Abstract
In Gram-positive bacteria, the functional role of surface polysaccharides (PS) that are not of capsular nature remains poorly understood. Here, we report the presence of a novel cell wall PS pellicle on the surface of Lactococcus lactis. Spontaneous PS-negative mutants were selected using semi-liquid growth conditions, and all mutations were mapped in a single chromosomal locus coding for PS biosynthesis. PS molecules were shown to be composed of hexasaccharide phosphate repeating units that are distinct from other bacterial PS. Using complementary atomic force and transmission electron microscopy techniques, we showed that the PS layer forms an outer pellicle surrounding the cell. Notably, we found that this cell wall layer confers a protective barrier against host phagocytosis by murine macrophages. Altogether, our results suggest that the PS pellicle could represent a new cell envelope structural component of Gram-positive bacteria.
Keywords: Carbohydrate/Bacterial, Cell/Surface/Bacteria, Cell/Wall/Bacteria, Gene/Mapping, Organisms/Bacteria, Atomic Force Microscopy, Lactococcus lactis, Peptidoglycan, Phagocytosis
Introduction
Polysaccharides (PS)5 are omnipresent components of Gram-positive bacteria surfaces, which may be divided into the following three major groups: (i) capsular polysaccharides (CPS), which are in most cases covalently bound to peptidoglycan (PG) and form a thick outer layer, (ii) wall polysaccharides (WPS), which may be attached to the cell wall covalently or not but without forming a capsule, and (iii) extracellular polysaccharides (EPS), which are released into the environment and thus are not attached to the cell surface. In some cases, these different PS are produced by the same bacterium (1, 2). However, there is a controversy regarding the differences between CPS, WPS, and EPS. CPS may be released into the growth medium as a consequence of handling conditions or because of the instability of the polysaccharide linkage, and consequently, they may be mistaken as EPS. Conversely, certain EPS could be tightly associated to the thick PG layer even in the absence of covalent anchoring (2, 3).
Polysaccharides, often exposed at the outermost layer of bacterial cells, have a wide range of important functions such as adhesion to abiotic surfaces leading to biofilm formation or specific interactions with other microorganisms or with eukaryotic host cells (3, 4). It was also reported that surface-exposed carbohydrates may serve as bacteriophage receptors (5). The presence of CPS confers resistance to nonspecific host defense mechanisms (3). Capsules have been extensively studied as virulence factors of many pathogens, which are able to proliferate in the bloodstream, such as Streptococcus pneumoniae, group B streptococci, or Staphylococcus aureus. CPS present large structural diversity. For example, S. pneumoniae produces a range of different CPS, leading to more than 90 different serotypes (3). The significant diversity of PS is determined by their structure, which may differ not only in the nature of sugar monomers composing repeating units, but also in their mode of linkage, branching, and substitution. These polysaccharides can be homo- or heteropolymers substituted by both organic and inorganic molecules (3, 4).
The biosynthesis pathways of CPS, WPS, and EPS share several common steps with other cell wall polymers (i.e. PG and teichoic acids), which include the use of undecaprenyl phosphate as lipid carrier and the polymerization of nucleotide sugars, most often containing glucose, galactose, and rhamnose (2). The enzymes responsible for CPS, WPS, and EPS synthesis are encoded by large gene clusters that are chromosomally or plasmid located; they often show unidirectional organization and are co-transcribed as single polycistronic mRNAs. These operons contain genes encoding glycosyltransferases together with genes that are responsible for chain polymerization and export (6). These loci are genetically heterogeneous within bacterial species, thus creating a diversity of polysaccharides that affect their function, as for example immune recognition (4).
Ubiquitous and recognized as virulence factors in pathogenic bacteria, CPS are not a common feature of food bacteria such as Lactococcus lactis. Instead, certain L. lactis strains are known to produce EPS that are of industrial importance because they contribute to the texture of fermented dairy products (1, 2). The plasmid-encoded genes responsible for EPS production in the dairy strains of L. lactis were characterized, and their organization was found to be similar to that of CPS biosynthesis gene clusters of Gram-positive pathogens (6, 7). The presence of chromosomally encoded cell surface-associated polysaccharides was suggested for some lactococcal strains (8). Nevertheless, the ability of PS to form an outer layer was never demonstrated in this species.
Here, we show that L. lactis MG1363 synthesizes a thin compact outer layer that we named pellicle. This chromosomally encoded pellicle is composed of PS of a novel structure, not previously described for L. lactis and other bacteria. The pellicle protects the bacteria from phagocytosis in vitro.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
Bacterial strains used in this study are listed in Table 1. L. lactis was grown at 30 °C in M17 medium (BD Biosciences) supplemented with 0.5% glucose (GM17). For long-chain mutant selection, overnight cultures were diluted and inoculated into 40-ml flasks of GM17 medium, containing 0.03% of agar (Difco), to produce ∼10–50 colonies per flask and incubated for 12 days. The nonsedimenting mutants were then removed from the top of the colonies and purified by re-streaking to single colonies, and the ability to form chains was observed microscopically.
TABLE 1.
