Abstract
Cornelia de Lange syndrome (CdLS) (OMIM #122470, #300590 and #610759) is a dominant genetic disorder with multiple organ system abnormalities which is classically characterized by typical facial features, growth and mental retardation, upper limb defects, hirsutism, gastrointestinal and other visceral system involvement. Mutations in three cohesin proteins, a key regulator of cohesin, NIPBL, and two structural components of the cohesin ring SMC1A and SMC3, etiologically account for about 65% of individuals with CdLS. Cohesin controls faithful chromosome segregation during the mitotic and meiotic cell cycles. Multiple proteins in the cohesin pathway are also involved in additional fundamental biological events such as double-strand DNA break repair and long-range regulation of transcription. Moreover, chromosome instability was recently associated with defective sister chromatid cohesion in several cancer studies, and an increasing number of human developmental disorders is being reported to result from disruption of this pathway. Here, we will discuss the human disorders caused by alterations of cohesin function (termed ‘cohesinopathies’), with an emphasis on the clinical manifestations of CdLS and mechanistic studies of the CdLS-related proteins.
Keywords: cohesion, cohesinopathy, Cornelia de Lange syndrome, epigenetics, gene expression, genetics, NIPBL, SMC1A
Cornelia de Lange syndrome (CdLS; OMIM #122470, #300590, and #610759) (also known as Brachmann–de Lange syndrome) is a rare, genetically heterogeneous disorder affecting multiple organs and systems during development. Vrolik and Brachmann reported severely affected cases in 1849 and 1916, respectively (1, 2), and de Lange subsequently proposed diagnostic criteria in 1933 and described two unrelated individuals (3). It has been estimated to occur in about 1:10,000 individuals; however, the actual incidence may be higher as the clinical presentations are quite variable and milder cases are likely to be underestimated (4). Almost all cases are sporadic and dominant, although recurrence in siblings due to parental mosaicism has been reported (5, 6). Central nervous system, craniofacial, musculoskeletal, and gastrointestinal systems are the most commonly affected. The diagnosis is made both clinically and molecularly (7), and management and treatment are generally symptomatic and supportive therapy based (7, 8). Mutations in three genes, NIPBL on chromosome 5p13 (9, 10), SMC1A on chromosome Xp11, and SMC3 on chromosome 10q25, can be identified in 65% of individuals with clinically diagnosed CdLS (11, 12). All of these three genes are involved in sister chromatid cohesion events.
Clinical presentations of CdLS
Dysmorphia
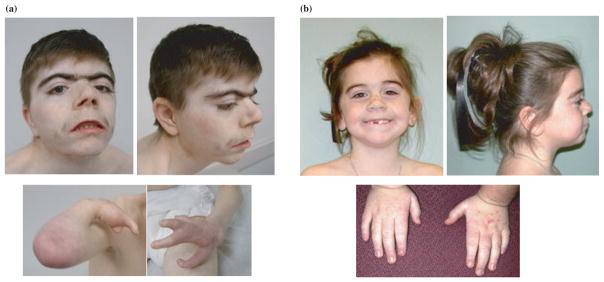
The facial features are the most clinically consistent and recognizable findings in CdLS (Fig. 1). Most individuals have a short neck, low posterior hairline, hirsute forehead, arched eyebrows and synophrys sometimes with severe ptosis (13). Thick and long eyelashes, low-set ears, flattened midface, short nose, and long philtrum are also commonly seen (13). Typical oral features include a thin upper lip with down-turned corners, a high palate, widely spaced teeth and micrognathia (13). Cleft palate, most of which are submucous clefts, appears in approximately 20% of the probands (13, 14). Typical extremity findings range from small hands and small feet to more severe reduction defects of the upper limbs (Fig. 1). Disproportional shortening of the first metacarpal, proximally placed thumb, brachydactyly and clinodactyly can be seen in majority of the cases and single palmar creases in over half (13). Nearly one-third of the probands have upper limb malformations with severity ranging from various forms of oligodactyly to ulnar deficiency to absent forearm. The lower extremities are less likely to be significantly involved (13). Probands can also have radial head dislocation with radioulnar synostosis and incomplete elbow extension (13). Hirsutism is mainly on the face, back and extremities. Cutis marmorata can also be seen in half of the probands (13).
Fig. 1.
Clinical characteristics in Cornelia de Lange syndrome. (a) A severely affected 19-year-old man with protein truncating mutation in NIPBL. Note the characteristic facial features (arched eyebrows, synophrys, ptosis, anteverted nares, long philtrum, thin upper lip with down-turned corners and micrognathia) and severe asymmetrical defects of the forearms. (b) A mildly affected 9-year-old girl with a missense mutation in SMC1A. Note the more mild facial features and hand involvement (small with mild fifth finger clinodactyly).
Growth and development
Probands have proportionate small stature that occurs prenatally, usually manifesting late in the second trimester. At birth, the measurement parameters tend to be below the 10th percentiles, and fall to below the 5th percentiles by early childhood, although the growth parallels the standard growth curves (15). CdLS-specific growth curves are available (15). In adulthood, both the average heights and weights are below the third percentiles, with a mean head circumference of 49 cm that is consistent with significant microcephaly (15). Developmental delay and mental retardation are typically observed. Speech and language are most significantly affected, while perceptual organization and visual-spatial memory are more preserved. The average IQ ranges from mild to moderate mental retardation; however, both borderline normal and severe mental retardation are commonly reported. Learning continues throughout life without evidence of regression (15). Early intervention has been proven to be helpful in improving developmental outcomes and should be continued for as long as possible (16).
Visceral defects
Multiple organ systems are involved in CdLS. Feeding problems are typical in infancy and young childhood and contribute to the universal presence of gastroesophageal reflux disease (GERD). Medical treatment is almost always indicated and surgical intervention is often required (17). Pyloric stenosis, diaphragmatic hernia, malrotation and increased risk for volvulus formation have also been frequently reported (18). A quarter of probands suffer from congenital heart defects most commonly represented as a ventriculoseptal defect or an atrial septal defect, although other lesions are also seen (19, 20). Renal malformations and dysfunction can be seen as well, commonly represented by vesiculoureteral reflux, pelvic dilatation and renal dysplasia (21). Causes of death in CdLS are primarily due to gastrointestinal complications including diaphragmatic hernia in infancy, followed by aspiration pneumonia complicated by GERD and volvulus at older ages (13).
Behavior and neurological problems
Almost every proband has behavioral issues that usually are caused or aggravated by physical complications (17, 22–24). Self-injurious behavior, obsessive-compulsive behaviors, attention deficit disorder with or without hyperactivity, short attention span, depression and autistic features have all been consistently reported. Social and environmental interactions can be achieved at variable degrees (17, 22–24). Seizures are the primary neuropathological manifestation. No specific EEG pattern has been described and the seizures can generally be well managed with standard medical intervention. Sleep disturbances are often seen (22). Neuroradiological findings include enlarged ventricles particularly at the basal cisterns, atrophy of white matter particularly at the frontal lobes, and hypoplasia of the brainstem and cerebellar vermis (25). Gyral structural abnormalities, myelination defects, and neurofibrillary tangles inside neurons were also seen on autopsy (26, 27). Both hypertonicity and hypotonia occur. Probands tend to have a high pain threshold probably due to poorly characterized peripheral neuropathy (25).
