Abstract
Timely acquisition of cell fates and the elaborate control of growth in numerous organs depend on Notch signaling. Upon ligand binding, the core transcription factor RBP-J activates transcription of Notch target genes. In the absence of signaling, RBP-J switches off target gene expression, assuring the tight spatiotemporal control of the response by a mechanism incompletely understood. Here we show that the histone demethylase KDM5A is an integral, conserved component of Notch/RBP-J gene silencing. Methylation of histone H3 Lys 4 is dynamically erased and re-established at RBP-J sites upon inhibition and reactivation of Notch signaling. KDM5A interacts physically with RBP-J; this interaction is conserved in Drosophila and is crucial for Notch-induced growth and tumorigenesis responses.
Keywords: Histone demethylase, RBP-J, Notch target gene repression, T cells, Drosophila, tumorigenesis
Despite the simple signal transduction cascade of the Notch pathway, this pathway can precisely control multiple binary cell fate decisions, cell proliferation and differentiation, and stem cell maintenance during embryogenesis and postnatal development (Bray 2006; Ilagan and Kopan 2007; Koch and Radtke 2007; Kopan and Ilagan 2009). Crucial to the dynamics of Notch signaling responses is the dual activity of its nuclear effector RBP-J {short for RBP-Jκ, also known as CSL (CBF1, Su[H], Lag-1)} (Borggrefe and Oswald 2009). RBP-J activates the expression of target genes in cells receiving the Notch signal and represses target expression in the nonreceiving cells. This dual role of RBP-J allows a fine spatial and temporal control of Notch-regulated transcription (Kopan and Ilagan 2009), but the mechanism is not fully understood.
Control of gene activation and repression programmed by Notch signaling involves histone acetylation and deacetylation (Kao et al. 1998; Hsieh et al. 1999; Oswald et al. 2002, 2005; Wallberg et al. 2002). Recently, a role of histone methylation in the regulation of Notch target genes has been suggested in Drosophila based on genetic interactions of histone methyltransferases and Notch in tumorigenesis (Ferres-Marco et al. 2006; Krejci and Bray 2007). Particularly, an enrichment of trimethylated histone H3 Lys 4 (H3K4me3), an active chromatin mark (Santos-Rosa et al. 2002; Schneider et al. 2004), was observed at target genes in response to Notch activation (Ferres-Marco et al. 2006; Krejci and Bray 2007) and observations in tissue culture. The regulation of H3K4me3 levels at Notch target genes by Drosophila histone chaperones ASF1, NAP1, and the histone demethylase Lid (little imaginal disc) has been reported very recently (Moshkin et al. 2009). This study, like previous studies, highlights a key unresolved issue of how histone methylation/demethylation enzymes are recruited at specific genes.
Genome-wide studies using ChIP sequencing (ChIP-Seq) have revealed that the H3K4me3 mark can be found at regulatory elements as well as transcriptional start sites (TSSs) (Barski et al. 2007; Mikkelsen et al. 2007), hinting at a closer than previously suspected role for histone lysine methylation/demethylation in the regulation of pathway-specific target genes. Here we report that the histone H3K4me3 demethylase KDM5A (Christensen et al. 2007; Eissenberg et al. 2007; Klose et al. 2007; Secombe et al. 2007) is an integral part of the Notch/RBP-J gene repression mechanism. KDM5A (also known as JARID1A) was originally discovered as the retinoblastoma-binding protein RBP2 (Fattaey et al. 1993), and was implicated in regulation of retinoblastoma target genes (Benevolenskaya et al. 2005). KDM5A was found subsequently to interact physically and functionally with the Polycomb complex (Pasini et al. 2008) and the Sin3 corepressor complex (van Oevelen et al. 2008). In this study, we show that the histone demethylase KDM5A associated with RBP-J protein and is essential for Notch/RBP-J target gene silencing in vitro. This association is conserved in Drosophila, and is crucial for Notch-mediated patterning, growth, and tumorigenesis responses in vivo.