L. lactis strains and plasmids used in this study and their relevant genetic properties
Mapping of Mutations in a Polysaccharide Synthesis Gene Cluster in MG1363 Chromosome
Two parts of the 24.27-kb cluster of the VES5748 chromosome were PCR-amplified using two couples of primers: rpmB (5′-TAGACATTATATTTACCTCCC), rgpF (5′-AGTATCATTCATAATTGTCACC) and rgpeF (5′-AAAGTGATACATCCCTTAGG); and 229 (5′- GAATTTGATTGATTACTTTAGG) and the Expand Long Template PCR System (Roche Diagnostics) according to manufacturer's instructions. The DNA nucleotide sequence of the amplified fragments (11.8 and 12.7 kb, respectively) was determined by GATC Biotech AG, using several internal primers, located approximately every 600 bp. Comparison of the obtained DNA sequence with the corresponding one of the MG1363 genome sequence (9) allowed the identification of a mutation in the llmg_0226 gene. To map other mutations, a 3175-bp fragment containing llmg_0226 gene was amplified by PCR with Phusion TM DNA Polymerase (Ozyme), using primers 226 (5′-GTTACTTCTTCAGGAGATCC) and 229. The DNA sequence of the obtained fragments was determined by using internal primers.
Microscopy
Microscopy images were taken with a phase contrast microscope (Leica Microsystems) equipped with an image analysis system Induscam (Andersa). Transmission electron microscopy was performed as described previously (10).
AFM Imaging
AFM images were obtained in acetate buffer, at room temperature, using a Nanoscope IV Multimode AFM and MCST tips (Veeco Metrology Group). Cells were immobilized by mechanical trapping into porous polycarbonate membranes (Millipore) with a pore size similar to the bacterial cell size. After filtering a concentrated cell suspension, the filter was gently rinsed with the adapted buffer, carefully cut (1 × 1 cm), and attached to a steel sample puck (Veeco Metrology Group) using a small piece of double-face adhesive tape, and the mounted sample was transferred into the AFM liquid cell while avoiding dewetting (11).
Polysaccharide Preparation and Structural Analysis
Bacterial cells were collected by centrifugation and washed with 20 mm ammonium acetate buffer, pH 4.7 (12). Cell wall PS were then prepared using a method developed previously (13). Bacteria were suspended in 4% SDS in ammonium acetate buffer and boiled with intensive stirring for 1 h. Cells were collected by centrifugation, and the pellet was washed six times with ammonium acetate buffer to remove SDS and stirred with 5% trichloroacetic acid in the presence of glass beads for 48 h at 4 °C. Insoluble material was removed by centrifugation (12,000 × g, 10 min), and the supernatant was dialyzed and freeze-dried. The resulting product was further purified on a Sephadex G-50 column. The major fraction, positive for neutral sugars, amino sugars, and phosphate (average yield ∼12 mg/liter of culture), was used for further chemical analysis and NMR analysis.
For the estimation of relative quantities of PS in L. lactis MG1363 and its mutant VES5748, cells were washed with buffer and freeze-dried. Equal amounts (200 mg) of dry WT and mutant cells were treated with SDS and trichloroacetic acid as described above, and after dialysis of trichloroacetic acid, 1 μm inositol was added to each preparation. The lyophilized product was depolymerized with 48% hydrofluoric acid (Acros Organic) for 48 h at 4 °C, hydrolyzed with 4 m trifluoroacetic acid for 3 h at 110 °C, converted into alditol acetates by conventional methods, and analyzed by gas chromatography.
For the preparation of oligosaccharides OS1, OS2, and OS3, PS (20 mg) was treated with 48% hydrofluoric acid (10 °C, 48 h), dried under a stream of air, and reduced with NaBH4. The resulting mixture of oligosaccharides was separated by high-performance anion-exchange chromatography, to give the reduced hexasaccharide OS1 as a major product.
General and Analytical Methods
Monosaccharides were identified as alditol acetates by gas chromatography on a Shimatzu GC-14 gas chromatograph equipped with a flame ionization detector and a Zebron ZB-5 capillary column (30 m × 0.25 mm), with hydrogen as carrier gas, using the following temperature gradient: 170 °C (3 min) and 260 °C at 5 °C/min. Gel permeation chromatography was carried out on a Sephadex G-50 column (1.6 × 90 cm) and irrigated with water. Fractions (5 ml) were assayed colorimetrically for aldose (14), amino sugars (15), and phosphate (16).
High-performance anion-exchange chromatography was performed on a CarboPac PA1 column (9 × 250 mm) with pulsed amperometric detection, with an isocratic flow of 0.1% NaOH. Fractions were collected and analyzed using Dionex system with an analytical CarboPac PA1 column (4 × 250 mm) at 1 ml/min. Products were desalted on a Sephadex G-15 column.
1H and 13C NMR spectra were recorded using a Varian Inova 500 MHz spectrometer for samples in D2O at 45 °C with acetone internal reference (δH = 2.23 ppm and δC = 31.5 ppm), using standard pulse sequences DQCOSY, TOCSY (mixing time 120 ms), nuclear Overhauser effect spectroscopy (mixing time 400 ms), and HSQC and HMBC (100-ms long range transfer delay). 1H-31P HMQC and HMQC-TOCSY were run with 1H-31P coupling set to 11 Hz, TOCSY mixing time of 100 ms.
L. lactis Phagocytosis Assay
Bacteria were grown to stationary phase (16–18 h; 1 × 109 colony-forming unit/ml) in GM17 at 37 °C without shaking. Bacterial cells were harvested by centrifugation at 3000 × g for 5 min and washed twice in phosphate-buffered saline solution before resuspension in RPMI 1640 medium (Invitrogen) to a final density of 1 × 109 colony-forming units/ml. The murine macrophage cell lines RAW 264.7 and J774 were propagated in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen) in the presence of penicillin and streptomycin. Infections were performed at a multiplicity of infection of 20:1 by centrifuging bacteria onto macrophages at 500 × g for 5 min. The plates were then incubated for 30 min at 37 °C under 10% CO2 atmosphere. Macrophages were extensively washed with RPMI 1640 medium and incubated for an additional hour in medium supplemented with 100 mg/ml streptomycin and 100 units/ml penicillin to kill extracellular bacteria. To monitor L. lactis phagocytosis, macrophages were washed with phosphate-buffered saline and lysed with 0.1% Triton X-100 in H2O. Serial dilution were immediately plated on M17-Glc agar plates for colony count.