Findings in other organ systems
Ophthalmologically, almost all the probands have peripapillary pigmentation. High myopia, ptosis, blepharitis and mild forms of microcornea are the most common. Nasolacrimal duct obstruction and nystagmus are less common. Cataract, glaucoma and other eye malformations are rare (28). Auditory and vestibular anomalies include both sensorineural and conductive hearing loss, recurrent otitis media and sinusitis (29, 30). Orthopedic manifestations, beyond the upper limb deficiencies, include hip dislocation or dysplasia, scoliosis, tight achilles tendons and delayed maturation of bone (31, 32). Genitalia are in general hypoplastic with cryptorchidism, micropenis and hypospadias being commonly seen in males and small labia majora in females. Normal puberty occurs with slight delay in some cases (13, 14). Behavioral issues, particularly self-injury and anxiety, usually increase during adolescence. Fertility is normal amongst less severely affected probands (33). Little is known about the natural history of CdLS and few reports describe large cohorts of affected adults; however, premature aging has been suggested (14). There is no obvious increased risk of cancer, although rare cases of liver hemangioendothelioma and Wilm’s tumor were reported in autopsies from individuals with features of CdLS (34). Thrombocytopenia has also been consistently reported (35).
Diagnostic criteria and clinical particularities of CdLS due to NIPBL, SMC1A or SMC3 mutations
Certain diagnostic criteria of CdLS have been proposed previously (36, 37), mainly based on clinical presentations with specific facial features, hand profile and neurodevelopmental characteristics. Recently, Kline et al. have proposed minimal criteria for the diagnosis of CdLS, and a scoring system to evaluate the severity (7). Molecular identification of mutations in NIPBL, SMC1A and SMC3 is considered as the decisive index in that individuals with a pathogenic mutation in any one of the associated genes are defined as having CdLS (7). As defined by Kline et al. (7), in order to diagnose an individual clinically (e.g. without a gene mutation) there are specific criteria that should be met. A severity scoring system based on developmental milestones, malformations (particularly of the upper limb) and hearing loss was created, which was found to correlate well with specific brain malformations, IQ levels and mutations in NIPBL (7).
About 60% of CdLS probands have a heterozygous mutation in NIPBL. Genotype–phenotype correlations amongst a large number of probands indicate that presumably haploinsufficient NIPBL mutations (protein truncating mutations such as nonsense mutations, splice site mutations, and out-of-frame deletions or insertions) usually result in a more severe cognitive and structural phenotype than missense mutations (38). Approximately 5% of probands with a clinical diagnosis of CdLS were found to have missense or small in-frame deletion mutations in SMC1A, and one individual was found to have an in-frame 3 bp deletion in the SMC3 gene (12). The SMC1A and SMC3 cases have mild to moderate mental retardation without significant impairments in growth or structural abnormalities of the limb or other organ systems (12). Notably, probands with SMC1A or SMC3 mutations demonstrated some clinical features that are in contrast to the ‘classical’ form of CdLS (12). This cohort tend to have a more prominent nasal bridge than is typically seen in CdLS (11), and the majority of them had birth weights within normal parameters with normal head circumferences and growth measures later in life as well (Fig. 1). For the most walking and speech are often acquired, and overall, they exhibit a much milder level of cognitive involvement (12).
Approximately 65% of CdLS probands with a confident clinical diagnosis have mutations in one of cohesin-associated genes (NIPBL, SMC1A or SMC3). The molecular etiology of the remaining 35% of probands is unknown at this time. Mutations in the regulatory sequences of these three genes may account for a small percentage of cases and are not routinely screened for in clinical testing. Still additional genes are yet to be identified to account for these molecularly uncharacterized individuals, and with over 20 known genes implicated in the cohesin complex and its regulation (and many more yet to be implicated), there are plenty of potential candidates.
Cohesin biology
Cohesin is a dynamic complex regulated at various cell cycle stages by multiple mechanisms (Fig. 2) (39). The primary biological role identified for cohesin is to control sister chromatid segregation during both mitosis and meiosis. Four evolutionarily conserved subunits form the core structural components of the cohesin complex: two SMC (structural maintenance of chromosomes) proteins SMC1A and SMC3, one kleisin protein RAD21 (also called MCD1 or SCC1), and SA1/SA2 (also called STAG1/STAG2). SMC1B, REC8 and STAG3 are cohesin subunits in meiosis (40). Homologs of the cohesin complex and its regulatory genes have been identified in several eukaryotic model systems such as Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Xenopus laevis, mouse and humans (41).
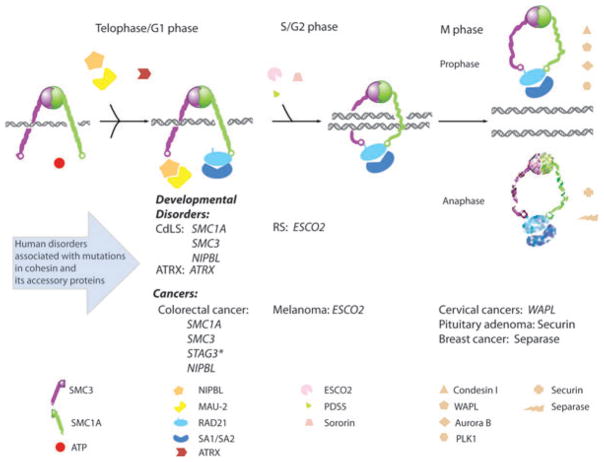
Fig. 2.
Cohesinopathies. Human disorders associated with mutations in cohesin subunits and accessory proteins of cohesin complex. SMC1A, SMC3, RAD21 and either SA1 or SA2 are the four major subunits of cohesin in somatic vertebrate cells. RAD21 crosslinks the head domains of SMC1A and SMC3 in an ATP-dependent manner, whereas RAD21 also binds to the fourth cohesin subunit SA1/SA2. NIPBL and MAU-2 form a complex and facilitate cohesin loading and unloading. Sister chromatid cohesion is established during S phase, mediated by sororin and ESCO1/ESCO2, after which PDS5 maintains cohesion through G2 phase. The removal of cohesin from the chromosome arms starts at prophase and is regulated by PLK1, Aurora B kinase, condensin I, and WAPL. During the metaphase-to-anaphase transition, the separase inhibitor securin is degraded by APC (anaphase-promoting complex), thereby activating separase which in turn cleaves centromeric cohesin as well as residual cohesin on the chromosome arms. This enables sister chromatid separation. Human disorders are listed corresponding to their disease causing genes in the cohesion pathway and the point in the cell cycle at which these respective genes are active. These disorders are further subclassified as developmental disorders and cancers. STAG3* is a cohesin subunit normally found in meiosis.