Results
Dynamic histone H3K4 methylation at endogenous Notch target promoters
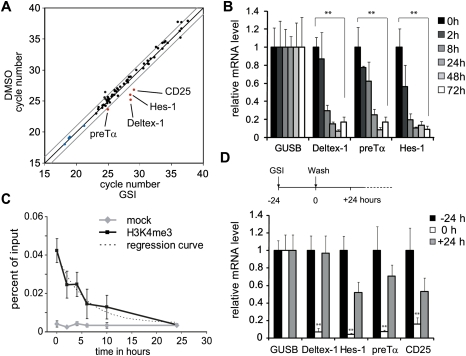
To study the dynamics of Notch target gene regulation, we used a spontaneous T-cell lymphoma cell line derived from TCRβ-deficient mice called Beko (T-cell receptor β knockout) as our experimental model system. Comparing untreated Beko T cells with Beko cells treated with the γ-secretase inhibitor (GSI) DAPT {N-[N-(3,5-difluorophenylacetyl-L-alanyl)]-S-phenylglycine t-butyl ester} for 24 h, several Notch target genes were down-regulated rapidly, including Deltex-1, pre-Tα, Hes-1, Hey-1, and CD25 (Fig. 1A). Similar results were obtained after introduction of dominant-negative Mastermind-like 1 fused to estrogen receptor (ER) ligand-binding domain into Beko T cells. Upon tamoxifen treatment, the same Notch target genes were down-regulated (Supplemental Table 1). In time-course experiments, the GSI effect was evident at several Notch target genes already after 8 h (Fig. 1B), and could be reversed after removal of the inhibitor (Fig. 1D). To look at changes of histone modifications, chromatin immunoprecipitation (ChIP) experiments were performed before and at different time points after GSI treatment. The active mark H3K4me3 disappeared upon GSI treatment at the RBP-J-binding site of the Deltex-1 (Fig. 1C), Hes-1, and CD25 enhancer, as well as at the promoter of preTα (Supplemental Fig. S3).
Figure 1.
Transcriptional regulation of Notch target genes after GSI treatment. (A) cDNA was synthesized from Beko cells before and 24 h after treatment with GSI (DAPT). A miniarray of Notch-relevant genes shows that only very few genes (Deltex-1, Hes-1, preTα, and CD25) are down-regulated upon GSI treatment (red, in blue housekeeping genes). The gray lines indicate a twofold change. (B) Expression of Deltex-1, Hes-1, and preTα is down-regulated rapidly after GSI treatment, as measured by RT–PCR comparing GSI versus control (DMSO-treated) cells (mean ± SD, n = 3; [**] P < 0.01, Student's t-test). (C) In ChIP experiments, H3K4 trimethylation at the conserved RBP-J-binding site (−1.2-kb upstream of the TSS) (see Fig. 2) at the Deltex-1 promoter disappears after GSI treatment with a half-life of 4 h. In the mock control, only the beads were used. Values are mean ± SD for triplicate samples from a representative experiment. (D) The transcriptional down-regulation by GSI treatment is reversible: Washing away GSI after 24 h leads to a rapid up-regulation of Deltex-1, Hes-1, preTα, and CD25. (Mean ± SD, n = 3, [**] P < 0.01, Student's t-test.)
