Abstract
The estrogen receptor α (ERα) is activated as a transcription factor by both estrogen and a large variety of other extracellular signals. The mechanisms of this ligand-independent activation, notably by cAMP signaling, are still largely unknown. We now close the gap in the signaling pathway between cAMP and ERα. Whereas the direct phosphorylation of ERα by the cAMP-activated protein kinase A (PKA) is dispensable, the phosphorylation of the coactivator-associated arginine methyltransferase 1 (CARM1) by PKA at a single serine is necessary and sufficient for direct binding to the unliganded hormone-binding domain (HBD) of ERα, and the interaction is necessary for cAMP activation of ERα. Sustained PKA activity promoting a constitutive interaction may contribute to tamoxifen resistance of breast tumors. Binding and activation involve a novel regulatory groove of the ERα HBD. As a result, depending on the activating signal, ERα recruits different coactivator complexes to regulate alternate sets of target genes.
Keywords: Steroid receptor, signaling, coactivator, protein kinase A, breast cancer, endocrine resistance
The estrogen receptors (ERs) are ligand-regulated transcription factors and belong to the nuclear receptor superfamily. The two isoforms ERα and ERβ mediate most of the physiological effects of estrogens. Moreover, ERα plays a pivotal role in promoting the proliferation of several types of estrogen-stimulated carcinomas, including breast cancer (Ali and Coombes 2002). How estrogen regulates ERα activity has been studied extensively. It induces a conformational change in the hormone-binding domain (HBD) that results in the release of the Hsp90 molecular chaperone complex that is associated with the unliganded ERα (Picard 2006), and promotes the recruitment of a host of coactivator proteins that mediate the transcriptional effects of ERα (Rosenfeld et al. 2006; Green and Carroll 2007; Lonard and O'Malley 2007).
Only a few years after the first steroid receptors were cloned, it was discovered that the progesterone receptor (PR) can be activated by elevated levels of cAMP even in the absence of progesterone (Denner et al. 1990). The phenomenon of ligand-independent activation was soon extended to ERα when it was found that the neurotransmitter dopamine, which signals through cAMP, could activate both PR and ERα (Power et al. 1991), and that ERα could also be turned on by epidermal growth factor (EGF) (Ignar-Trowbridge et al. 1992). Since then, a large variety of signals that do not function as direct ligands of these steroid receptors have been shown to induce the transcriptional activities of the four sex steroid receptors ERα, ERβ, PR, and androgen receptor (for review, see Picard 2003; for an updated list, see http://www.picard.ch/downloads/downloads.htm). There is growing evidence for the relevance of this extreme form of signaling cross-talk. For example, (1) the stimulation of uterine growth by EGF and IGF-I depends on ERα (Curtis et al. 1996; Klotz et al. 2002), and persists in mice with an ERα knock-in mutant that severely compromises the response to endogenous estrogens (Sinkevicius et al. 2008); (2) the regulation of sexual behavior in rodents by growth factors may be mediated by ligand-independent activation of ERα (Apostolakis et al. 2000); (3) the social behavior of neonatal female rats is shaped by dopamine in an ERα-dependent manner (Olesen et al. 2005; Olesen and Auger 2008); (4) the anti-estrogen tamoxifen used for endocrine therapy of breast cancer can be switched to an agonist by signaling cross-talk (Fujimoto and Katzenellenbogen 1994; Lee et al. 2000; Michalides et al. 2004), an effect that may underly the development of tamoxifen resistance in some tumors (discussed by Johnston et al. 2003); and (5) cyclin D1 overexpression, often associated with breast cancer, activates ERα (Neuman et al. 1997; Zwijsen et al. 1997).
The characterization of the molecular mechanisms of ligand-independent activation of ERα has proven to be difficult. The activation of ERα by growth factors involves the direct phosphorylation by MAPK of a serine within the N-terminal activation function 1 (AF1) of ERα (Bunone et al. 1996). Although this is not sufficient for activation, it is necessary for the recruitment of the splicing factor SF3a120 as a coactivator (Masuhiro et al. 2005), as well as the subsequent attenuation of ERα activity by SPBP (Gburcik et al. 2005). Cyclin D1 associates with the hinge region of apo-ERα and recruits the coactivator SRC1 (Zwijsen et al. 1998). In contrast, how cAMP functions to activate unliganded ERα has remained largely enigmatic. Early on, inhibitor studies already suggested that cAMP signals to ERα through protein kinase A (PKA) (Aronica and Katzenellenbogen 1993; see also Al-Dhaheri and Rowan 2007), and limited mutational analyses pointed to the HBD as the target domain (Smith et al. 1993; Coleman et al. 2003), rather than AF1, as for growth factors, or the hinge, as for cyclin D1. Whether the direct phosphorylation of ERα by PKA, which does indeed occur and modulates its activity (Aronica and Katzenellenbogen 1993; Chen et al. 1999b; Cui et al. 2004; Michalides et al. 2004; Al-Dhaheri and Rowan 2007; Zwart et al. 2007), plays any role has not been clarified.
It has been recognized that transcriptional coactivators can act as integrators of signaling pathways (Wu et al. 2005; Rosenfeld et al. 2006). They are substrates for a variety of kinases, and their post-translational modifications modify their coactivator activities. Although certain coactivators can potentiate ligand-independent activation of ERα (for example, see Zwijsen et al. 1998; Coleman et al. 2003; Fenne et al. 2008; see also Rowan et al. 2000), it has not been determined which one, if any, is necessary. Nevertheless, they have remained good candidates as mediators of ligand-independent pathways, including the activation of ERα by cAMP. We therefore undertook to revisit the molecular determinants on ERα that are required for activation by cAMP, and to close the gap in this particular signaling cross-talk pathway between cAMP and ERα.
Results
cAMP activates endogenous ERα through PKA
A literature survey and pilot experiments suggested that experimental parameters might strongly influence signaling cross-talk between cAMP and ERα. We therefore wished to ensure that we could recapitulate and study the phenomenon in our own setting. The response of endogenous ERα was assessed by a transactivation assay with human MCF7 breast cancer cells. A typical result obtained with a transfected luciferase reporter gene for ERα is shown in Supplemental Figure S1. As expected from previous studies, ERα is activated by both the cognate ligand estradiol (E2) and a cell-permeable analog of cAMP. Since the response is blocked by the PKA inhibitor H89 and mimicked by overexpression of the catalytic subunit of PKA (Fig. 1; Supplemental Fig. S1), we conclude that cAMP activates ERα through PKA, and that they can be used interchangeably despite quantitative differences in the response. Intriguingly, in early experiments, PKA overexpression occasionally failed to turn on ERα. Prompted by a report that the PKA phosphorylation of S236 in the DNA-binding domain of ERα inhibits dimerization and, as a consequence, DNA binding (Chen et al. 1999b), we analyzed the effects of PKA more carefully by performing a dose response experiment (Supplemental Fig. S1B). The bimodal response of ERα to increasing PKA concentrations suggests that low and intermediate levels of PKA can activate ERα, and that the inhibitory phosphorylation of S236 becomes dominant at high PKA levels.
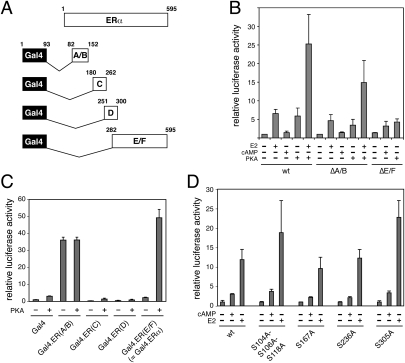
Figure 1.