The protein structures of SMC1A and SMC3 are very similar, each spanning 1000–1500 amino acids, and both contain globular domains at the N - and C-terminal ends and a globular hinge domain in the middle which separates the alpha helical structures (42). N and C termini fold at the hinge domain to form an antiparallel coiled coil by bringing together the two halves of the alpha helix, which also subsequently form the head domain. Three highly conserved motifs, Walker A, Walker B, and a signature motif, were identified in the ATPase domain in the cohesin head formed by the N and C termini (42, 43). SMC1A and SMC3 dimerize at the hinge domains forming a V-shaped structure most likely through a hydrophobic interaction (44). The N terminus of the RAD21 subunit interacts with the head domain of SMC3 and the C terminus interacts with the head domain of SMC1A, with ATP being required for this association. RAD21 was therefore suggested to be a central regulator of cohesin function because it simultaneously crosslinks the SMC heterodimer, lies in close proximity to the ATPase active sites in cohesin head domains, and binds to the fourth known cohesin subunit, SA1/SA2 (45). The maturely formed cohesin is a four-subunit, ring-like complex with globular hinge and head domains separated by a 45 nm of coiled coil. Several working models have been proposed for the molecular basis of sister chromatid cohesion, with ‘topological’ and ‘handcuff’ being the most representative (44, 46). The topological model proposes no physical contact between chromatin and cohesin. The cohesin ring opens up at the head domain followed by the sister chromatids entering the ring and becoming topologically entrapped when RAD21 re-locks the ring. Alternatively, the handcuff model proposes specific interactions of DNA–protein and protein–protein. One cohesin ring binds one single chromatid after which inter-complex oligomerization occurs. Both hinge domain and coiled-coil domain have been suggested to be able to serve as the tethering surfaces; however, the prominent role of the hinge in cohesin dynamics implies that it could be the target of regulatory factors (46–48). At present, a lot of current biological observations support that cohesin binds to chromatin, and none of the models is able to rule out interactions between cohesin and DNA (46).
Cohesin binds to chromatin during the G1/S phase in budding yeast; however, in vertebrates binding happens during telophase of the preceding cell division (49). Additionally, cohesin binds at G2/M phase when a double-strand DNA break is created. Removal of cohesin from chromosome arms commences in prophase and is completed by anaphase (Fig. 2). In yeast, cohesin-associated regions (CARs) are distributed on average at 15-kb intervals on chromosome arms at transcriptional convergent sites which are usually AT-rich (50, 51). Although no consensus DNA sequence for cohesin binding has been identified, cohesin binding is enriched at heterochromatin (52) and the chromatin surrounding a DNA double-strand break (DSB) (53). Several factors that could affect cohesin–chromatin association have been demonstrated. First, all four subunits of cohesin and a well assembled complex are essential for chromatin binding in vivo (54, 55). Second, normal structure and function of all three motifs in the SMC head domains are indispensable (56, 57). Third, specific residues in the coiled-coil regions that are close to the heads are required. Fourth, the hinge domains of both SMC proteins also have a role in cohesin binding to chromatin (58, 59).
The heterodimer NIPBL/hSCC4 complex is evolutionarily conserved and required for loading of cohesin onto chromatin in both mitosis and meiosis, and to all chromosome regions currently under study such as heterochromatin, CARs, centromeres, and DSBs (53, 60). Our understanding of how NIPBL loads cohesin onto DNA is poor and no such study has been reported yet. The yeast homolog of NIPBL, Scc2, was suggested to be a kinase through sequence homology analysis (61), and mutations in Scc2 have greatly reduced Rad21 phosphorylation (62). NIPBL/SCC4 seems to be involved in all cohesin activities including SMC ATPase activation, hinge dimerization, chromatin binding, and chromatin remodeling (46). In addition to NIPBL/SCC4, certain histone modifications and local chromosome structures are also targeted by cohesin (52, 63, 64). Distinct protein complexes such as MRX, CEN complexes and Rep which are distinct from NIPBL/SCC4 can also recruit cohesin to specific chromosome regions (53, 65, 66). In summary, cells elaborately regulate chromatin binding of cohesin temporally and spatially, but most of the mechanisms are unknown at present. This regulation of the complex enables cohesin to perform diverse biological functions. Mutations in those associated major genes are expected to contribute to multiple human diseases, both known and unknown. Cohesin-independent mechanisms of cohesion have also been suggested in that complete unpairing of sister chromatids cannot be achieved by knocking down cohesin function alone. Condensin complexes (67), origin recognition complexes (68), centromere complexes (69) and DNA catenation (70) have all been reported to have a role in mediating cohesin-independent sister chromatid cohesion.
The removal of cohesin from chromosomes is another field that has been extensively studied. Cohesin begins to disassociate from the chromosome arms during prophase whereas the pericentric cohesin is protected until the onset of anaphase (71–73). In vertebrates, both cohesin subunits Stag2 and Rad21 are substrates of Polo-like kinase 1 (Plk1). A functioning Wapl is also required for prophase removal. At the same time, Shugoshin/MEIS-322/Sgo1 (74) and sororin (75) are able to protect centromere cohesins. An evolutionally conserved protein Pds5/BimD/Spo76 interacting with Wapl, sororin and Eco1 is also involved in maintaining pericentric cohesion in prophase (76). Inactivation of cohesion and the complete dissolution of cohesin at the onset of anaphase enable faithful sister chromatid segregation. Securin and separase were identified to regulate this event. Securin binds and inhibits protease activity of separase (Esp1) before anaphase (77). At the beginning of anaphase, securin is degraded by anaphase-promoting complex (APC) and separase is activated (78). The active separase cleaves Rad21 and the cohesin complex is further degraded (77). Mechanisms other than cohesin cleavage to ensure the complete inactivation of all cohesion have also been proposed (79).
Cohesin function and the etiologies of CdLS
CdLS is a genetically heterogeneous dominant multi-system developmental disorder that can be caused by mutations in one of at least three genes identified to date: the human Scc2 ortholog NIPBL and cohesin subunits SMC1A and SMC3 (9–12). All of these genes encode proteins that have been implicated to play roles in chromosome segregation as well as in DNA repair, gene expression and chromosome conformation (80). Cohesion or cell cycle defects were first speculated to cause developmental deficits in CdLS. Until now, there is no clear answer. One early study in a large cohort of CdLS samples reported that precocious sister chromatid separation was identified in 41% of probands’ lymphoblastoid cell lines vs 9% in healthy controls (81); however, another study with limited specimens found CdLS cells had no obvious cohesion defects (82). More experiments, larger groups of probands with clearly identified gene mutations, as well as additional assays able to sensitively evaluate chromosome segregation are required to clarify this question.