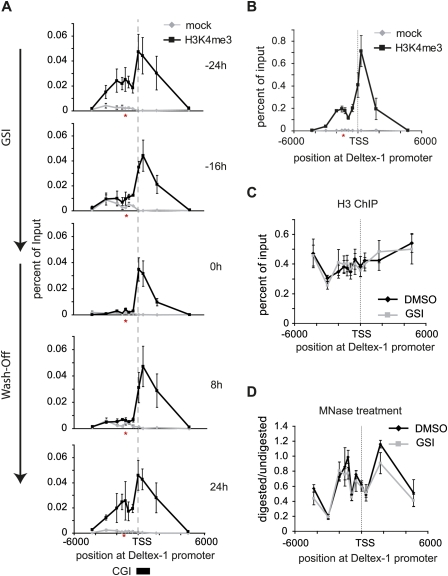
To look at the dynamic nature of H3K4me3, we scanned the Deltex-1 promoter region by ChIP. Two major peaks were observed: one at the TSS, and one at the RBP-J-binding site, 1.2 kb upstream of the TSS (Fig. 2A, −24 h). The peak at the TSS correlates with a CpG island (CGI) (Fig. 2A, bottom), which is consistent with previous genome-wide findings that H3K4me3 is found frequently at CGIs (Barski et al. 2007; Heintzman et al. 2007; Mikkelsen et al. 2007). Time-course ChIP experiments after GSI treatment showed a drop of H3K4me3 at the RBP-J-binding site, but, surprisingly, no changes occurred at the TSS (Fig. 2A, −16 and 0 h). After removal of GSI, the peak of H3K4me3 at the RBP-J-binding site reappeared (Fig. 2A, 8 h and 24 h). The same reduction of H3K4me3 could be observed after induction of a tamoxifen-regulated form of dominant-negative Mastermind (dn-MAML-ER) (Supplemental Fig. S1A,B). To further test the specificity of the GSI treatment, a rescue experiment was performed by introducing Notch-IC fused to ER (Notch-IC-ER, see Supplemental Fig. S2A–D). In the absence of nuclear Notch-IC (no tamoxifen treatment) and presence of GSI, H3K4me3 was erased (Supplemental Fig. S2C, time point 24 h). Upon tamoxifen treatment, H3K4me3 levels were restored also in the presence of GSI (Supplemental Fig. S2C, time point 48 h +Tam). Furthermore, the two H3K4me3 peaks at the Deltex-1 promoter are also found in primary T-cell progenitors from RAG1 (recombination-activating gene 1) knockout mice (Fig. 2B). Due to an arrest in T-cell development, RAG1-deficient mice have more T-cell precursors that express Notch target genes. H3K4 trimethylation was also erased at Notch target genes Hes-1 and CD25 at the RBP-J-binding sites, and for preTα in part at the promoter (Supplemental Fig. S3). In primary pre-T cells, the same pattern of H3K4me3 is observed for Hes-1 and preTα, but not CD25 (Supplemental Fig. S4). Taken together, dynamic histone H3K4me3 methylation occurs at Notch target genes—specifically at RBP-J-binding sites—and is dependent on Notch activity.
Figure 2.
H3K4 trimethylation at Notch target gene Deltex-1 is dynamic only at the RBP-J-binding site. (A) ChIP experiments were performed using anti-H3K4me3 antibodies and the pre-T-cell line Beko treated as in Figure 1D. When Notch signaling is switched off by treatment with GSI, H3K4 trimethylation is lost specifically at the conserved RBP-J-binding site (−1.2-kb upstream, red asterisk) but not at the TSS. The sequence of the RBP-J-binding site information can be found in Supplemental Figure S10. H3K4 trimethylation is re-established at the RBP-J-binding site upon removal of GSI (24 h). Shown are the values as mean ± SD for duplicate samples from a representative experiment; the experiment was repeated three times. H3K4me3 is significantly enriched with P < 0.05 (Student's t-test) at the RBP-J-binding site (−24 h, 24 h) and at the TSS. (B) Primary CD4−/CD8− precursor T cells from RAG1 knockout mice have a similar H3K4me3 profile at the Deltex-1 promoter compared with Beko T cells (shown in A); H3K4 trimethylation is observed at the RBP-J-binding (−1.2-kb upstream) site and at the TSS. H3K4me3 is significantly enriched with P < 0.05 (Student's t-test) at the RBP-J-binding site and at the TSS. (C) The nucleosome occupancy does not change before or after GSI treatment, as measured by ChIP with antibodies directed against the core of Histone H3. (D) Nucleosomal structure, indicated by the ratio MNase-digested versus undigested DNA, remains unaltered in presence of GSI. For B–D, the data points are mean ± SD for triplicate samples from representative experiments.