The role of ERα domains and phosphorylation sites. These luciferase reporter assays were carried out in ERα-negative cells. Data shown are averages of at least triplicate samples, expressed relative to that of the very first sample in each panel. (A) Schematic representation of ERα domains and corresponding Gal4 DNA-binding domain fusion proteins. (B) Response of ERα deletion mutants to cAMP/PKA. (C) PKA response of ERα domains expressed as chimeras with the DNA-binding domain of Gal4. (D) cAMP responses of ERα phosphorylation mutants. SKBr3 (B,C) and 293T (D) cells were used for the experiments. (wt) Wild-type; (cAMP) 8-Br-cAMP.
Domain requirements for signaling cross-talk
To dissect the ERα domain requirements in transfection experiments (Fig. 1), we switched to ER-negative cell lines such as SKBr3 or 293T. The N-terminal A/B and the C-terminal E/F domains are associated with the transcriptional activation functions AF1 and AF2, respectively. Interestingly, deletion of either domain does not abolish the response to cAMP or PKA (Fig. 1A,B). The N-terminally truncated receptor can still be activated in the absence of E2, and the C-terminal truncation mutant, only poorly active in this experimental system, is further stimulated. Whereas the constitutively active A/B domain by itself does not seem to be sufficient to respond to cAMP/PKA as a fusion protein with the Gal4 DNA-binding domain, the HBD responds robustly in this context, suggesting that the HBD may be the primary target domain (Fig. 1C). The very C-terminal and poorly conserved F domain, which was present in Gal4 construct Gal4.ERα but is not part of the HBD itself, is dispensable for this response (Supplemental Fig. S2A). The role of known ERα phosphorylation sites was explored with a panel of point mutants. Apart from S236, S305 was of particular interest because its phosphorylation by p21-activated kinase 1 and PKA had been demonstrated to mediate ligand-independent activation (Wang et al. 2002) and tamoxifen resistance (Michalides et al. 2004), respectively. Phosphorylation of S305 had also been shown to increase the hormone sensitivity of ERα by blocking acetylation of K303 (Cui et al. 2004). Although there are quantitative effects of mutating these serine residues, none of these mutations abolishes the response to cAMP/PKA (Fig. 1D). These findings are consistent with the idea that the direct phosphorylation of ERα by PKA, which may occur, is not a major determinant for this type of signaling cross-talk.
Signaling cross-talk depends on AF2 and on a novel ERα-specific surface of the HBD
An AF2 mutant of ERα or a dominant-negative mutant coactivator block cAMP activation (Supplemental Fig. S2). Although these findings indicated that AF2 is required, this did not exclude the possibility that other surfaces of the HBD might also be involved. In this context, the ERα mutant G400V provided a valuable hint. It has a slightly lower affinity for E2, but still achieves full activation at higher E2 concentrations (Tora et al. 1989). In contrast, it had been reported that its response to the neurotransmitter dopamine, most likely mediated by cAMP/PKA, is completely abolished (Smith et al. 1993). We therefore revisited this result with a Gal4-ERα HBD fusion protein, which provides a larger dynamic range than wild-type ERα. Whereas E2 induces G400V almost 40-fold, PKA is completely ineffective (Supplemental Fig. S2B).
Residue G400 is not part of AF2. In fact, in the dimeric HBD, it is located on the opposite side of the subunit as well as far from AF2 of the other HBD monomer (Supplemental Fig. S3). G400 forms part of a turn between helix 5 (H5) and the only β sheet in the HBD (Fig. 2A; Supplemental Fig. S3). According to the crystal structure, a valine substitution would result in the isopropyl group of the side chain protruding outside of the structure into a solvent-accessible area, which appears to be part of a larger groove-like structure in the HBD dimer. In addition to the loop, several amino acids from H6 and H7 belonging to one subunit, and from H8 to H9 from the other subunit, contribute to the formation of this groove. Among this group, the amino acids R412, S433, S463, and S464, which are most proximal to G400 and carry charges or hydrophilic side chains, were selected for mutagenesis (Fig. 2). In addition, L429 was also selected because its side chain appears to form part of the base of the groove and is most proximal to G400. Interestingly, G400, R412, S463, and S464 are highly conserved in ERα sequences from a large range of vertebrate species (Supplemental Fig. S3A).
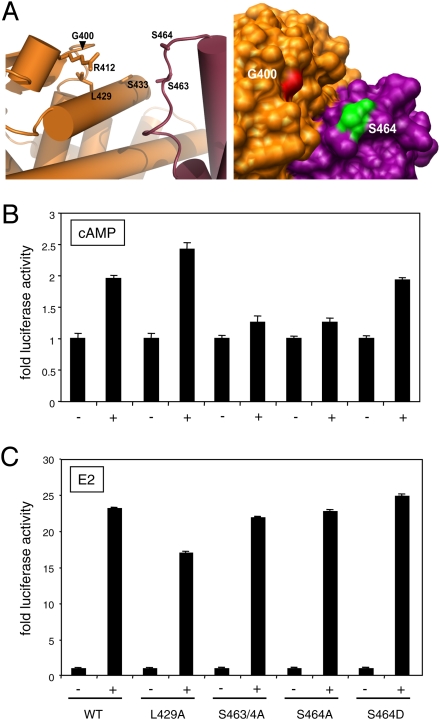
Figure 2.
A new surface of the ERα HBD is required for the cAMP response. (A) G400-centered views of a portion of the ERα HBD dimer structure. The model was generated with the data of PDB file 1A52. The left panel is a cartoon view showing the relative positions of the mutagenized amino acids (G400, R412, L429, S433, S463, and S464). The panel on the right shows a surface view, which is tilted slightly to the right relative to the view in the left panel. The two subunits were colored in orange and purple. (B,C) Ser 464 is a key determinant of the cAMP response of full-length ERα. Graphs of luciferase reporter assays in B and C show cAMP and E2 responses, respectively, of wild-type (WT) and mutant ERα in transfected 293T cells. Where indicated with a plus sign (+), cells were treated with 8-Br-cAMP (cAMP) or E2.
Initially, our panel of alanine substitution mutants were tested as Gal4-ERα HBD chimeras (Supplemental Fig. S4). The key results were confirmed in the context of full-length ERα by a transactivation assay in ER-negative 293T cells (Fig. 2). Whereas the full-length version of L429A also yielded a higher maximal response in response to cAMP, it remains to be further examined whether it is activated by lower concentrations of E2 as its Gal4 fusion variant (Supplemental Fig. S4). Since ERβ, which is completely refractory to cAMP (Supplemental Fig. S5), contains an alanine in the position corresponding to S464, we speculated that the double mutant S463/4A might be defective because of the substitution of S464. As can be seen in Figure 2, an alanine substitution abolishes the cAMP response but not the response to E2, whereas S464D behaves like wild type for both responses. While we cannot formally exclude that S464 is phosphorylated by PKA, we consider this very unlikely in view of our other findings (see below), and the fact that two different groups have shown that S305 is by far the major PKA phosphorylation site (Cui et al. 2004; Michalides et al. 2004). The functionality of S464D suggests that it is the specific nature of the substitution in S464A that leads to the cAMP signaling defect. Thus, the specific cAMP response defects of G400V and S464A suggest that the groove that these residues line might represent a novel regulatory or interaction surface of ERα for the recruitment of a coregulator in response to cAMP.
Coactivator-associated arginine methyltransferase 1 (CARM1) is a mediator of signaling cross-talk
Since our attempts to identify such proteins by an unbiased biochemical approach proved unsuccessful, we turned to a candidate protein approach. We were intrigued by a report that apo-ERα cycles on and off a target promoter along with several known coregulators, but in the notable absence of AF2 coactivators of the p160 family (Métivier et al. 2004). It seemed possible that one or several of these cycling factors might engage in transcriptionally productive complexes following cAMP/PKA stimulation. We decided to examine more closely the histone acetyltransferases CBP/p300, and CARM1 (for review, see Bedford and Clarke 2009; Lee and Stallcup 2009).