Increasingly, however, mounting evidence suggests that the dysregulation of gene expression that results from mutation in cohesin genes is more likely to represent the underlying pathogenesis of CdLS. The first key evidence of cohesin mediated gene regulation arose from screens in Drosophila for genes that regulate gene expression in a dosage-sensitive manner (83, 84). In Drosophila, the homeobox genes cut and Ultrabithorax (Ubx) control multiple aspects of development and the Drosophila Scc2 homolog, Nipped-B, is required for long-range activation of these two genes. The effect of reducing cohesin dosage on cut expression is contradictory to the effect of reducing Nipped-B leading to the hypothesis that cohesin might function as an insulator factor interfering with long-range communication between the wing margin enhancer and the cut promoter, and that Nipped-B facilitates long-range gene activation by controlling cohesin dynamics (85). Of note, although heterozygous Nipped-B mutant flies had reduced cut and Ubx expression without detectable cohesion defects, the heterozygous Nipped-B null alleles only reduced Nipped-B mRNA by some 25%, and that further reduction to 50% wild-type levels by in vivo RNAi would be lethal; however, cohesion defects were not observed (84). Whole genome mapping of cohesin and Nipped-B chromatin binding sites in Drosophila has revealed that the loading factor Nipped-B co-localize with cohesin, suggesting that Nipped-B and cohesin still stay together on the chromatin after the loading process (86). These data revealed very different cohesin binding patterns in yeast and flies, and possibly very different targeting mechanisms which may be related to different roles for cohesin and Nipped-B either in cohesion formation or in gene regulation (87).
Recently, homozygous Drosophila mutants of Smc1 or Scc3 have shown defective axon pruning in the post-mitotic mushroom body (88, 89). The ecdysone steroid hormone receptor (EcR), an important factor for axon pruning, was found to have reduced protein expression in these mutants (89). Overexpression of EcR in the post-mitotic neurons in those knockout flies could rescue the pruning defect indicating that reduced EcR expression is primarily responsible for the pruning defect (89). The block of pruning could also be induced by knocking out another cohesin subunit Rad21 in the same neurons indicating a complete cohesin complex is essential for pruning event (88). As all of these above observations occurred in post-mitotic (non-dividing) cells, chromosome segregation and cell cycle regulation are clearly not involved. In addition, cholinergic neurons’ lack of Rad21 caused abnormal larva locomotion without obvious effects on mitosis (88). In other model organisms, such as zebrafish, the functions of Rad21 and Smc3 are needed for proper expression of runx1 and runx3 genes in early embryonic development (90). Genome-wide chromatin immunoprecipitation experiments in human and mouse cells have revealed co-localization of cohesin and CTCF, a zinc-finger protein with enhancer blocking/boundary activities (91–93). Knockdown of either cohesin or CTCF would influence genome-wide gene expression levels, and knockdown of cohesin alone resulted in dysregulated gene expression of CTCF targets (91, 92). Genome-wide gene expression and ChIP array studies in lymphoblastoid cell lines have revealed a highly conserved transcriptional profile in CdLS probands which tightly associates to cohesin binding status (94).
CdLS probands with NIPBL missense mutations are usually more mildly affected, whereas severely affected probands are much more likely to carry protein truncating alleles (38). In Drosophila, the expression of cut and Ubx is less affected in mutants with missense Nipped-B alleles similar to some CdLS-causing mutations than those with truncating or null alleles (95). Thus the milder effects of NIPBL missense mutations in humans could reflect milder effects on gene expression. Moreover, CdLS probands who carry heterozygous SMC1A or SMC3 mutations tend to be more mildly affected and have a much lower prevalence of overt structural defects (12). The mutations in SMC1A or SMC3 are all either missense or small in-frame deletions that preserve the protein reading frame, and cell lines from probands with cohesin subunit mutations lack cohesion defects and produce normal levels of SMC1A (4). More interestingly, the level of NIPBL mRNA was only 30% reduced in CdLS probands’ cell lines and heterozygous NIPBL knockout mice as well as in Nipped-B mutant flies (4). This slight impairment in NIPBL expression (<30%) or a single amino acid changes in SMC1A or SMC3 can lead to enormous effects on human development without considerable defects in sister chromatid cohesion. These findings further support the hypothesis that it is likely the cohesin’s role in regulating gene expression that is being impacted in CdLS and causative of the resulting phenotype. Several lines of evidence support this hypothesis. First, none of the CdLS-causing mutations in SMC1A is predicted to inactivate the ATPase activity, interfere with interactions with other cohesin subunits, or disrupt the coiled-coil arm (12). This is consistent with the normal sister chromatid cohesion seen in probands. Second, SMC1A is an X-linked gene and reported to escape X-inactivation (96); however, there are both male and female probands affected with heterozygous missense mutations, and no female proband was found to carry a loss-of-function mutation such as a nonsense or frame shift mutation (12). It is therefore presumed that SMC1A dose reduction is not likely to be the cause of CdLS, but rather the mutations are more likely to result in a slightly altered cohesin dynamic function that interferes with its ability to regulate (94). Third, naturally occurring SMC1A missense mutations in CdLS probands have been shown to have an enhanced DNA binding affinity than wild-type proteins in vitro. This is, to date, the most direct evidence that cohesin dynamics are changed by gene mutations (97).
CTCF is the only well characterized insulator protein in vertebrates. In addition to its enhancer blocking and barrier activities, CTCF can function as a transcription factor at a subset of genomic sites and can directly bind to RNA polymerase II, indicating a potential function in directly regulating gene transcription both positively and negatively (98, 99). Additionally, several known chromatin modifications, including DNA methylation, occur near CTCF binding sites. Formation of higher order chromatin structure was also mediated by CTCF (100). Cohesin and CTCF were found not only to significantly co-localize but also to be functionally associated with each other in the mammalian genome (91, 92, 101). It was therefore suggested that cohesin could regulate gene expression through multiple mechanisms that are quite similar to the regulatory mechanisms of CTCF. Cohesin and CTCF may act either cooperatively or independently (91, 92). As a result of these findings, it will be of interest to investigate if the phenotypic manifestations of CdLS could result from epigenetic alterations in the genome such as loss imprinting at specific loci.
Currently, 35% of clinically diagnosed CdLS probands do not carry an identifiable mutation in one of the three implicated genes. This would indicate that either new disease causing genes remain to be discovered or non-coding regions of the known genes require re-screening. Additional candidate genes would include the other components of the cohesin ring: RAD21 and STAG1/STAG2, or other proteins that interact with or regulate cohesin and modulate its chromatin association dynamics, for example, PDS5, WAPL and Sororin. Knockout animals for one of the two genes encoding a vertebrate Pds5 protein, Pds5b, were recently reported (102, 103). Mice with homozygous null Pds5b alleles die soon after birth and presented some congenital anomalies found in probands of the CdLS.