For microscopic observation, macrophages cultivated on glass coverslips were infected as described above and extensively washed with phosphate-buffered saline. Monolayers were washed three times with phosphate-buffered saline, fixed with MeOH 100% for 2 min, and colored with May-Grünwald stain (Sigma). Specimens were observed with a Nikon Eclipse E600 microscope equipped with a Nikon DXM1200 FCCD camera.
RESULTS
Isolation of Spontaneous Mutations in the Chromosomal Gene Cluster, Encoding Polysaccharide Biosynthesis of L. lactis
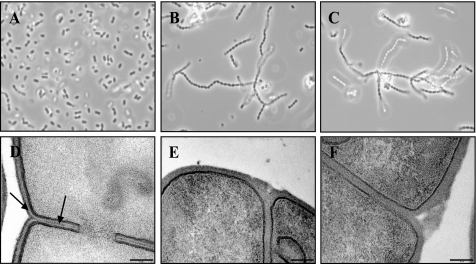
To isolate long-chain L. lactis mutants as nonsedimenting derivatives, semi-liquid growth conditions were used (17, 18). Long-chain derivatives of L. lactis MG1363 (Fig. 1A), exemplified here by mutant strains VES5748 and VES5751 (Fig. 1, B and C), were obtained in these conditions. In most cases, lactococci form long chains when cell separation is affected by reduced activity of autolytic enzymes (17, 19) or by increased PG resistance to hydrolysis (20, 21). Surprisingly, the activity of the main lactococcal autolysin AcmA was unaffected in more than 100 of such mutants (data not shown), albeit the long-chain phenotype of the acmA mutant allows us to expect the isolation of such mutants in the screen that we used (19).
FIGURE 1.
Phase contrast (A–C) and transmission electron micrographs (D–F) of WT L. lactis control strain MG1363 (A and D) and of two long-chain mutants VES5748 (B and E) and VES5751 (C and F). The outer pellicle indicated by arrows (D) is not visible in the mutants (E and F). The scale bar represents 0.1 μm.
After different phenotypical tests, we found that part of these mutants appeared to be resistant to the lactococcal virulent small isometric headed phage sk1, specific to MG1363 (22). The phage resistance phenotype was due to a phage adsorption defect and not to an inhibition of intracellular development (data not shown). It was previously established that genes important for the adsorption of phages belonging to this group are located within a 24.2-kb chromosomal gene cluster, which is putatively involved in cell wall polysaccharide (PS) biosynthesis in L. lactis strain IL1403 (5). We therefore determined the nucleotide sequence of the corresponding cluster in one of the mutants, VES5748 (Table 1), and found that it carries a C to T transition in the llmg_0226 gene (gene annotation as in Ref. 9), resulting in the appearance of a stop codon. Other long-chain sk1-resistant (sk1-r) mutants carried point mutations or insertions of IS981 in llmg_0226, probably because of the intrachromosomal transposition event. Similar mutants were also isolated based on phage sk1 resistance. In total, 20 sk1-r long-chain derivatives were selected, all of which carried mutations in the gene llmg_0226 (4 point mutations and 16 IS insertions). Although gene cluster, harboring all isolated mutations, putatively encodes cell wall PS (23), this compound was never isolated or characterized.
Isolation and Structural Characterization of the Cell Wall Polysaccharide of L. lactis
To gain further molecular insights into the PS produced by WT L. lactis MG1363, the carbohydrate-containing polymers of its cell wall were analyzed. The cell wall PS was prepared by trichloroacetic acid extraction, followed by purification by gel filtration chromatography. The purified material was positive in colorimetric reactions for neutral sugars, amino sugars, and phosphate. Monosaccharide analysis revealed the presence of Rha, Glc, Gal, and GlcN in an approximate molar ratio of 0.8:2:1.4:2.
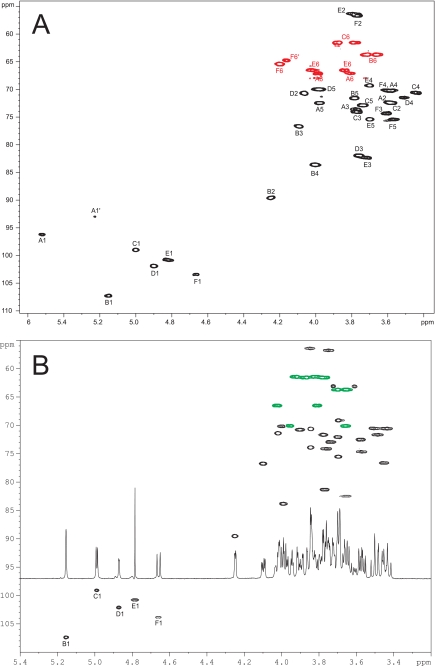
NMR analysis of the preparation indicated a homogeneous phosphate-containing PS. To elucidate its detailed chemical structure, the 1H and 13C spectra of the PS were fully assigned using two-dimensional homo- and heteronuclear correlation techniques (Table 2 and Fig. 2A). The structure of the PS is schematically shown in Fig. 3. The PS is composed of the hexasaccharide repeating units, containing two Glc (A and C), two GlcNAc (E and F), one Rha (D), and one Galf (B) residues, linked via a phosphodiester bond (Table 2 and Fig. 2A and Fig. 3). To confirm this structure, the PS was depolymerized with hydrofluoric acid and reduced, and the generated oligosaccharide fragments were purified by preparative high-performance anion-exchange chromatography and analyzed by two-dimensional NMR. One main product, the hexasaccharide OS1, was formed by cleavage of the phosphodiester bonds; two other oligosaccharides were produced by additional cleavage of galactofuranosyl (OS2) or rhamnosyl (OS3) linkages (Fig. 3). 1H and 13C NMR spectra of all oligosaccharides were fully assigned (Table 2; Fig. 2B, and data not shown). Altogether, our data unambiguously establish the structure of the PS of L. lactis MG1363 as presented in Fig. 3.