The overexpression of p300, CBP, or CARM1 in MCF7 cells markedly increased the response of the endogenous ERα to cAMP signaling (Fig. 3A; Supplemental Fig. S6A). It had no effect on the activity of apo-ERα in the absence of cAMP. Whereas CBP/p300 can interact directly with ERα through both AF1 and AF2 even in the absence of ligand (Kobayashi et al. 2000; Dutertre and Smith 2003), CARM1 is known to be recruited indirectly to liganded ERα; for example, through the p160 coactivator GRIP1 (Chen et al. 1999a, 2000). We therefore investigated the recruitment of CBP and CARM1 to ERα in response to cAMP (Supplemental Fig. S6B). As expected, endogenous ERα could be constitutively coprecipitated with HA-tagged CBP from transfected MCF7 cells. In contrast, under our experimental conditions, HA-CARM1 only immunoprecipitated ERα after stimulation with cAMP. The cAMP dependence for this interaction was confirmed by a reciprocal immunoprecipitation (IP) of endogenous CARM1 with endogenous ERα (Fig. 3B), and, using a chromatin IP (ChIP) experiment, we found that CARM1 is recruited to the ERα target gene pS2 (officially known as TFF1) in response to both E2 and cAMP (Fig. 3C). Thus, regarding the response to cAMP, IP and ChIP results correlate, including with earlier reports showing the recruitment of ERα itself to the pS2 gene (Al-Dhaheri and Rowan 2007; Fenne et al. 2008), and support the conclusion that endogenous CARM1 is recruited to ERα upon cAMP signaling.
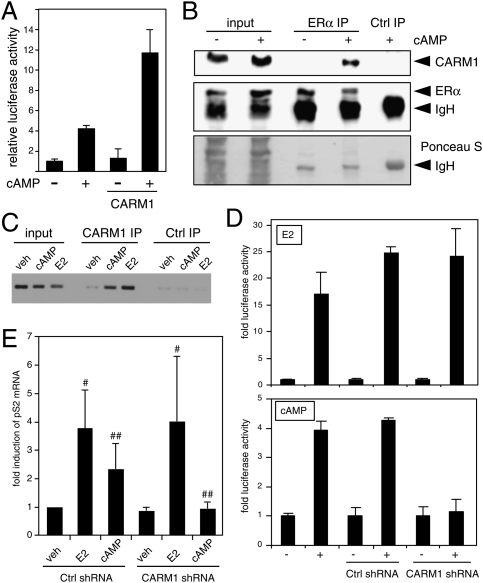
Figure 3.
CARM1 interacts with and is required for the cAMP response of ERα. (A) The effect of overexpression of CARM1 on the activity of endogenous ERα was determined with a luciferase reporter by transient transfection of MCF7 cells. (B) CARM1 interacts with ERα in MCF7 cells in a cAMP-dependent manner. Co-IP experiment of endogenous proteins. Cells were stimulated with 8-Br-cAMP 2 h before lysis. Equal amounts of extracts (bottom panel: Ponceau S-stained filter) were immunoprecipitated with an antibody against ERα or control (Ctrl) antibodies, and immunoblots were probed with antibodies to the endogenous CARM1 (top panel) or ERα (middle panel). (IgH) Heavy chain of antibodies. (C) E2 and cAMP induce the recruitment of CARM1 to the pS2 promoter. DNA gel visualizing the PCR products of a ChIP experiment with CARM1 and control (Ctrl) antisera. (D,E) CARM1 is necessary for the cAMP response of endogenous ERα in MCF7-SH cells. (D) Luciferase assay of cells cotransfected with the indicated shRNA constructs. Graphs show averages of the means of three independent experiments, each with triplicate samples. (E) Quantitative RT–PCR analysis of the endogenous ERα target gene pS2 with EEF1A1 internal standard. In this case, stable RNAi was obtained by infection with corresponding lentiviral preparations. Each data point represents the average of the data of three independent experiments, standardized to the value of the control shRNA sample, arbitrarily set to 1. In contrast to the induction by E2 ([#] P > 0.37), the cAMP induction is significantly reduced by the CARM1 knockdown ([##] P < 0.057).
We next used an RNAi experiment (see Supplemental Fig. S7 for validation) to determine whether CARM1 is also necessary. Knocking down CARM1 expression in MCF7-SH cells with a specific shRNA construct reduced the cAMP-induced activity of endogenous ERα, as assayed with a transfected reporter gene or by analyzing expression of the endogenous pS2 gene (Fig. 3D,E), whereas it did not affect the E2-induced activity. Note that the latter is consistent with previous reports (Chen et al. 1999a, 2000) that also indicated that E2-induced ERα activity may be largely CARM1-independent under these experimental conditions, and even for some endogenous target genes (Frietze et al. 2008). The fact that CARM1 is required for some E2-induced responses (see also Yadav et al. 2003) supports the notion that coregulator requirement can be highly tissue- and context-dependent.
To facilitate the next series of experiments, we verified that the cAMP response of the Gal4-ERα HBD chimera (Gal4.ERα), like that of endogenous ERα in MCF7 cells, can be further augmented by overexpression of CARM1, and that there is no effect of CARM1 overexpression on Gal4.ERα activity in the absence of cAMP (Fig. 4A).
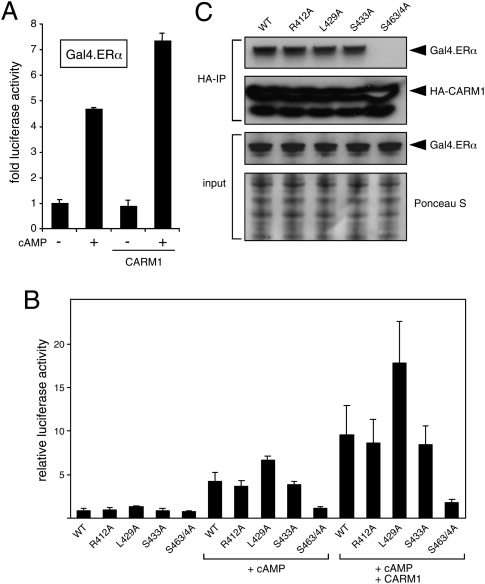
Figure 4.
Potentiation of ERα mutants by CARM1 and interaction correlate. (A) cAMP-induced but not basal activities of Gal4.ERα are potentiated by CARM1 overexpression. Luciferase reporter gene assays in 293T cells. (B) cAMP responses of mutant Gal4.ERα chimeras without and with coexpressed CARM1. (C) Co-IP of mutant Gal4.ERα chimeras with CARM1. HA-tagged CARM1 and the indicated wild-type (WT) or mutant versions of Gal4.ERα were coexpressed in 293T cells. Cells were stimulated with 8-Br-cAMP 1 h before lysis. Equal amounts of extracts (bottom panel: Ponceau-stained filter of gel run in parallel; bottom middle panel: immunoblot probed with an anti-Gal4 antibody) were immunoprecipitated with an anti-HA antibody. Immunoblots were probed with antibodies to Gal4 (top panel) or to the HA tag (top middle panel).
First, we tested whether CARM1 activity is required by comparing the augmentation by wild-type CARM1 and by a methyltransferase mutant CARM1. To our surprise, the enzymatic activity turned out to be dispensable (Supplemental Fig. S8A), as reported recently for the coactivation of NFκB by CARM1 (Jayne et al. 2009). Next, we speculated that the cAMP-induced recruitment of CARM1 to ERα is nevertheless necessary for the transcriptional response, and that it might involve a new regulatory or interaction surface on the ERα HBD. We tested this notion by using our set of HBD point mutants (Fig. 4). They were coexpressed as Gal4-ERα HBD chimeras along with CARM1 in 293T cells. The experiment showed that, with the notable exceptions of the single-point mutant G400V and the double mutant S463/4A, all other mutants can be potentiated (Fig. 4B; Supplemental Fig. S8B). L429A again was the most active one. S463/4A was not only refractory to cAMP and/or CARM1 overexpression, it also failed to interact with it, as judged by an IP experiment (Fig. 4C).