Cohesin pathway and other human disorders
Roberts/SC phocomelia syndrome
Roberts/SC phocomelia syndrome (RBS OMIM #268300) is an autosomal recessive genetic disorder caused by homozygous or compound heterozygous mutations in the ESCO2 gene (104, 105). The clinical features of this syndrome are distinct from CdLS but with some overlap (85). The yeast homologous gene Eco1/Ctf7 acetyltransferase is required for the establishment and stabilization of sister chromatid cohesion but not for the binding of cohesin to chromosomes (106, 107). In fission yeast, Eco1 physically interacts with Pds5 and counteracts Pds5’s block of cohesion (107). In budding yeast, Eco1 acetylates Smc3’s head domain in a cell cycle dependent manner to promote sister chromatid cohesion (108, 109). The Drosophila Eco1 ortholog specifically establishes cohesion at centromeres, and not along the chromosome arms (110). In humans, much similar to the chromosome cohesion defects seen in Drosophila Eco1 mutants, cell lines derived from Roberts probands show ‘heterochromatic repulsion’ which was demonstrated by premature sister chromatid separation primarily at the heterochromatic regions on prophase and metaphase chromosomes (111). A new study has shown that most mutations in the ESCO2 gene identified in RBS probands result in disruption of the acetyltransferase domain. This results in faulty cohesion, and other cellular events in RBS cell lines, indicating that acetyltransferase activity contributes to the development of the major organ systems affected in RBS (112).
α-Thalassemia/mental retardation syndrome, X-linked
α-Thalassemia/mental retardation syndrome, X-linked (ATRX) (OMIM #301040), is a multi-system disorder characterized by postnatal growth and mental deficiency, microcephaly, dysmorphic craniofacial features (hypertelorism, midface hypoplasia, anteverted nares and full lips with protruding tongue), lack of speech, seizures and abnormal genitalia in males (113). Affected individuals usually have a mild form of hemoglobin H (Hb H) disease. ATRX is caused by mutations in the ATRX gene on the X chromosome (113). The ATRX gene encodes a chromatin remodeling enzyme that associates with the chromoshadow domain of HP1α (as does NIPBL) and preferentially localizes to the pericentromeric heterochromatin in mouse and human cells (114). ATRX was suggested to have a role in loading cohesin onto chromatin during S phase and recruiting cohesin to specific chromosome loci (114). Both defective sister chromatid cohesion and impaired chromosome congression were seen in cultured human cells with depleted ATRX, indicating a disruption of mitotic progression. Similar findings were seen in embryonic mouse brains with no ATRX protein (114). The impaired cohesin targeting or transportation due to mutations in the ATRX gene may therefore contribute to the clinical phenotypes in ATRX syndrome.
Human malignancies
There is increasing evidence that links disruption of the cohesin complex or the cohesion pathway to many forms of human cancer. The tumor suppressor gene BRCA1 associates with many factors that function in the sister chromatid cohesion pathway, indicating a role in BRCA1-tumorigenesis (115–117). BRCA1 and Eco1/Ctf7 family members share overlapping partners, and cells harboring mutations in either BRCA1- or ESCO-related pathways exhibit similar chromosomal abnormalities including cohesion defects especially along heterochromatic and centromeric regions (118, 119).
Human WAPL protein overexpression was found in cervical cancers and significantly correlated to the grades of the malignancy (120, 121). NIH3T3 cells overexpressing WAPL produced tumors in 100% of injected nude mice (121). Human papillomavirus E6 and E7 oncoproteins are able to induce the expression of human WAPL (122). Downregulated WAPL inhibited the growth of tumors derived from cervical cancer cell lines; therefore, WAPL was proposed as a therapeutic target for cervical cancer. In addition, a splice variant of WAPL may also associate with other types of human neoplasia because it interacts with the Epstein–Barr virus transformation-related protein EBNA2 in human cells (120, 123). The contribution of dysregulated WAPL to cervical carcinogenesis may be partially due to its resulted chromosomal instability (CIN) (124).
Separase digests RAD21 at the beginning of anaphase to release cohesin from the paired sister chromatids, and it has been suggested as a tumor suppressor gene in zebrafish (90). Heterozygous mutations of separase contribute to the initiation and progression of epithelial tumors partially due to its generated genome instability (125). In Drosophila, epithelial organization and integrity seems to be the most affected by the loss of separase (126). On the other hand, a different group has proposed that separase is an oncogene (127). Significant overexpression of separase was detected in human breast tumors, most of which are infiltrating ductal carcinomas. Overexpression of separase alone is sufficient to induce aneuploid tumors in mouse mammary epithelial cells under a p53 mutant background. Cohesion defects represented by premature sister chromatid separation were manifested in separase induced cell lines as well.
Recently, defective sister chromatid cohesion has been suggested to play a major role in human colorectal cancers (128). A systematic study to identify somatic mutations in potential CIN genes in 132 colorectal cancer samples has identified 11 somatic mutations distributed among five genes SMC1A, NIPBL, SMC3, STAG3 and RNF20. Except RNF20, which is not involved in sister chromatid cohesion, SMC1, SMC3 and STAG3 are subunits of the cohesin complex, whereas NIPBL is the protein that facilitates loading and unloading of cohesin. Knockdown of the expression of SMC1A, SMC3, STAG2 by siRNA and disruption of the expression of two previously well studied major candidate CIN genes (MRE11A and CDC4) have all resulted in CIN and unambiguous chromatid cohesion defects in human cells, although MRE11A and CDC4 have been postulated to play a different biological role. These results suggest that mutations in cohesin subunit or cohesin regulatory genes are common pathways leading to CIN in colorectal cancers. The other solid tumors may also harbor mutations in cohesin-associated genes as CIN appears to be a common event among them.
Many other regulatory factors of cohesin complex have been discussed in cancer research as well. Human securin which inhibits separase’s enzyme activity before the onset of anaphase is actually the human proto-oncogene pituitary tumor-transforming gene (PTTG) (129). The protein levels of securin were reported to correlate to the invasiveness of pituitary tumors. Securin is able to transform cultured cells and its expression is elevated in human cancer cell lines (130). Human cancer cells with securin loss-of-function mutations show high levels of CIN (131), and cells overexpressing securin produce tumors in nude mice (132). The cohesion establishment factor EFO2/ESCO2 is highly upregulated in aggressive melanoma cells (133).
Future directions
Cohesin’s role in mammalian development was not appreciated until genes involved in structural components and in the regulation of this complex were positionally cloned and found to be causative of multi-system disorders when disrupted. Since the identification of the first cohesin gene to be mutated in CdLS, additional developmental disorders and diseases have been implicated in this pathway (summarized in Fig. 2). The term ‘cohesinopathy’ has been used to describe human conditions due to mutations in cohesin subunits or regulatory proteins of cohesin complex. Pathogenic mutations in new genes involved in cohesinopathies are expected to be identified. Extra effort is needed to understand how cohesin functions in meiosis and human malignancy, and how cohesion pathway is involved in gene expression and epigenetic regulation.
Acknowledgments
The authors would like to acknowledge the support of the NICHD (PO1HD052860) (IDK) and the CdLS Foundation Fellowship Grant (JL).
