvi M. M., Keller B. O., Plummer G., Prescher J. A., Hangauer M. J., Bertozzi C. R., Rajaiah G., Falck J. R., Berthiaume L. G. (2008) FASEB J. 22, 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hang H. C., Geutjes E. J., Grotenbreg G., Pollington A. M., Bijlmakers M. J., Ploegh H. L. (2007) J. Am. Chem. Soc. 129, 2744–2745 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.