Direct phosphorylation of CARM1 by PKA and interaction with ERα
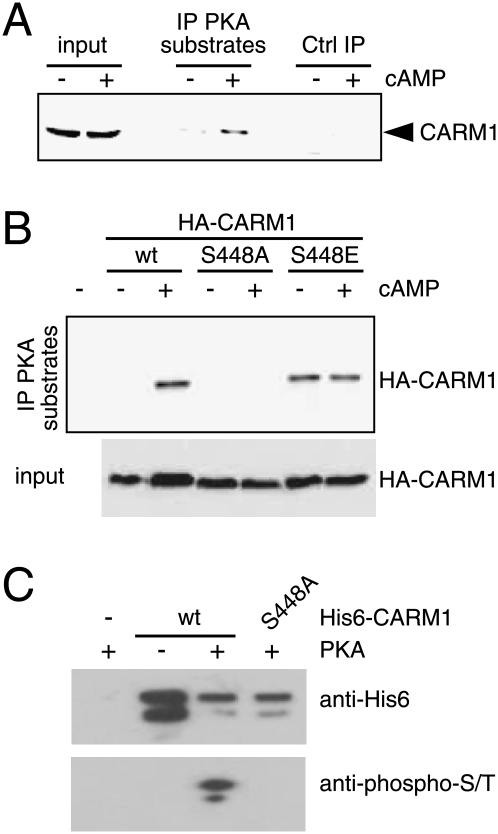
With CARM1 established as a necessary mediator, it seemed plausible that it might be the target of PKA. We used an antibody directed against the PKA-phosphorylated motif RRX-phospho-S/T to immunoprecipiate PKA substrates from MCF7 extracts, and probed for the presence of CARM1 in the immunoprecipitate. The result shows that endogenous CARM1 becomes phosphorylated in cells treated with cAMP (Fig. 5A). We then looked for potential PKA phosphorylation sites in CARM1 using Scansite 2.0 (http://scansite.mit.edu). At medium stringency, the motif scan yielded only one hit: S448 within the sequence KRQS. The importance of this residue for in vivo phosphorylation of HA-CARM1 by PKA was assessed by expressing both the wild-type and S448A mutant versions in transfected 293T cells. The IP was done as before, except that this time the presence of exogenous HA-CARM1 in the immunoprecipitates was revealed with an anti-HA antibody (Fig. 5B). In contrast to wild-type CARM1, the mutant S448A appears to be completely refractory to PKA-induced phosphorylation. The phosphoserine mimic S448E is constitutively recognized by the antibody, further corroborating the prediction of this residue as a PKA target site. To confirm that CARM1 is phosphorylated directly by PKA, we incubated purified recombinant CARM1 with a commercial preparation of PKA. Again, only wild-type, but not the S448A mutant, could be phosphorylated (Fig. 5C). These results argue very strongly that S448 is phosphorylated by PKA, and that it is the major, if not the only, PKA phosphorylation site of CARM1.
Figure 5.
CARM1 is phosphorylated by PKA in vivo and in vitro. (A) CARM1 immunoblot of an IP with an antibody against substrates phosphorylated by PKA (“IP PKA substrates”). MCF7 cells were treated with 8-Br-cAMP or vehicle 2 h before lysis. (B) CARM1 S448 is the main target of PKA in vivo. Wild-type (wt) and mutant HA-CARM1 were expressed in 293T cells and analyzed as in A, except that the membrane was probed with an anti-HA antibody. (C) In vitro phosphorylation of CARM1 by PKA. Recombinant purified His6-tagged CARM1 (wt or S448A as indicated) was phosphorylated with the catalytic β subunit of PKA and analyzed by immunoblotting with a monoclonal against phospho-Ser/Thr.
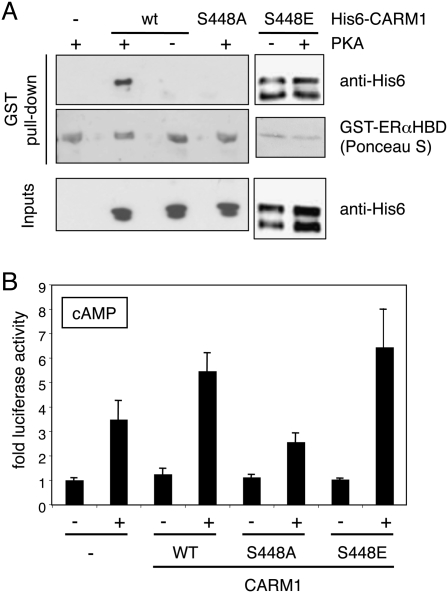
The impact of CARM1 phosphorylation on the interaction with ERα was investigated with a GST pull-down experiment using purified recombinant interaction partners (Fig. 6A). CARM1 was first phosphorylated with PKA before incubation with a GST fusion of the ERα HBD. Unlike the mutant S448A, which could not be phosphorylated, wild-type CARM1 was able to interact with the ERα fusion protein. Note that the GST-ERαHBD fusion protein does not get detectably phosphorylated by PKA that remains present during the GST pull-down (data not shown). Thus, the phosphorylation of CARM1 by PKA promotes the direct interaction with apo-ERα in the absence of other cellular proteins. The mutant S448E, which resembles the phosphorylated form, interacts constitutively with ERα. These results argue that phosphorylation of S448 by PKA is sufficient to promote the interaction with the ERα HBD.
Figure 6.
Direct interaction of CARM1 with ERα depends on direct phosphorylation of CARM1 by PKA. (A) In vitro phosphorylated CARM1 interacts directly with the ERα HBD. Following phosphorylation of His6-tagged CARM1 (wild type, S448A, or S448E) by PKA, its interaction with the ERα HBD was assessed by a GST pull-down experiment. Inputs represent 20% of each reaction. (B) The S448A CARM1 phosphorylation mutant is defective for stimulation of the cAMP response of ERα. Luciferase reporter gene assays showing stimulation of cAMP responses of ERα by wild-type and mutant CARM1 in cotransfected 293T cells. Note that expression levels of the three CARM1 versions were similar (data not shown).
Since the mutant S448A fails to be phosphorylated by PKA and to interact with ERα, it was to be expected that it would also be defective for stimulating the cAMP response of ERα in vivo (Fig. 6B). This clearly demonstrates that S448 is necessary for this response. Interestingly, the phosphoserine mimic S448E behaves like wild type in that it does not stimulate ERα transcriptional activity without stimulation by PKA. As will be discussed further below, this suggests that the phosphorylation of S448 may not be sufficient for stimulation of transcriptional activity.
Elevated CARM1 phosphorylation in tamoxifen-resistant cells promotes ERα binding and tamoxifen-induced activity
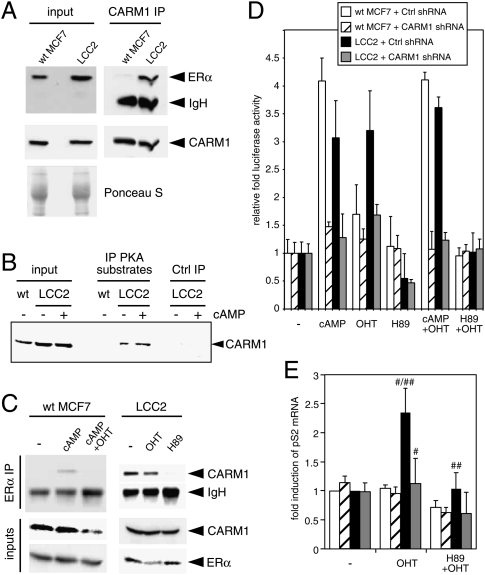
One setting where our findings might be physiologically relevant is in the context of breast cancer progression. There is already evidence that tamoxifen resistance correlates with and may be due to increased PKA activity (Michalides et al. 2004; Zwart et al. 2007). Using the tamoxifen-resistant MCF7 subclone LCC2 (Brünner et al. 1993), we explored the possibility that its altered behavior correlates with changes in the CARM1–ERα relationship. While ERα and CARM1 levels are comparable between wild-type and LCC2 cells, a CARM1 IP revealed that endogenous CARM1 is constitutively associated with ERα in LCC2 cells (Fig. 7A). This may be explained by the fact that PKA phosphorylation of CARM1 displays a high basal level in LCC2 cells (Fig. 7B), which in turn may be due to elevated levels of PKA in LCC2 relative to MCF7 cells (data not shown). Indeed, the PKA inhibitor H89 abolishes the constitutive interaction in LCC2 cells (Fig. 7C). Wild-type MCF7 and LCC2 cells also differ in the effect of (hydroxy-)tamoxifen (OHT) on the CARM1–ERα interaction. The interaction of the endogenous proteins is sensitive and resistant to OHT in wild-type MCF7 and LCC2 cells, respectively.