References
- 1.Brachmann W. Ein fall von symmetrischer monodaktylie durch Ulnadefekt, mit symmetrischer flughautbildung in den ellenbeugen, sowie anderen abnormitaten (zwerghaftogkeit, halsrippen, behaarung) Jarb Kinder Phys Erzie. 1916;84:225–235. [Google Scholar]
- 2.Vrolik W. Tabulae ad illustrandam embryogenesin hominis et mammalium tam naturalem quam abnormem. London, Amsterdam: 1849. [Google Scholar]
- 3.de Lange C. Sur un type nouveau de dégénération (typus Amstelodamensis) Arch Med Enfants. 1933;36:713–719. [Google Scholar]
- 4.Dorsett D, Krantz ID. On the molecular etiology of Cornelia de Lange syndrome. Ann N Y Acad Sci. 2009;1151:22–37. doi: 10.1111/j.1749-6632.2008.03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeScipio C, Kaur M, Yaeger D, et al. Chromosome rearrangements in cornelia de Lange syndrome (CdLS): report of a der(3)t(3;12)(p25.3;p13.3) in two half sibs with features of CdLS and review of reported CdLS cases with chromosome rearrangements. Am J Med Genet. 2005;137:276–282. doi: 10.1002/ajmg.a.30857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu DM, Huang JY, Li HY, et al. Paternal gonadal mosaicism of NIPBL mutation in a father of siblings with Cornelia de Lange syndrome. Prenat Diagn. 2006;26:1054–1057. doi: 10.1002/pd.1554. [DOI] [PubMed] [Google Scholar]
- 7.Kline AD, Krantz ID, Sommer A, et al. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet. 2007;143:1287–1296. doi: 10.1002/ajmg.a.31757. [DOI] [PubMed] [Google Scholar]
- 8.FitzPatrick D, Kline AD. Cornelia de Lange syndrome. In: Cassidy SB, Allanson JE, editors. Management of genetic syndromes. New York: Wiley-Liss; 2005. pp. 139–149. [Google Scholar]
- 9.Krantz ID, McCallum J, DeScipio C, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tonkin ET, Wang TJ, Lisgo S, et al. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 11.Musio A, Selicorni A, Focarelli ML, et al. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- 12.Deardorff MA, Kaur M, Yaeger D, et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson L, Kline AD, Barr MA, et al. de Lange syndrome: a clinical review of 310 individuals. Am J Med Genet. 1993;47:940–946. doi: 10.1002/ajmg.1320470703. [DOI] [PubMed] [Google Scholar]
- 14.Kline AD, Grados M, Sponseller P, et al. Natural history of aging in Cornelia de Lange syndrome. Am J Med Genet C Semin Med Genet. 2007;145C:248–260. doi: 10.1002/ajmg.c.30137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kline AD, Barr M, Jackson LG. Growth manifestations in the Brachmann-de Lange syndrome. Am J Med Genet. 1993;47:1042–1049. doi: 10.1002/ajmg.1320470722. [DOI] [PubMed] [Google Scholar]
- 16.Kline AD, Stanley C, Belevich J, et al. Developmental data on individuals with the Brachmann-de Lange syndrome. Am J Med Genet. 1993;47:1053–1058. doi: 10.1002/ajmg.1320470724. [DOI] [PubMed] [Google Scholar]
- 17.Luzzani S, Macchini F, Valade A, et al. Gastroesophageal reflux and Cornelia de Lange syndrome: typical and atypical symptoms. Am J Med Genet. 2003;119A:283–287. doi: 10.1002/ajmg.a.20191. [DOI] [PubMed] [Google Scholar]
- 18.Masumoto K, Izaki T, Arima T. Cornelia de Lange syndrome associated with cecal volvulus: report of a case. Acta Paediatr. 2001;90:701–703. [PubMed] [Google Scholar]
- 19.Mehta AV, Ambalavanan SK. Occurrence of congenital heart disease in children with Brachmann-de Lange syndrome. Am J Med Genet. 1997;71:434–435. doi: 10.1002/(sici)1096-8628(19970905)71:4<434::aid-ajmg12>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 20.Tsukahara M, Okamoto N, Ohashi H, et al. Brachmann-de Lange syndrome and congenital heart disease. Am J Med Genet. 1998;75:441–442. doi: 10.1002/(sici)1096-8628(19980203)75:4<441::aid-ajmg20>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 21.Selicorni A, Sforzini C, Milani D, et al. Anomalies of the kidney and urinary tract are common in de Lange syndrome. Am J Med Genet. 2005;132:395–397. doi: 10.1002/ajmg.a.30445. [DOI] [PubMed] [Google Scholar]
- 22.Berney TP, Ireland M, Burn J. Behavioural phenotype of Cornelia de Lange syndrome. Arch Dis Child. 1999;81:333–336. doi: 10.1136/adc.81.4.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hyman P, Oliver C, Hall S. Self-injurious behavior, self-restraint, and compulsive behaviors in Cornelia de Lange syndrome. Am J Ment Retard. 2002;107:146–154. doi: 10.1352/0895-8017(2002)107<0146:SIBSRA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 24.Moss J, Oliver C, Hall S, et al. The association between environmental events and self-injurious behaviour in Cornelia de Lange syndrome. J Intellect Disabil Res. 2005;49:269–277. doi: 10.1111/j.1365-2788.2005.00649.x. [DOI] [PubMed] [Google Scholar]
- 25.Kline AD, Krantz ID, Sommer A, et al. Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet. 2007;143A:1287–1296. doi: 10.1002/ajmg.a.31757. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi K, Ishitobi F. Brain dysgenesis in Cornelia de Lange syndrome. Clin Neuropathol. 1999;18:99–105. [PubMed] [Google Scholar]
- 27.Vuilleumier N, Kovari E, Michon A, et al. Neuropathological analysis of an adult case of the Cornelia de Lange syndrome. Acta Neuropathol (Berl) 2002;104:327–332. doi: 10.1007/s00401-002-0562-4. [DOI] [PubMed] [Google Scholar]
- 28.Wygnanski-Jaffe T, Shin J, Perruzza E, et al. Ophthalmologic findings in the Cornelia de Lange Syndrome. J Aapos. 2005;9:407–415. doi: 10.1016/j.jaapos.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Sataloff RT, Spiegel JR, Hawkshaw M, et al. Cornelia de Lange syndrome. Otolaryngologic manifestations. Arch Otolaryngol Head Neck Surg. 1990;116:1044–1046. doi: 10.1001/archotol.1990.01870090060008. [DOI] [PubMed] [Google Scholar]
- 30.Kaga K, Tamai F, Kitazumi E, et al. Auditory brainstem responses in children with Cornelia de Lange syndrome. Int J Pediatr Otorhinolaryngol. 1995;31:137–146. doi: 10.1016/0165-5876(94)01078-c. [DOI] [PubMed] [Google Scholar]
- 31.Roposch A, Bhaskar AR, Lee F, et al. Orthopaedic manifestations of Brachmann-de Lange syndrome: a report of 34 patients. J Pediatr Orthop. 2004;13:118–122. doi: 10.1097/00009957-200403000-00011. [DOI] [PubMed] [Google Scholar]
- 32.Kousseff BG, Thomson-Meares J, Newkirk P, et al. Physical growth in Brachmann-de Lange syndrome. Am J Med Genet. 1993;47:1050–1052. doi: 10.1002/ajmg.1320470723. [DOI] [PubMed] [Google Scholar]
- 33.Russell KL, Ming JE, Patel K, et al. Dominant paternal transmission of Cornelia de Lange syndrome: a new case and review of 25 previously reported familial recurrences. Am J Med Genet. 2001;104:267–276. doi: 10.1002/ajmg.10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maruiwa M, Nakamura Y, Motomura K, et al. Cornelia de Lange syndrome associated with Wilms’ tumour and infantile haemangioendothelioma of the liver: report of two autopsy cases. Virchows Arch. 1988;413:463–468. doi: 10.1007/BF00716995. [DOI] [PubMed] [Google Scholar]
- 35.Froster UG, Gortner L. Thrombocytopenia in the Brachmann-de Lange syndrome. Am J Med Genet. 1993;46:730–731. doi: 10.1002/ajmg.1320460629. [DOI] [PubMed] [Google Scholar]
- 36.Allanson JE, Hennekam RC, Ireland M. De Lange syndrome: subjective and objective comparison of the classical and mild phenotypes. J Med Genet. 1997;34:645–650. doi: 10.1136/jmg.34.8.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halal F, Preus M. The hand profile on de Lange syndrome: diagnostic criteria. Am J Med Genet. 1979;3:317–323. doi: 10.1002/ajmg.1320030402. [DOI] [PubMed] [Google Scholar]