TABLE 2.
1H and 13C chemical shifts of WPS of L. lactis MG1363 and the hexosaccharide OS1
Spectra were recorded in D2O at 45 °C. Chemical shifts are given relative to acetone (δH = 2.23 ppm and δC = 31.5 ppm). NAc, CH3 signals at 2.03 (1H); 23.1 (13C) and 2.10 (1H); 23.3 ppm (13C); 31P at ∼0 ppm.
| Unit | 1 | 2 | 3 | 4 | 5 | 6a/b |
|---|---|---|---|---|---|---|
| α-Glc A, PS | ||||||
| 1H | 5.52 | 3.59 | 3.77 | 3.57 | 3.97 | 3.80/3.98 |
| 13C | 96.2 | 72.3 | 73.5 | 70.1 | 72.4 | 67.0 |
| β-Galf B, PS | ||||||
| 1H | 5.15 | 4.25 | 4.09 | 4.00 | 3.78 | 3.66/3.72 |
| 13C | 107.3 | 89.5 | 76.6 | 83.5 | 71.5 | 63.7 |
| α-Glc C, PS | ||||||
| 1H | 5.00 | 3.57 | 3.77 | 3.44 | 3.74 | 3.78/3.88 |
| 13C | 99.0 | 72.4 | 74.0 | 70.5 | 72.8 | 61.5 |
| α-Rha D, PS | ||||||
| 1H | 4.90 | 4.06 | 3.76 | 3.50 | 3.98 | 1.25 |
| 13C | 101.9 | 70.6 | 81.8 | 71.4 | 69.9 | 17.5 |
| β-GlcNAc E, PS | ||||||
| 1H | 4.82 | 3.79 | 3.71 | 3.70 | 3.70 | 3.84/4.02 |
| 13C | 100.8 | 56.3 | 82.3 | 69.2 | 75.4 | 66.5 |
| β-GlcNAc F, PS | ||||||
| 1H | 4.66 | 3.77 | 3.60 | 3.60 | 3.57 | 4.20/4.20 |
| 13C | 103.4 | 56.6 | 74.3 | 70.1 | 75.4 | 65.3 |
| Glc-ol A, OS1 | ||||||
| 1H | 3.61/3.72 | 3.84 | 3.84 | 3.70 | 3.90 | 3.65/3.96 |
| 13C | 63.1 | 73.9 | 70.6 | 72.1 | 70.8 | 70.1 |
| β-Galf B, OS1 | ||||||
| 1H | 5.15 | 4.25 | 4.10 | 3.99 | 3.77 | 3.65/3.70 |
| 13C | 107.4 | 89.5 | 76.7 | 83.8 | 71.7 | 63.7 |
| α-Glc C, OS1 | ||||||
| 1H | 4.99 | 3.57 | 3.76 | 3.43 | 3.74 | 3.77/3.88 |
| 13C | 99.1 | 72.5 | 74.1 | 70.5 | 72.9 | 61.6 |
| α-Rha D, OS1 | ||||||
| 1H | 4.87 | 4.02 | 3.77 | 3.48 | 3.99 | 1.24 |
| 13C | 102.1 | 71.4 | 81.3 | 71.7 | 70.1 | 17.5 |
| β-GlcNAc E, OS1 | ||||||
| 1H | 4.78 | 3.85 | 3.65 | 3.69 | 3.69 | 3.81/4.02 |
| 13C | 100.8 | 56.4 | 82.5 | 69.1 | 75.5 | 66.5 |
| β-GlcNAc F, OS1 | ||||||
| 1H | 4.66 | 3.75 | 3.57 | 3.50 | 3.45 | 3.81/3.93 |
| 13C | 103.9 | 56.8 | 74.7 | 70.5 | 76.6 | 61.4 |
FIGURE 2.
Partial heteronuclear 1H-13C chemical shift correlation (HSQC) spectrum of the L. lactis MG1363 polysaccharide (A) and OS1 fragment (B). The corresponding one-dimensional 1H NMR spectrum of OS1 is also shown (B). C-H signals appear in black; CH2 signals are in red (A) and green (B). Residues are indicated as in Table 2.
FIGURE 3.
Structure of L. lactis MG1363 PS and the oligosaccharide products of its partial hydrolysis (OS1 to OS3). A and C, Glc residues; B, Galf; E and F, GlcNAc, and D, Rha; see also Table 2.