Figure 7.
Increased PKA activity promotes the constitutive and tamoxifen-resistant interaction of ERα and CARM1 in LCC2 cells. (A) Immunoblot showing constitutive interaction. Equal amounts of MCF7 and LCC2 cell extracts (bottom panel: Ponceau staining) were immunoprecipitated with an anti-CARM1 antibody, and immunoblots were probed with antibodies to the endogenous ERα (top panel) or CARM1 (middle panel). (B) CARM1 is constitutively phosphorylated in LCC2 cells. The experiment was done as described in the legend for Figure 5A. (C) The constitutive interaction between CARM1 and ERα in LCC2 cells depends on PKA activity and is not affected by OHT. Wild-type (wt) MCF7 and LCC2 cells were treated with 8-Br-cAMP, OHT, 8-Br-cAMP + OHT, or H89 4 h before lysis. Extracts were immunoprecipitated with an anti-ERα antibody, and immunoblots were probed with antibodies to endogenous CARM1 (top and middle panels) or ERα (bottom panel). (D) OHT activates ERα in LCC2 in a PKA- and CARM1-dependent fashion. Luciferase reporter gene assays of endogenous ERα in cells cotransfected with shRNA constructs and treated as indicated. Activities are expressed relative to that of the control sample for each transfection. (E) Quantitative RT–PCR analysis of the endogenous ERα target gene pS2 with EEF1A1 internal standard. In this case, stable RNAi was obtained by infection with corresponding lentiviral preparations. The color code of the bars is the same as in D. Each data point represents the average of the data of three independent experiments, standardized to the values of the control samples of the two cell lines that were arbitrarily set to 1. The differences of the highlighted comparisons are highly significant ([#] P < 0.01; [##] P < 0.03).
The functional impact on the activity of endogenous ERα became apparent with a luciferase reporter gene assay (Fig. 7D). OHT activates ERα as well as cAMP in LCC2 but not wild-type MCF7 cells, and this activity depends on both PKA activity and CARM1. It is important to note that the endogenous ERα target gene pS2 responds exactly the same way (Fig. 7E). Its activation by OHT in LCC2 cells depends on both CARM1 and PKA activity. This OHT response, and the resistance to OHT, can be recapitulated in ER-negative 293T cells by either overexpressing wild-type CARM1 and stimulating with cAMP, or overexpressing the CARM1 phosphoserine mimic S448E even in the absence of cAMP (Supplemental Fig. S9).
Discussion
Our demonstration that the direct phosphorylation of CARM1 by PKA leads to its recruitment to a novel regulatory surface of the unliganded ERα represents a fundamental leap in our understanding of the molecular mechanism of the extreme form of signaling cross-talk that allows ERα to be activated by cAMP. It explains previous failures to unravel the mechanism, and sets the stage for further investigations—notably of the pathological and physiological consequences of this mode of ERα activation.
Earlier efforts to unravel the mechanism of ligand-independent activation of ERα by cAMP
Past efforts to unravel the mechanism were hampered by various difficulties inherent to this system. There is no doubt that PKA phosphorylates ERα directly and does modulate its activity in various ways (Aronica and Katzenellenbogen 1993; Chen et al. 1999b; Cui et al. 2004; Michalides et al. 2004; Al-Dhaheri and Rowan 2007). One obvious complication is that the phosphorylation of S236 in the DNA-binding domain inhibits even DNA binding of ERα (Chen et al. 1999b). Hence, the level and, perhaps, the duration, of activation become important parameters that are difficult to control. It had been shown that cAMP/PKA stimulates the recruitment of cyclin D1 to unliganded ERα (Lamb et al. 2000) and of the coactivator SRC-1 to the unliganded chicken PR (Rowan et al. 2000), but whether the latter is true for ERα and whether either cofactor is necessary for the response were not determined. In fact, a mutational analysis indicated that none of the known phosphorylation sites of SRC-1 is necessary (Coleman et al. 2003).
CARM1 is the link between PKA and ERα
Once we established that none of the known PKA phosphorylation sites on ERα are necessary for ligand-independent activation of ERα by cAMP/PKA, it was clear that the target had to be a cofactor. With CARM1, we believe to have closed the gap between PKA and ERα. CARM1 is phosphorylated directly by PKA at S448, and both CARM1 and this phosphorylation are necessary for the response. S448 resides within the PKA consensus phosphorylation site R/KXXS/T, and this whole motif (KRQS) is highly conserved in evolution, but not in other protein arginine methyltransferases such as PRMT1. The known structure of CARM1 (Troffer-Charlier et al. 2007; Yue et al. 2007) does not allow one to predict the molecular effects of the phosphorylation of S448, which is located in the catalytic core domain, for binding to ERα and/or coactivator activity. The discovery of this phosphorylation site further emphasizes that CARM1 is subject to regulatory inputs. As-yet-unknown kinase(s) phosphorylate(s) human CARM1 at two different serine residues—S217 and S229—to inhibit the methyltransferase activity (Higashimoto et al. 2007; Feng et al. 2009). Whereas the phosphorylation of S448, which can be mimicked by a phosphoserine mutant, proved to be sufficient to promote the binding to the ERα HBD, it is insufficient to activate ERα in the absence of ligand. The most likely explanation is that there is yet another factor that PKA signaling must modify to allow the transcriptional activation of unliganded ERα. Future work will address these additional mechanistic questions, as well as the questions of whether CARM1 also mediates the activation of PR by cAMP, and whether the phosphorylation of CARM1 modulates the activity of other CARM1-dependent transcription factors.
A novel interaction groove in the ERα HBD
Our mutational analysis of the ERα HBD revealed that a novel regulatory surface is involved in mediating the response to cAMP. We speculate that a groove that is formed at the interface of the two subunits of the HBD homodimer might serve as the binding surface for CARM1. Determining the structure of the complex will be necessary to prove this hypothesis, and to determine whether this groove accommodates CARM1 through phosphorylated S448. Interestingly, this groove is located on the “back” side of the ERα HBD with respect to AF2, and has not received much attention. Other ERα coregulators might use this surface as well. ERα S464, which is located at the edge of the groove, seems to be a critical residue for this response. While this residue is highly conserved in ERα across vertebrates, it is an alanine in human ERβ. Considering that the ERα S464A mutant cannot be activated by cAMP/PKA, this sequence divergence between the two ER isoforms provides an explanation for why unliganded ERβ cannot be activated by cAMP/PKA. This does not exclude that cAMP/PKA could modulate certain ERβ target genes through alternate mechanisms. It was demonstrated that activation of ERβ by cAMP/PKA could be monitored in transfections with reporter genes containing promoter-proximal cryptic AP1 sites (Coleman et al. 2003), but it should be emphasized that all of our reporter genes were devoid of these. Our results suggest that cAMP signaling would help to discriminate, at least at certain target genes, between ERα and ERβ effects in cells that coexpress the two isoforms, and thereby to fine-tune estrogenic responses.