- 38.Gillis LA, McCallum J, Kaur M, et al. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet. 2004;75:610–623. doi: 10.1086/424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ. Chromosome cohesion - rings, knots, orcs and fellowship. J Cell Sci. 2008;121:2107–2114. doi: 10.1242/jcs.029132. [DOI] [PubMed] [Google Scholar]
- 40.Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 41.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 42.Hopfner KP, Karcher A, Shin DS, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 43.Walker JE, Saraste M, Runswick MJ, et al. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- 45.Haering CH, Lowe J, Hochwagen A, et al. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 46.Onn I, Heidinger-Pauli JM, Guacci V, et al. Sister chromatid cohesion: a simple concept with a complex reality. Annual review of cell and developmental biology. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- 47.Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos Trans R Soc London. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitajima TS, Yokobayashi S, Yamamoto M, et al. Distinct cohesin complexes organize meiotic chromosome domains. Science. 2003;300:1152–1155. doi: 10.1126/science.1083634. [DOI] [PubMed] [Google Scholar]
- 49.Haering CH, Schoffnegger D, Nishino T, et al. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 50.Glynn EF, Megee PC, Yu HG, et al. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259, 1325–1339. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lengronne A, Katou Y, Mori S, et al. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang CR, Wu CS, Hom Y, et al. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 2005;19:3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unal E, Arbel-Eden A, Sattler U, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 54.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 55.Hornig NC, Uhlmann F. Preferential cleavage of chromatin-bound cohesin after targeted phosphorylation by Polo-like kinase. EMBO J. 2004;23:3144–3153. doi: 10.1038/sj.emboj.7600303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arumugam P, Gruber S, Tanaka K, et al. ATP hydrolysis is required for cohesin’s association with chromosomes. Curr Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- 57.Weitzer S, Lehane C, Uhlmann F. A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol. 2003;13:1930–1940. doi: 10.1016/j.cub.2003.10.030. [DOI] [PubMed] [Google Scholar]
- 58.Gruber S, Arumugam P, Katou Y, et al. Evidence that loading of cohesin onto chromosomes involves opening of its SMC hinge. Cell. 2006;127:523–537. doi: 10.1016/j.cell.2006.08.048. [DOI] [PubMed] [Google Scholar]
- 59.Milutinovich M, Unal E, Ward C, et al. A multi-step pathway for the establishment of sister chromatid cohesion. PLoS Genet. 2007;3:e12, 146–157. doi: 10.1371/journal.pgen.0030012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 61.Jones S, Sgouros J. The cohesin complex: sequence homologies, interaction networks and shared motifs. Genome Biol. 2001;2:RESEARCH0009. doi: 10.1186/gb-2001-2-3-research0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomonaga T, Nagao K, Kawasaki Y, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hakimi MA, Bochar DA, Schmiesing JA, et al. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature. 2002;418:994–998. doi: 10.1038/nature01024. [DOI] [PubMed] [Google Scholar]
- 64.Partridge JF, Scott KS, Bannister AJ, et al. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002;12:1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- 65.Weber SA, Gerton JL, Polancic JE, et al. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260, 1340–1353. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang XM, Mehta S, Uzri D, et al. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol Cell Biol. 2004;24:5290–5303. doi: 10.1128/MCB.24.12.5290-5303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lam WW, Peterson EA, Yeung M, et al. Condensin is required for chromosome arm cohesion during mitosis. Genes Dev. 2006;20:2973–2984. doi: 10.1101/gad.1468806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shimada K, Gasser SM. The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell. 2007;128:85–99. doi: 10.1016/j.cell.2006.11.045. [DOI] [PubMed] [Google Scholar]
- 69.Monje-Casas F, Prabhu VR, Lee BH, et al. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diaz-Martinez LA, Gimenez-Abian JF, Azuma Y, et al. PIASgamma is required for faithful chromosome segregation in human cells. PLoS ONE. 2006;1:e53, 1–14. doi: 10.1371/journal.pone.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sumara I, Vorlaufer E, Stukenberg PT, et al. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- 72.Kueng S, Hegemann B, Peters BH, et al. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 73.Hauf S, Roitinger E, Koch B, et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:e69, 419–432. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McGuinness BE, Hirota T, Kudo NR, et al. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86, 433–449. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmitz J, Watrin E, Lenart P, et al. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr Biol. 2007;17:630–636. doi: 10.1016/j.cub.2007.02.029. [DOI] [PubMed] [Google Scholar]
- 76.Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- 77.Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 78.Ciosk R, Zachariae W, Michaelis C, et al. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- 79.Guacci V. Sister chromatid cohesion: the cohesin cleavage model does not ring true. Genes Cells. 2007;12:693–708. doi: 10.1111/j.1365-2443.2007.01093.x. [DOI] [PubMed] [Google Scholar]
- 80.Gause M, Schaaf CA, Dorsett D. Cohesin and CTCF: cooperating to control chromosome conformation? Bioessays. 2008;30:715–718. doi: 10.1002/bies.20787. [DOI] [PubMed] [Google Scholar]
- 81.Kaur M, DeScipio C, McCallum J, et al. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet. 2005;138:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, et al. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum Mol Genet. 2007;16:1478–1487. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- 83.Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rollins RA, Korom M, Aulner N, et al. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Misulovin Z, Schwartz YB, Li XY, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Misulovin Z, Schwartz YB, Li XY, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma. 2008;117:89–102. doi: 10.1007/s00412-007-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pauli A, Althoff F, Oliveira RA, et al. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schuldiner O, Berdnik D, Levy JM, et al. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev Cell. 2008;14:227–238. doi: 10.1016/j.devcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Horsfield JA, Anagnostou SH, Hu JK, et al. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- 91.Wendt KS, Yoshida K, Itoh T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 92.Parelho V, Hadjur S, Spivakov M, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 93.Stedman W, Kang H, Lin S, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu J, Zhang Z, Bando M, Itoh T, et al. Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 2009;7:e1000119, 1–16. doi: 10.1371/journal.pbio.1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gause M, Webber HA, Misulovin Z, et al. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2008;117:51–66. doi: 10.1007/s00412-007-0125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brown CJ, Miller AP, Carrel L, et al. The DXS423E gene in Xp11.21 escapes X chromosome inactivation. Hum Mol Genet. 1995;4:251–255. doi: 10.1093/hmg/4.2.251. [DOI] [PubMed] [Google Scholar]