To confirm that llmg_0226 mutation in sk1-r long-chain derivatives indeed affected the PS biosynthesis, the quantities of PS synthesized by MG1363 and VES5748 mutant were compared. The relative quantities of Glc and GlcNAc, which are characteristic for the PS of L. lactis MG1363, were measured using gas chromatography analysis with inositol as internal standard. We found that the VES5748 mutant contained about 20-fold less Glc per mg of dry cells than MG1363, and no detectable GlcNAc. We therefore conclude that the VES5748 mutant lacks the PS that is present in the wild type L. lactis MG1363.
Detection of an Outer Polysaccharide Pellicle by Transmission Electron Microscopy (TEM) and AFM
We chose two long-chain mutants VES5748 and VES5751 (Table 1) for microscopy studies. As observed from TEM micrographs, an outer pellicle is present in WT L. lactis MG1363 (Fig. 1D), which is absent in the two long-chain mutants (Fig. 1, E and F). This pellicle is clearly visible around the cells and intercalated between the peptidoglycan of two dividing cells (Fig. 1D). We therefore conclude that WT lactococci contain an outer layer made of PS.
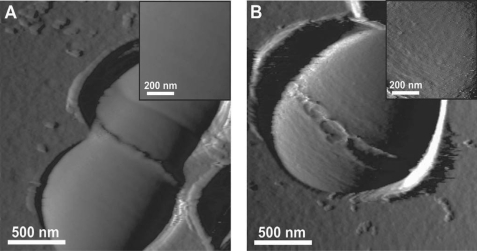
Because TEM micrographs could be biased by the fixation and staining procedures, the presence of the polysaccharide pellicle was examined by live cell AFM imaging (24), a microscopy technique that was reportedly used for EPS and CPS visualization (25, 26). For this purpose, living bacterial cells were immobilized in porous membranes for noninvasive imaging. Fig. 4 shows AFM deflection images of single L. lactis NZ3900 (WT) and VES5748 mutant cells. The surface of WT cells showed well defined division septum and ring-like structures presumably reflecting “equatorial rings” of newly synthesized PG (27). Besides these structures, the cell surface was featureless and very smooth, the root mean square roughness (Rr.m.s.) on 200 × 200 nm height images being 0.6 ± 0.1 nm. By contrast, mutant VES5748 exhibited a much rougher surface, with an Rr.m.s. of 2.9 ± 0.7 nm. The rough surface may reflect another cell wall constituent, such as peptidoglycan, lying underneath the pellicle. The increased surface roughness in PS-negative mutants correlates with TEM observations revealing that mutations lead to the disappearance of an outer layer.
FIGURE 4.
AFM deflection images for single L. lactis WT (A) and its pellicle-negative derivative VES5748 (B) cells during the course of the division process.
Antiphagocytic Properties of the L. lactis Polysaccharide Pellicle
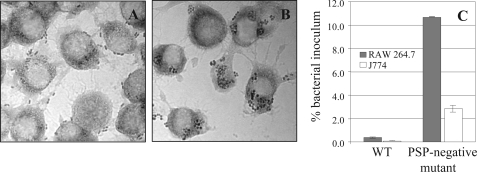
The CPS of Gram-positive pathogens play important roles during bacterial infection and survival in the blood stream and thus affect their virulence (3). In these bacteria, the presence of a capsule is often associated with antiphagocytic properties (28). We hypothesized that the outer PS pellicle of L. lactis MG1363 could display a similar property. A murine macrophage phagocytosis assay, which is routinely used for pathogenic bacteria, was thus performed using lactococcal cells (29, 30). The obtained results showed that PS-negative bacteria (strain VES5748) are more efficiently trapped inside the murine phagocyte lines RAW 264.7 and J774 than the WT strain (Fig. 5). The yield of internalized mutant bacteria was about 10-fold higher than that of WT, indicating a capacity of the lactococcal PS pellicle (PSP) to provide antiphagocytic properties (Fig. 5). Analysis of phagocyte micrographs confirmed the increased susceptibility of the PS-negative mutant to phagocytosis by showing large clusters of bacteria accumulating inside macrophages, whereas WT bacteria were essentially located at the macrophage surface (Fig. 5, A and B). Similar results were obtained using different bacteria/macrophage ratios (data not shown). These results indicate that the PSP can protect L. lactis against host phagocytosis by macrophages and in this way plays, at least in vitro, a role similar to CPS of pathogenic bacteria.
FIGURE 5.
Phagocytosis of L. lactis WT (A) and its pellicle-negative derivative VES5748 (B) by murine macrophages RAW 264.7 and J774 is shown. Micrographs of bacteria after 30 min of incubation with RAW 264.7 (A and B) and phagocytosis efficiency (C) are shown.
DISCUSSION
In this work, we identified a cell wall PS produced by the food bacterium L. lactis that forms a compact pellicle located on the outer surface of the cell. Isolation of long-chain mutants and their examination by complementary microscopy techniques (TEM and AFM) allowed us to unambiguously demonstrate the existence of this outer PSP. To the best of our knowledge, this compact pellicle-like layer was never studied in detail in L. lactis nor in other Gram-positive bacteria. The absence of a “traditional” capsule, which could hide or replace the PSP, turned L. lactis into a particularly suitable model organism to reveal and characterize this novel cell envelope structure. Noteworthy, a similar outer layer could be observed in a number of TEM micrographs of L. lactis isolates of different origins, although its existence was not discussed by the authors (31–33). Interestingly, such a layer is also visible in TEM micrographs of other Gram-positive bacteria such as Enterococcus faecalis (34), S. pneumoniae (35), and Streptococcus mutans (36). This pellicle may thus represent a novel cell envelope structural component in a number of Gram-positive bacteria. The identification of this layer in L. lactis now opens the possibility to confirm the presence of similar pellicles in these bacteria, which possess chromosomal clusters, putatively encoding cell wall polysaccharide synthesis.