Building signal-specific coactivator complexes for signal-specific responses
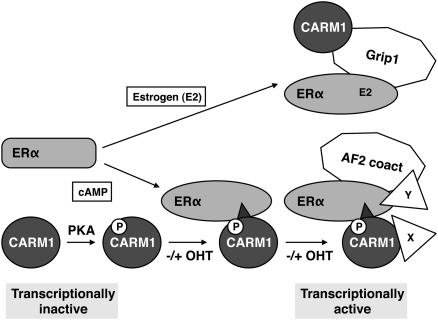
With CARM1 binding the ERα HBD on the “back” side, this potentially leaves the AF2 surface available for binding other coactivators. To allow this, one must assume that CARM1 binding somehow results in the release or reorganization of the Hsp90 complex and in the repositioning of helix 12 of the ERα HBD to form AF2. This is indeed supported by our finding that AF2 is required for cAMP/PKA activation of ERα. Whether CARM1 and AF2 coactivators are bound simultaneously—and, if so, which AF2 coactivator(s)—remains to be established, but it is intriguing to speculate that CARM1 might act as a pioneer factor (Fig. 8). Its main function and mode of action might be to allow and to promote the recruitment of yet other factors. In addition to those that might be recruited to the “CARM1 remodeled” ERα HBD, the C-terminal CARM1 transactivation domain (Teyssier et al. 2002) and interactors such as Flightless I (Lee et al. 2004) might contribute.
Figure 8.
Model of CARM1-mediated activation of ERα by cAMP signaling. For comparison, the E2-induced indirect Grip1-mediated recruitment of CARM1 is illustrated. Irrespective of the presence of OHT, PKA-phosphorylated CARM1 interacts with ERα and may function as a pioneer factor, allowing the subsequent binding of others. (AF2 coact) Transcriptional coactivators that are recruited to AF2 (Grip1 being one of them). The scheme also highlights that the two signals induce the assembly of different complexes, resulting in distinct transcriptional programs.
Interestingly, although prolonged treatment with cAMP leads to the proteasome-dependent degradation of GRIP1 (Hoang et al. 2004), early during induction, cAMP stimulates the recruitment of GRIP1 to the ERα target gene pS2 (Fenne et al. 2008). Although it was not determined whether this is necessary for cAMP activation of ERα, it is intriguing to speculate that, in this case, the recruitment of GRIP1 to ERα may be CARM1-dependent, rather than the other way around, as in the presence of E2 (Fig. 8).
Conversely, CARM1 may also be recruited in two different ways (Fig. 8). Following phosphorylation by PKA, it can bind unliganded ERα directly, whereas in the presence of estrogen, it can be incorporated into ERα complexes indirectly through a p160 coactivator such as GRIP1. Even if CARM1 is built into the complex in both cases, the functional requirements, topology, stoichiometry, and dynamics of these complexes are likely to be different. For example, the arginine methyltransferase activity of CARM1, and thus its ability to methylate histone H3 and other proteins, is necessary for CARM1 function in the context of E2-induced ERα–coactivator complexes (Lee et al. 2002), but it is dispensable for the cAMP response. Such differences may affect the selection of additional CARM1 and/or ERα partners through other protein surfaces.
Thus, depending on how ERα is activated, it will recruit different coregulator complexes. This prediction is supported by a recent study that examined an analogous situation in which the recruitment of several coregulators to ERα target genes was shown to be different depending on whether ERα was activated by E2 or IGF-1 (Cascio et al. 2007). As a consequence, one might expect that the activating signal of ERα determines target gene selection and/or regulation. We recently carried out a gene expression profiling analysis with MCF7 cells that shows just that: ERα regulates considerably different sets of genes depending on how it is activated (Dudek and Picard 2008).
Pathological and physiological significance
With the CARM1–ERα connection, we add another twist to the already established link between cAMP/PKA signaling and tamoxifen resistance of breast cancer cells (Michalides et al. 2004; Zwart et al. 2007). Our results obtained with the tamoxifen-resistant MCF7 variant LCC2 indicate that increased PKA activity also preserves ERα activity in the presence of OHT through the recruitment of CARM1. The finding that OHT abolishes the cAMP-induced interaction in wild-type MCF7 cells but not the constitutive interaction in tamoxifen-resistant LCC2 cells argues that high and, perhaps, persistent PKA activities may be required for tamoxifen resistance. The fact that the OHT stimulation of ERα activity could be recapitulated in ER-negative cells by overexpression of the CARM1 phosphoserine mimic mutant lends further support to these notions. Unfortunately, we could not directly test the CARM1 requirement for tamoxifen resistance in a proliferation assay, since a prolonged knockdown of CARM1 proved to be lethal (data not shown). Manipulating the CARM1–ERα interaction may yield interesting insights and turn out to be a promising therapeutic avenue in the future.
A large number of extracellular signals lead to the activation of adenylate cyclase, and the ensuing increase in cAMP and activation of PKA contribute to their cellular and physiological effects. Considering that cAMP can activate ERα, it is therefore conceivable that far more of these extracellular factors than previously thought also signal through ERα. We now have the tools to explore the pathological and physiological consequences of ligand-independent activation of ERα by cAMP.
Materials and methods
Plasmids
Details about the plasmids used can be found in the Supplemental Material.
Cell culture, transfection, RNAi, and luciferase assays
Where indicated, E2 (to 10–100 nM), the PKA inhibitor H89 (to 5 μM), and 8-Br-cAMP (to 1 mM from a 100 mM stock in H2O) were added. Firefly luciferase activities were standardized to a Renilla luciferase transfection control. Unless indicated otherwise, the data shown are averages of triplicate samples (error bars indicate standard deviations). Additional details are given in the Supplemental Material.
Antibodies and recombinant proteins
The following primary antibodies were used: the mouse monoclonals 2GV10 against the Gal4 DNA-binding domain (White et al. 1992), HA.11 against the HA tag (Babco), H90-10 against Hsp90 (a gift from David O. Toft), His-1 against the His6 tag (Sigma), anti-phospho-Ser/Thr (Upstate Biotechnologies), the rabbit monoclonal 100G7E against substrates phosphorylated by PKA (Cell Signaling), the rabbit polyclonal sera CARM1-421A against CARM1 (Bethyl Laboratories), and HC-20 against the ERα C terminus (Santa Cruz Biotechnologies). Recombinant proteins were expressed in Escherichia coli and purified on glutathione-Sepharose (Amersham) or TALON Metal Affinity Resin (Clontech) as directed by the manufacturers. After elution from the beads, His6-tagged proteins were dialyzed against 10 mM Tris-HCl (pH 7.02), 6.25 mM MgCl2, and 10% glycerol, and stored at −80°C.
IP experiments
The standard procedure for all IP experiments was as follows (except for those with the anti-HA monoclonal) (see the Supplemental Material): Cells were lysed by sonication in 20 mM Tris-HCl (pH 7.5), 1 mM EDTA, 10% glycerol, 20 mM Na-molybdate, and protease inhibitors. Five-hundred micrograms of each extract were first incubated with the primary antibody for 2 h at 4°C, and then with 20 μL of protein G-sepharose beads for an additional 16 h at 4°C. Beads were washed four times with a buffer containing 20 mM Tris-HCl (pH 7.5), 1 mM EDTA, 10% glycerol, 20 mM Na-molybdate, 50 mM NaCl, and 0.1% Tween.
ChIP experiments
The recruitment of CARM1 to the pS2 promoter upon stimulation of MCF7-SH cells with 100 nM E2, 1 mM 8-Br-cAMP, or vehicle for 45 min was determined by ChIP with the polyclonal CARM1 antiserum. Further details are given in the Supplemental Material.