- 97.Revenkova E, Focarelli ML, Susani L, et al. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum Mol Genet. 2009;18:418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chernukhin I, Shamsuddin S, Kang SY, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27:1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 100.Ling JQ, Li T, Hu JF, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 101.Yusufzai TM, Tagami H, Nakatani Y, et al. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 102.Zhang B, Chang J, Fu M, et al. Dosage effects of cohesin regulatory factor PDS5 on mammalian development: implications for cohesinopathies. PLoS ONE. 2009;4:e5232, 1–17. doi: 10.1371/journal.pone.0005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang B, Jain S, Song H, et al. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development. 2007;134:3191–3201. doi: 10.1242/dev.005884. [DOI] [PubMed] [Google Scholar]
- 104.Vega H, Waisfisz Q, Gordillo M, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 105.Schule B, Oviedo A, Johnston K, et al. Inactivating mutations in ESCO2 cause SC phocomelia and Roberts syndrome: no phenotype-genotype correlation. Am J Hum Genet. 2005;77:1117–1128. doi: 10.1086/498695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Toth A, Ciosk R, Uhlmann F, et al. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tanaka K, Hao Z, Kai M, et al. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Unal E, Heidinger-Pauli JM, Kim W, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- 109.Ben-Shahar TR, Heeger S, Lehane C, et al. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- 110.Williams BC, Garrett-Engele CM, Li Z, et al. Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr Biol. 2003;13:2025–2036. doi: 10.1016/j.cub.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 111.German J. Roberts’ syndrome. I. Cytological evidence for a disturbance in chromatid pairing. Clin Genet. 1979;16:441–447. doi: 10.1111/j.1399-0004.1979.tb01354.x. [DOI] [PubMed] [Google Scholar]
- 112.Gordillo M, Vega H, Trainer AH, et al. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum Mol Genet. 2008;17:2172–2180. doi: 10.1093/hmg/ddn116. [DOI] [PubMed] [Google Scholar]
- 113.Gibbons RJ, Picketts DJ, Villard L, et al. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 114.Ritchie K, Seah C, Moulin J, et al. Loss of ATRX leads to chromosome cohesion and congression defects. J Cell Biol. 2008;180:315–324. doi: 10.1083/jcb.200706083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kobayashi J, Antoccia A, Tauchi H, et al. NBS1 and its functional role in the DNA damage response. DNA Repair (Amst) 2004;3:855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 116.Mayer ML, Pot I, Chang M, et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell. 2004;15:1736–1745. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Petronczki M, Chwalla B, Siomos MF, et al. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J Cell Sci. 2004;117:3547–3559. doi: 10.1242/jcs.01231. [DOI] [PubMed] [Google Scholar]
- 118.Skibbens RV. Unzipped and loaded: the role of DNA helicases and RFC clamp-loading complexes in sister chromatid cohesion. J Cell Biol. 2005;169:841–846. doi: 10.1083/jcb.200503129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Skibbens RV, Maradeo M, Eastman L. Fork it over: the cohesion establishment factor Ctf7p and DNA replication. J Cell Sci. 2007;120:2471–2477. doi: 10.1242/jcs.011999. [DOI] [PubMed] [Google Scholar]
- 120.Oikawa K, Akiyoshi A, Tanaka M, et al. Expression of various types of alternatively spliced WAPL transcripts in human cervical epithelia. Gene. 2008;423:57–62. doi: 10.1016/j.gene.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 121.Oikawa K, Ohbayashi T, Kiyono T, et al. Expression of a novel human gene, human wings apart-like (hWAPL), is associated with cervical carcinogenesis and tumor progression. Cancer Res. 2004;64:3545–3549. doi: 10.1158/0008-5472.CAN-03-3822. [DOI] [PubMed] [Google Scholar]
- 122.Kuroda M, Kiyono T, Oikawa K, et al. The human papillomavirus E6 and E7 inducible oncogene, hWAPL, exhibits potential as a therapeutic target. Br J Cancer. 2005;92:290–293. doi: 10.1038/sj.bjc.6602329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kwiatkowski BA, Ragoczy T, Ehly J, et al. Identification and cloning of a novel chromatin-associated protein partner of Epstein-Barr nuclear protein 2. Exp Cell Res. 2004;300:223–233. doi: 10.1016/j.yexcr.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 124.Ohbayashi T, Oikawa K, Yamada K, et al. Unscheduled over-expression of human WAPL promotes chromosomal instability. Biochem Biophys Res Commun. 2007;356:699–704. doi: 10.1016/j.bbrc.2007.03.037. [DOI] [PubMed] [Google Scholar]
- 125.Shepard JL, Amatruda JF, Finkelstein D, et al. A mutation in separase causes genome instability and increased susceptibility to epithelial cancer. Genes Dev. 2007;21:55–59. doi: 10.1101/gad.1470407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pandey R, Heidmann S, Lehner CF. Epithelial re-organization and dynamics of progression through mitosis in Drosophila separase complex mutants. J Cell Sci. 2005;118:733–742. doi: 10.1242/jcs.01663. [DOI] [PubMed] [Google Scholar]
- 127.Zhang N, Ge G, Meyer R, et al. Overexpression of Separase induces aneuploidy and mammary tumorigenesis. Proc Natl Acad Sci U S A. 2008;105:13033–13038. doi: 10.1073/pnas.0801610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barber TD, McManus K, Yuen KW, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang X, Horwitz GA, Heaney AP, et al. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J Clin Endocrinol Metab. 1999;84:761–767. doi: 10.1210/jcem.84.2.5432. [DOI] [PubMed] [Google Scholar]
- 130.Zou H, McGarry TJ, Bernal T, et al. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]
- 131.Jallepalli PV, Waizenegger IC, Bunz F, et al. Securin is required for chromosomal stability in human cells. Cell. 2001;105:445–457. doi: 10.1016/s0092-8674(01)00340-3. [DOI] [PubMed] [Google Scholar]
- 132.Zhang X, Horwitz GA, Prezant TR, et al. Structure, expression, and function of human pituitary tumor-transforming gene (PTTG) Mol Endocrinol. 1999;13:156–166. doi: 10.1210/mend.13.1.0225. [DOI] [PubMed] [Google Scholar]
- 133.Ryu B, Kim DS, Deluca AM, et al. Comprehensive expression profiling of tumor cell lines identifies molecular signatures of melanoma progression. PLoS ONE. 2007;2:e594, 1–13. doi: 10.1371/journal.pone.0000594. [DOI] [PMC free article] [PubMed] [Google Scholar]