The composition of the L. lactis pellicle was established by determining the structure of the PS and showing that it is not produced in the PS-negative mutant strains. In contrast to EPS, which are usually purified from culture supernatants, the PS of MG1363 was extracted by a trichloroacetic acid treatment, preceded by boiling of bacteria in SDS solution (see under “Experimental Procedures”). Such a harsh procedure, necessary for the isolation of the PS, strongly suggests that it is covalently linked to PG and therefore can be qualified as a cell wall PS. Moreover, the extraction procedure also allows us to speculate about the ratio between PG and PS in the L. lactis cell envelope. Indeed, for PG extraction, we routinely use SDS, protease, and nuclease digestion, followed by strong acid treatment (20). Following this procedure, we obtain ∼50% mass reduction after acid treatment, suggesting that PG and PS are present in comparable amounts in the cell wall preparation (data not shown). Nevertheless, the PSP on TEM micrographs is thinner than the PG layer, indicating that this PS layer should be more compact in comparison with PG.
To our knowledge, the PS structure that was determined here is unique among the previously described bacterial PS structures. The monosaccharide sequence constituting the PS repeating unit was not found by searching the carbohydrate structural data bases (37) Noteworthy, the Glc residue branched on GlcNAc was previously found in the O-antigen from Escherichia coli lipopolysaccharide (38). Interestingly, the PS characterized in this study is composed of hexasaccharide repeating units linked by phosphodiester bonds. Although this structure with phosphate in the polymer backbone is reminiscent of the teichoic acid structures (39), it does not contain typical polyols (i.e. glycerol, ribitol, and glucitol) found in the “classical” teichoic acids (2, 39) or in the teichoic acid-like polymer (40). Consequently, the PS characterized in this study can be qualified as a sugar-phosphate cell-wall PS (41, 42).
The resistance to small isometrically headed bacteriophage sk1 phenotype of long-chain PS-negative mutants greatly facilitated further characterization and mapping of responsible mutations. It was previously shown that lactococcal receptors for the phages of 936 species, to which sk1 belongs, are encoded in a gene cluster, potentially responsible for PS synthesis (5). This chromosomally located gene cluster was first identified in the closely related L. lactis subsp. lactis strain IL1403 (23). The clusters of IL1403 and MG1363 strains are similar but not identical (9). However, both clusters harbor genes identified as putatively responsible for the synthesis of rhamnose precursors (rmlA, -B, -C, and -D), rhamnosyltransferases (rgpA and -B), and several glycosyltransferases (9). The presence of an Rha residue in the repeating unit of the PS and the absence of other genes encoding the same functions in the genome of MG1363 unequivocally suggests that this cluster is involved in the synthesis of the PS pellicle. The presence of a pellicle-like layer, visible in TEM micrographs of IL1403,6 also indicates that a similar PS is produced by the latter strain.
Surprisingly, all isolated mutations were IS insertions or frameshifts located in llmg_0226, which is absent from the orthologous gene cluster of IL1403. Consequently, we cannot rule out that these mutations have a polar effect on the downstream-located and closely linked llmg_0227 gene (9). The proximal part of Llmg_0227 protein (117 amino acids) shows homology to Llmg_0215 of the same cluster and to YcaG of the corresponding cluster in IL1403. This indicates that these three proteins may belong to the same family. Noteworthy, the mutations leading to resistance of IL1403 to bacteriophage bIL170, belonging to 936 species, were located in the ycaG gene (5). Outside of the Lactococcus genus, homologs of llmg_0226 or llmg_0227 were only identified in the genome of the opportunistic pathogen Streptococcus agalactiae (group B streptococcus) (43). Interestingly, the corresponding genes in group B streptococcus are located in a locus that could be involved in the synthesis of the group B antigen, a cell wall-anchored polysaccharide used to type this species (40).
Sequence analysis and topology prediction indicate the presence of a signal peptide and of a transmembrane domain in the C-terminal part of the Llmg_0226 protein and of seven transmembrane domains in Llmg_0227. These features suggest that Llmg_0226 is a secreted membrane-associated protein, and Llmg_0227 is an integral membrane protein, but the exact functions of these two proteins cannot be inferred from homology searches. We speculate that the products of llmg_0226 and/or llmg_0227 fulfill a nonstructural but essential role in the synthesis of the PS pellicle.
Intriguingly, we observed an elevated frequency of mutation events in the llmg_0226 gene in comparison with the acmA gene, although inactivation of both genes leads to a similar long-chain phenotype. This may indicate the existence of an IS981 transposition-based mechanism dedicated to affect PS-related surface diversity that could play a role in survival of lactococci in changing environmental conditions. Mutations due to IS981 transposition were previously reported and were shown to be involved in fitness improvement of L. lactis (44).
In Gram-positive pathogens, the CPS are considered as virulence factors, acting by preventing phagocytosis. Because the lactococcal pellicle may have similar properties, we performed phagocytosis experiments and showed that the lactococcal PSP confers resistance to ingestion by two cell lines of murine macrophages. In our nonopsonic phagocytosis assay, the pellicle could act by hindering cell surface components, preventing their recognition by macrophage pattern recognition receptors (28). The susceptibility to phagocytosis of PS-negative mutant strains indicates that the PSP can help lactococci to escape innate immune defenses, for instance to protect themselves against mucosal immunity of the gut (45). Also, the protection against host immunity could be relevant to the rare human systemic dissemination cases (46).