Quantitative RT–PCR analysis of pS2 expression
MCF7 and LCC2 cells were induced for 4 h, and gene expression was analyzed by standard procedures as detailed in the Supplemental Material.
In vitro phosphorylation and interaction assays
Five-hundred nanograms of His6-tagged CARM1 were incubated with 70 ng of the catalytic β subunit of PKA (Stressgen) and 20 mM ATP in 20 μL of PKA buffer (10 mM Tris-HCl at pH 7.02, 6.25 mM MgCl2) for 1 h at 30°C. The reaction was then added to 10 μL of glutathione-sepharose beads loaded with GST-ERαHBD in 200 μL of pull-down buffer (20 mM HEPES-KOH at pH 7.9, 4.3 mM Na-phosphate, 10% glycerol, 100 mM NaCl, 100 mM KCl, 5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.2 mM PMSF) and incubated overnight at 4°C. The beads were washed four times with pull-down buffer.
Acknowledgments
S.C. wishes to dedicate this work to the memory of Thomas. We are very grateful to Olivier Donzé for his help early on in this project. We also thank Pierre Chambon, Daniel Metzger, Robert Clarke, Gilles Freiss, Richard H. Goodman, Steven R. Grossman, Jan-Åke Gustafsson, Natasha Kralli, David Livingston, Marc Montminy, Günther Schütz, Mike Stallcup, and David O. Toft for reagents, and Valérie Jean for her help with the quantitative PCR. This work was supported by the Canton de Genève, the Swiss National Science Foundation, and the Fondation Medic.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.568410.
Supplemental material is available at http://www.genesdev.org.
References
- Al-Dhaheri MH, Rowan BG. Protein kinase A exhibits selective modulation of estradiol-dependent transcription in breast cancer cells that is associated with decreased ligand binding, altered estrogen receptor α promoter interaction, and changes in receptor phosphorylation. Mol Endocrinol. 2007;21:439–456. doi: 10.1210/me.2006-0059. [DOI] [PubMed] [Google Scholar]
- Ali S, Coombes RC. Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer. 2002;2:101–112. doi: 10.1038/nrc721. [DOI] [PubMed] [Google Scholar]
- Apostolakis EM, Garai J, Lohmann JE, Clark JH, O'Malley BW. Epidermal growth factor activates reproductive behavior independent of ovarian steroids in female rodents. Mol Endocrinol. 2000;14:1086–1098. doi: 10.1210/mend.14.7.0490. [DOI] [PubMed] [Google Scholar]
- Aronica SM, Katzenellenbogen BS. Stimulation of estrogen receptor-mediated transcription and alteration in the phosphorylation state of the rat uterine estrogen receptor by estrogen, cyclic adenosine monophosphate, and insulin-like growth factor-I. Mol Endocrinol. 1993;7:743–752. doi: 10.1210/mend.7.6.7689695. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünner N, Frandsen TL, Holst-Hansen C, Bei M, Thompson EW, Wakeling AE, Lippman ME, Clarke R. MCF7/LCC2: A 4-hydroxytamoxifen resistant human breast cancer variant that retains sensitivity to the steroidal antiestrogen ICI 182,780. Cancer Res. 1993;53:3229–3232. [PubMed] [Google Scholar]
- Bunone G, Briand P-A, Miksicek RJ, Picard D. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
- Cascio S, Bartella V, Garofalo C, Russo A, Giordano A, Surmacz E. Insulin-like growth factor 1 differentially regulates estrogen receptor-dependent transcription at estrogen response element and AP-1 sites in breast cancer cells. J Biol Chem. 2007;282:3498–3506. doi: 10.1074/jbc.M606244200. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999a;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Chen D, Pace PE, Coombes RC, Ali S. Phosphorylation of human estrogen receptor α by protein kinase A regulates dimerization. Mol Cell Biol. 1999b;19:1002–1015. doi: 10.1128/mcb.19.2.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Huang SM, Stallcup MR. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- Coleman KM, Dutertre M, El-Gharbawy A, Rowan BG, Weigel NL, Smith CL. Mechanistic differences in the activation of estrogen receptor-α (ERα)- and ERβ-dependent gene expression by cAMP signaling pathway(s) J Biol Chem. 2003;278:12834–12845. doi: 10.1074/jbc.M212312200. [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhang M, Pestell R, Curran EM, Welshons WV, Fuqua SA. Phosphorylation of estrogen receptor α blocks its acetylation and regulates estrogen sensitivity. Cancer Res. 2004;64:9199–9208. doi: 10.1158/0008-5472.CAN-04-2126. [DOI] [PubMed] [Google Scholar]
- Curtis SW, Washburn T, Sewall C, DiAugustine R, Lindzey J, Couse JF, Korach KS. Physiological coupling of growth factor and steroid receptor signaling pathways: Estrogen receptor knockout mice lack estrogen-like response to epidermal growth factor. Proc Natl Acad Sci. 1996;93:12626–12630. doi: 10.1073/pnas.93.22.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denner LA, Weigel NL, Maxwell BL, Schrader WT, O'Malley BW. Regulation of progesterone receptor-mediated transcription by phosphorylation. Science. 1990;250:1740–1743. doi: 10.1126/science.2176746. [DOI] [PubMed] [Google Scholar]
- Dudek P, Picard D. Genomics of signaling crosstalk of estrogen receptor α in breast cancer cells. PLoS ONE. 2008;3:e1859. doi: 10.1371/journal.pone.0001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M, Smith CL. Ligand-independent interactions of p160/steroid receptor coactivators and CREB-binding protein (CBP) with estrogen-α: Regulation by phosphorylation sites in the A/B region depends on other receptor domains. Mol Endocrinol. 2003;17:1296–1314. doi: 10.1210/me.2001-0316. [DOI] [PubMed] [Google Scholar]
- Feng Q, He B, Jung SY, Song Y, Qin J, Tsai SY, Tsai MJ, O'Malley BW. Biochemical control of CARM1 enzymatic activity by phosphorylation. J Biol Chem. 2009;284:36167–36174. doi: 10.1074/jbc.M109.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenne IS, Hoang T, Hauglid M, Sagen JV, Lien EA, Mellgren G. Recruitment of coactivator glucocorticoid receptor interacting protein 1 to an estrogen receptor transcription complex is regulated by the 3′,5′-cyclic adenosine 5′-monophosphate-dependent protein kinase. Endocrinology. 2008;149:4336–4345. doi: 10.1210/en.2008-0037. [DOI] [PubMed] [Google Scholar]
- Frietze S, Lupien M, Silver PA, Brown M. CARM1 regulates estrogen-stimulated breast cancer growth through up-regulation of E2F1. Cancer Res. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]
- Fujimoto N, Katzenellenbogen BS. Alteration in the agonist/antagonist balance of antiestrogens by activation of protein kinase A signaling pathways in breast cancer cells: Antiestrogen selectivity and promoter dependence. Mol Endocrinol. 1994;8:296–304. doi: 10.1210/mend.8.3.7517003. [DOI] [PubMed] [Google Scholar]
- Gburcik V, Bot N, Maggiolini M, Picard D. SPBP is a phosphoserine-specific repressor of estrogen receptor α. Mol Cell Biol. 2005;25:3421–3430. doi: 10.1128/MCB.25.9.3421-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KA, Carroll JS. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer. 2007;7:713–722. doi: 10.1038/nrc2211. [DOI] [PubMed] [Google Scholar]
- Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc Natl Acad Sci. 2007;104:12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T, Fenne IS, Cook C, Borud B, Bakke M, Lien EA, Mellgren G. cAMP-dependent protein kinase regulates ubiquitin-proteasome-mediated degradation and subcellular localization of the nuclear receptor coactivator GRIP1. J Biol Chem. 2004;279:49120–49130. doi: 10.1074/jbc.M409746200. [DOI] [PubMed] [Google Scholar]