The intercalation of PS between peptidoglycan layers in the septum of dividing lactococcal cells, as observed in Fig. 1D, could play a role in separation of daughter cells. The absence of such a septal pellicle could thus affect cell separation and consequently explain the long-chain phenotype of PS-negative mutants. Alternatively, the compact pellicle could serve as a barrier, preventing passage of major autolysin AcmA through the cell wall. Because the lack of AcmA activity in L. lactis results in formation of long chains (19), a decrease of PG hydrolase activity due to a leakage of AcmA in the absence of such external barrier could also be a reason for the long-chain formation of PS-negative mutants.
Besides its use in milk industry, L. lactis is considered as a convenient delivery vector for biological molecules for anti-infective and anti-allergic therapies in the gastro-intestinal tract (47). The potential capacity of the lactococcal pellicle to interfere with the immune system or to constitute a barrier for the extracellular delivery of proteins suggests its importance for such applications. Pellicle-negative derivatives of Gram-positive food and probiotic bacteria could thus display a strong applied potential. The use of PS-negative derivatives in the food industry could be greatly facilitated by the possibility to apply a food-grade procedure for their selection, based on differential sedimentation of spontaneous long-chain mutants in semi-liquid conditions.
Acknowledgments
We thank Alexandra Gruss and Veronique Monnet for support and helpful reading of the manuscript, Girbe Buist and Marie-Agnès Petit for valuable discussions, Sophie Chat for help with electronic photography, and Eric Guédon for sharing unpublished results.
This work was supported in part by INRA (to M.-P. C.-C., M.-Y. M., S. F., E. B., P. C., C. P., and S. K.).
E. Guédon, personal communication.
- PS
- polysaccharide
- PSP
- PS pellicle
- PG
- peptidoglycan
- CPS
- capsular polysaccharide
- WPS
- wall polysaccharide
- EPS
- extracellular polysaccharide
- TEM
- transmission electron microscopy
- AFM
- atomic force microscopy
- WT
- wild type.
REFERENCES
- 1.Cerning J. (1990) FEMS Microbiol. Rev. 7, 113–130 [DOI] [PubMed] [Google Scholar]
- 2.Delcour J., Ferain T., Deghorain M., Palumbo E., Hols P. (1999) Antonie Van Leeuwenhoek 76, 159–184 [PubMed] [Google Scholar]
- 3.Roberts I. S. (1996) Annu. Rev. Microbiol. 50, 285–315 [DOI] [PubMed] [Google Scholar]
- 4.Comstock L. E., Kasper D. L. (2006) Cell 126, 847–850 [DOI] [PubMed] [Google Scholar]
- 5.Dupont K., Janzen T., Vogensen F. K., Josephsen J., Stuer-Lauridsen B. (2004) Appl. Environ. Microbiol. 70, 5825–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Kranenburg R., Vos H. R., van Swam, Kleerebezem M., de Vos W. M. (1999) J. Bacteriol. 181, 6347–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Kranenburg R., Marugg J. D., van Swam I. I., Willem N. J., de Vos W. M. (1997) Mol. Microbiol. 24, 387–397 [DOI] [PubMed] [Google Scholar]
- 8.Dabour N., LaPointe G. (2005) Appl. Environ. Microbiol. 71, 7414–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wegmann U., O'Connell-Motherway M., Zomer A., Buist G., Shearman C., Canchaya C., Ventura M., Goesmann A., Gasson M. J., Kuipers O. P., van Sinderen D., Kok J. (2007) J. Bacteriol. 189, 3256–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veiga P., Piquet S., Maisons A., Furlan S., Courtin P., Chapot-Chartier M. P., Kulakauskas S. (2006) Mol. Microbiol. 62, 1713–1724 [DOI] [PubMed] [Google Scholar]
- 11.Dufrêne Y. F. (2008) Nat. Protoc. 3, 1132–1138 [DOI] [PubMed] [Google Scholar]
- 12.Fedtke I., Mader D., Kohler T., Moll H., Nicholson G., Biswas R., Henseler K., Götz F., Zähringer U., Peschel A. (2007) Mol. Microbiol. 65, 1078–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadovskaya I., Vinogradov E., Li J., Jabbouri S. (2004) Carbohydr. Res. 339, 1467–1473 [DOI] [PubMed] [Google Scholar]
- 14.Dubois M., Gilles K., Hamilton J. K., Rebers P. A., Smith F. (1956) Anal. Chem. 28, 350–356 [Google Scholar]
- 15.Enghofer E., Kress H. (1979) Carbohydr. Res. 76, 233–238 [DOI] [PubMed] [Google Scholar]
- 16.Chen P. S., Toribara T. Y., Warner H. (1956) Anal. Chem. 28, 1756–1758 [Google Scholar]
- 17.Mercier C., Domakova E., Tremblay J., Kulakauskas S. (2000) FEMS Microbiol. Lett. 187, 47–52 [DOI] [PubMed] [Google Scholar]
- 18.Mercier C., Durrieu C., Briandet R., Domakova E., Tremblay J., Buist G., Kulakauskas S. (2002) Mol. Microbiol. 46, 235–243 [DOI] [PubMed] [Google Scholar]
- 19.Buist G., Kok J., Leenhouts K. J., Dabrowska M., Venema G., Haandrikman A. J. (1995) J. Bacteriol. 177, 1554–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyrand M., Boughammoura A., Courtin P., Mézange C., Guillot A., Chapot-Chartier M. P. (2007) Microbiology 153, 3275–3285 [DOI] [PubMed] [Google Scholar]
- 21.Veiga P., Bulbarela-Sampieri C., Furlan S., Maisons A., Chapot-Chartier M. P., Erkelenz M., Mervelet P., Noirot P., Frees D., Kuipers O. P., Kok J., Gruss A., Buist G., Kulakauskas S. (2007) J. Biol. Chem. 282, 19342–19354 [DOI] [PubMed] [Google Scholar]
- 22.Chandry P. S., Moore S. C., Boyce J. D., Davidson B. E., Hillier A. J. (1997) Mol. Microbiol. 26, 49–64 [DOI] [PubMed] [Google Scholar]
- 23.Bolotin A., Wincker P., Mauger S., Jaillon O., Malarme K., Weissenbach J., Ehrlich S. D., Sorokin A. (2001) Genome Res. 11, 731–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dufrêne Y. F. (2008) Nat. Rev. Microbiol. 6, 674–680 [DOI] [PubMed] [Google Scholar]
- 25.Francius G., Lebeer S., Alsteens D., Wildling L., Gruber H. J., Hols P., De Keersmaecker S., Vanderleyden J., Dufrêne Y. F. (2008) ACS Nano 2, 1921–1929 [DOI] [PubMed] [Google Scholar]
- 26.Stukalov O., Korenevsky A., Beveridge T. J., Dutcher J. R. (2008) Appl. Environ. Microbiol. 74, 5457–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morlot C., Zapun A., Dideberg O., Vernet T. (2003) Mol. Microbiol. 50, 845–855 [DOI] [PubMed] [Google Scholar]
- 28.Areschoug T., Gordon S. (2008) Contrib. Microbiol. 15, 45–60 [DOI] [PubMed] [Google Scholar]
- 29.Chen P. M., Chen H. C., Ho C. T., Jung C. J., Lien H. T., Chen J. Y., Chia J. S. (2008) Microbes Infect. 10, 293–301 [DOI] [PubMed] [Google Scholar]
- 30.Segura M., Gottschalk M., Olivier M. (2004) Infect. Immun. 72, 5322–5330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapot-Chartier M. P., Deniel C., Rousseau M., Vassal L., Gripon J. C. (1994) Int. Dairy J. 4, 251–269 [Google Scholar]
- 32.Dabour N., Kheadr E., Benhamou N., Fliss I., LaPointe G. (2006) J. Dairy Sci. 89, 95–110 [DOI] [PubMed] [Google Scholar]
- 33.Mifune J., Grage K., Rehm B. H. (2009) Appl. Environ. Microbiol. 75, 4668–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pooley H. M., Shockman G. D., Higgins M. L., Porres-Juan J. (1972) J. Bacteriol. 109, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Le Gouëllec A., Roux L., Fadda D., Massidda O., Vernet T., Zapun A. (2008) J. Bacteriol. 190, 4501–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chia J. S., Chang L. Y., Shun C. T., Chang Y. Y., Tsay Y. G., Chen J. Y. (2001) Infect. Immun. 69, 6987–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toukach P., Joshi H. J., Ranzinger R., Knirel Y., von der Lieth C. W. (2007) Nucleic Acids Res. 35, D280–D286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenutz R., Weintraub A., Widmalm G. (2006) FEMS Microbiol. Rev. 30, 382–403 [DOI] [PubMed] [Google Scholar]
- 39.Neuhaus F. C., Baddiley J. (2003) Microbiol. Mol. Biol. Rev. 67, 686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutcliffe I. C., Black G. W., Harrington D. J. (2008) Microbiology 154, 1354–1363 [DOI] [PubMed] [Google Scholar]
- 41.Naumova I. B., Shashkov A. S. (1997) Biochemistry 62, 809–840 [PubMed] [Google Scholar]
- 42.Schäffer C., Messner P. (2005) Microbiology 151, 643–651 [DOI] [PubMed] [Google Scholar]
- 43.Glaser P., Rusniok C., Buchrieser C., Chevalier F., Frangeul L., Msadek T., Zouine M., Couvé E., Lalioui L., Poyart C., Trieu-Cuot P., Kunst F. (2002) Mol. Microbiol. 45, 1499–1513 [DOI] [PubMed] [Google Scholar]
- 44.van Hylckama Vlieg J. E., Rademaker J. L., Bachmann H., Molenaar D., Kelly W. J., Siezen R. J. (2006) Curr. Opin. Biotechnol. 17, 183–190 [DOI] [PubMed] [Google Scholar]
- 45.Corthésy B., Gaskins H. R., Mercenier A. (2007) J. Nutr. 137, 781S–790S [DOI] [PubMed] [Google Scholar]
- 46.Zechini B., Cipriani P., Papadopoulou S., Di Nucci G., Petrucca A., Teggi A. (2006) Diagn. Microbiol. Infect. Dis. 56, 325–328 [DOI] [PubMed] [Google Scholar]
- 47.Wells J. M., Mercenier A. (2008) Nat. Rev. Microbiol. 6, 349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gasson M. J. (1983) J. Bacteriol. 154, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Ruyter P. G., Kuipers O. P., Beerthuyzen M. M., van Alen-Boerrigter I., de Vos W. M. (1996) J. Bacteriol. 178, 3434–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]