- Ignar-Trowbridge DM, Nelson KG, Bidwell MC, Curtis SW, Washburn TF, McLachlan JA, Korach KS. Coupling of dual signaling pathways: Epidermal growth factor action involves the estrogen receptor. Proc Natl Acad Sci. 1992;89:4658–4662. doi: 10.1073/pnas.89.10.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayne S, Rothgiesser KM, Hottiger MO. CARM1 but not its enzymatic activity is required for transcriptional coactivation of NF-κB-dependent gene expression. J Mol Biol. 2009;394:485–495. doi: 10.1016/j.jmb.2009.09.032. [DOI] [PubMed] [Google Scholar]
- Johnston SR, Head J, Pancholi S, Detre S, Martin LA, Smith IE, Dowsett M. Integration of signal transduction inhibitors with endocrine therapy: An approach to overcoming hormone resistance in breast cancer. Clin Cancer Res. 2003;9:524S–532S. [PubMed] [Google Scholar]
- Klotz DM, Hewitt SC, Ciana P, Raviscioni M, Lindzey JK, Foley J, Maggi A, DiAugustine RP, Korach KS. Requirement of estrogen receptor-α in insulin-like growth factor-1 (IGF-1)-induced uterine responses and in vivo evidence for IGF- 1/estrogen receptor cross-talk. J Biol Chem. 2002;277:8531–8537. doi: 10.1074/jbc.M109592200. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kitamoto T, Masuhiro Y, Watanabe M, Kase T, Metzger D, Yanagisawa J, Kato S. p300 mediates functional synergism between AF-1 and AF-2 of estrogen receptor α and β by interacting directly with the N-terminal A /B domains. J Biol Chem. 2000;275:15645–15651. doi: 10.1074/jbc.M000042200. [DOI] [PubMed] [Google Scholar]
- Lamb J, Ladha MH, McMahon C, Sutherland RL, Ewen ME. Regulation of the functional interaction between cyclin D1 and the estrogen receptor. Mol Cell Biol. 2000;20:8667–8675. doi: 10.1128/mcb.20.23.8667-8675.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Stallcup MR. Protein arginine methylation of nonhistone proteins in transcriptional regulation. Mol Endocrinol. 2009;23:425–433. doi: 10.1210/me.2008-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Jiang F, Wang Q, Nicosia SV, Yang J, Su B, Bai W. MEKK1 activation of human estrogen receptor α and stimulation of the agonistic activity of 4-hydroxytamoxifen in endometrial and ovarian cancer cells. Mol Endocrinol. 2000;14:1882–1896. doi: 10.1210/mend.14.11.0554. [DOI] [PubMed] [Google Scholar]
- Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Synergy among nuclear receptor coactivators: Selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol. 2002;22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Campbell HD, Stallcup MR. Developmentally essential protein Flightless I is a nuclear receptor coactivator with actin binding activity. Mol Cell Biol. 2004;24:2103–2117. doi: 10.1128/MCB.24.5.2103-2117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard DM, O'Malley BW. Nuclear receptor coregulators: Judges, juries, and executioners of cellular regulation. Mol Cell. 2007;27:691–700. doi: 10.1016/j.molcel.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Masuhiro Y, Mezaki Y, Sakari M, Takeyama K, Yoshida T, Inoue K, Yanagisawa J, Hanazawa S, O'Malley BW, Kato S. Splicing potentiation by growth factor signals via estrogen receptor phosphorylation. Proc Natl Acad Sci. 2005;102:8126–8131. doi: 10.1073/pnas.0503197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier R, Penot G, Carmouche RP, Hubner MR, Reid G, Denger S, Manu D, Brand H, Kos M, Benes V, et al. Transcriptional complexes engaged by apo-estrogen receptor-α isoforms have divergent outcomes. EMBO J. 2004;23:3653–3666. doi: 10.1038/sj.emboj.7600377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalides R, Griekspoor A, Balkenende A, Verwoerd D, Janssen L, Jalink K, Floore A, Velds A, van't Veer L, Neefjes J. Tamoxifen resistance by a conformational arrest of the estrogen receptor α after PKA activation in breast cancer. Cancer Cell. 2004;5:597–605. doi: 10.1016/j.ccr.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Neuman E, Ladha MH, Lin N, Upton TM, Miller SJ, DiRenzo J, Pestell RG, Hinds PW, Dowdy SF, Brown M, et al. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen KM, Auger AP. Dopaminergic activation of estrogen receptors induces fos expression within restricted regions of the neonatal female rat brain. PLoS One. 2008;3:e2177. doi: 10.1371/journal.pone.0002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen KM, Jessen HM, Auger CJ, Auger AP. Dopaminergic activation of estrogen receptors in neonatal brain alters progestin receptor expression and juvenile social play behavior. Endocrinology. 2005;146:3705–3712. doi: 10.1210/en.2005-0498. [DOI] [PubMed] [Google Scholar]
- Picard D. SCOPE/IUPAC project on environmental implications of endocrine active substances: Molecular mechanisms of cross-talk between growth factors and nuclear receptor signaling. Pure Appl Chem. 2003;75:1743–1756. [Google Scholar]
- Picard D. Chaperoning steroid hormone action. Trends Endocrinol Metab. 2006;17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Power RF, Mani SK, Codina J, Conneely OM, O'Malley BW. Dopaminergic and ligand-independent activation of steroid hormone receptors. Science. 1991;254:1636–1639. doi: 10.1126/science.1749936. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Rowan BG, Garrison N, Weigel NL, O'Malley BW. 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol Cell Biol. 2000;20:8720–8730. doi: 10.1128/mcb.20.23.8720-8730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkevicius KW, Burdette JE, Woloszyn K, Hewitt SC, Sugg L, Temple KA, Wondisford FE, Korach KS, Woodruff TK, Greene GL. An estrogen receptor-α knock-in mutation provides evidence of ligand-independent signaling and allows modulation of ligand-induced pathways in vivo. Endocrinology. 2008;149:2970–2979. doi: 10.1210/en.2007-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Conneely OM, O'Malley BW. Modulation of the ligand-independent activation of the human estrogen receptor by hormone and antihormone. Proc Natl Acad Sci. 1993;90:6120–6124. doi: 10.1073/pnas.90.13.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teyssier C, Chen D, Stallcup MR. Requirement for multiple domains of the protein arginine methyltransferase CARM1 in its transcriptional coactivator function. J Biol Chem. 2002;277:46066–46072. doi: 10.1074/jbc.M207623200. [DOI] [PubMed] [Google Scholar]
- Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. The cloned human estrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 1989;8:1981–1986. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO J. 2007;26:4391–4401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RA, Mazumdar A, Vadlamudi RK, Kumar R. P21-activated kinase-1 phosphorylates and transactivates estrogen receptor-α and promotes hyperplasia in mammary epithelium. EMBO J. 2002;21:5437–5447. doi: 10.1093/emboj/cdf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992;11:2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Smith CL, O'Malley BW. Transcriptional regulation by steroid receptor coactivator phosphorylation. Endocr Rev. 2005;26:393–399. doi: 10.1210/er.2004-0018. [DOI] [PubMed] [Google Scholar]
- Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J. 2007;26:4402–4412. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart W, Griekspoor A, Berno V, Lakeman K, Jalink K, Mancini M, Neefjes J, Michalides R. PKA-induced resistance to tamoxifen is associated with an altered orientation of ERα towards co-activator SRC-1. EMBO J. 2007;26:3534–3544. doi: 10.1038/sj.emboj.7601791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- Zwijsen RM, Buckle RS, Hijmans EM, Loomans CJ, Bernards R. Ligand-independent recruitment of steroid receptor coactivators to estrogen receptor by cyclin D1. Genes & Dev. 1998;12:3488–3498. doi: 10.1101/gad.12.22.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]