ChIP experiments with an antibody directed against the core of histone H3 showed no change in nucleosome occupancy when comparing Beko untreated cells with Beko cells treated for 24 h with GSI (Fig. 2C). Thus, inhibition of Notch by the GSI does not generally lead to removal of nucleosomes at the RBP-J-binding site at the Deltex-1 promoter, but specifically leads to removal of the methylated active chromatin mark. Furthermore, we confirmed this result by mapping nucleosome occupancy comparing micrococcal nuclease (MNase) digested/nondigested, followed by genomic PCR, at different sites of the Deltex-1 promoter. Again, no changes in nucleosome occupancy before and after GSI treatment were detected (Fig. 2D).
Histone demethylase KDM5A forms a complex with RBP-J
From the time-course experiments (Fig. 1C), we hypothesized that H3K4 demethylation is an active process. Thus, we searched for a putative H3K4me3 demethylase involved in the transcriptional regulation of RBP-J/Notch target genes.
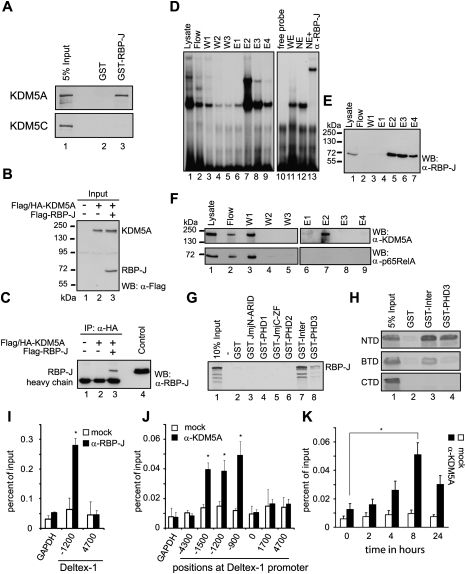
Organisms like Caenorhabditis elegans and Drosophila have only one described H3K4me3 demethylase. Functional inactivation of the C. elegans KDM5A homolog Rbr-2 results in undeveloped vulvas or a multivulval phenotype (Christensen et al. 2007). Mutations in the Drosophila KDM5A homolog lid results in severe defects in cell growth and differentiation, and are homozygous lethal (Gildea et al. 2000). In mice, there are four described H3K4me3 demethylases, named KDM5A–D. In our pre-T-cell line, Beko RT–PCR experiments showed that only KDM5A and KDM5C are expressed (data not shown). We reasoned that if KDM5A or KDM5C are required for RBP-J-dependent target gene regulation, the demethylase(s) might interact physically with RBP-J. Indeed, in GST pull-down experiments, KDM5A but not KDM5C bound strongly to GST-RBP-J, but was not retained by GST only (Fig. 3A).
Figure 3.
RBP-J and histone demethylase KDM5A physically interact in vitro and in vivo and colocalize at promoters. (A) RBP-J and KDM5A interact in GST pull-down experiments: Cell-free synthesized 35S-labeled KDM5A binds to GST-RBP-J immobilized to glutathione-Sepharose beads, but not with GST only. Cell-free synthesized 35S-labeled KDM5C binds to neither GST-RBP-J nor GST only. (B,C) RBP-J and KDM5A interact in coimmunoprecipitation experiments: HEK293 cells were cotransfected with expression plasmids for Flag-RBP-J, together with Flag/HA-tagged KDM5A; see expression and input controls in B. (B) The expression was detected by Western blotting (WB) using antibodies against the Flag tag. (C) RBP-J was coimmunoprecipitated with only KDM5A from lysates of cells cotransfected together with HA-KDM5A. Coimmunoprecipitated RBP-J was detected using an α-RBP-J antibody. (D–F) KDM5A is part of an endogenous RBP-J complex in Beko T cells: Purification of endogenous RBP-J complexes from Beko cells by DNA affinity chromatography. For purification, a DNA fragment containing 12 RBP-J-binding sites was biotinylated and immobilized on a streptavidin-Sepharose column. The column was incubated with whole-cell lysates from Beko cells. After incubation and washing, the DNA-bound RBP-J complexes were eluted with increasing concentrations of NaCl and were analyzed by EMSA (D). Western blot analysis (WB) of the lysate, the first wash step, and the eluted fractions (E1–E4) were performed using antibodies against RBP-J (E) and KDM5A (F, top panel) and control NF-κB (F, bottom panel). KDM5A, but not p65, is detected in the elution step E2 together with RBP-J. (G) Mapping of the KDM5A-interacting domain: Cell-free synthesized 35S-labeled RBP-J was incubated with different bacterially expressed GST fusion proteins of KDM5A (JmjN/ARID, PHD1, JmjC/Zn finger, PHD2, Inter, or PHD3) (see also Supplemental Fig. S6A,B). (Lanes 7,8) The Inter region and the PHD3 domain containing the C-terminal part of KDM5A interact with RBP-J. (H) Mapping of the RBP-J-interacting domain: Different cell-free synthesized 35S-labeled RBP-J constructs (RBP-J-NTD, RBP-J-β-treefoil [BTD], and RBP-J C-terminal [CTD]) (see Supplemental Fig. S6C) were incubated with bacterially expressed and immobilized GST-Inter or GST-PHD3 fusion proteins. The NTD and BTD of RBP-J interacts with the Inter-PHD2/3 of KDM5A, whereas the PHD3 of KDM5A binds to only the NTD of RBP-J. (I) ChIP analysis using an α-RBP-J antibody (black bars) or mock (IgG control, white bars) at the Deltex-1 promoter of control GAPDH promoter. As described previously (Kathrein et al. 2008), RBP-J is found at −1.2 kb. Data are mean ± SD of two independent experiments measured in triplicate ([*] P < 0.05, Student's t-test). (J) ChIP analysis with an α-KDM5A antibody (black bars) or mock (no antibody, white bars). KDM5A was found at the RBP-J-binding site (1.2 kb), but also at −1.5 kb and at −900 bp. Analysis was performed 8 h after GSI treatment (see also K). No occupancy was found at the GAPDH promoter. Values are mean ± SD for triplicate samples from a representative experiment ([*] P < 0.05, Student's t-test). (K) ChIP time-course experiment with α-KDM5A-specific antibody at 0, 2, 4, 6, 8, and 24 h after GSI treatment. KDM5A occupancy at the RBP-J-binding site increases upon GSI treatment and reaches its maximum after 8 h. Values are mean ± SD for triplicate samples from a representative experiment and were normalized to the levels of KDM5A at Notch-independent KDM5A target TOM22, measured with the same samples ([*] P < 0.05, Student's t-test) (see Fig. 4G).
The KDM5A/RBP-J interaction was examined further in vivo using coimmunoprecipitation experiments. Flag-tagged RBP-J was expressed either alone or together with Flag/HA-tagged KDM5A in HEK-293 cells (Fig. 3B). After the immunoprecipitation of KDM5A from cell lysates using a monoclonal α-HA antibody, RBP-J was detected with Western blotting using an α-RBP-J antibody. RBP-J was detected in coimmunoprecipitation experiments only when both proteins were expressed (Fig. 3C, lane 3).
In addition, we isolated endogenous KDM5A/RBP-J-containing complexes by DNA affinity chromatography (Fig. 3D–F). A DNA fragment containing 12 RBP-J-binding sites was biotinylated and immobilized with streptavidin Sepharose (Oswald et al. 2005). The column was incubated with cellular extracts from Beko T cells. After washing, the endogenous RBP-J complexes were eluted with increasing amounts of NaCl concentration as indicated in the Materials and Methods. The presence of RBP-J was verified by electromobility gel shift assays (EMSA) (Fig. 3D) and Western blotting experiments (Fig. 3E). Specific binding of RBP-J to the probe was assessed using an α-RBP-J antibody showing a supershift (Fig. 3D, lane 13). KDM5A, but not p65/RelA (NF-κB), eluted in the same fractions as RBP-J (Fig. 3F, top panel, lane 7). In parallel, we performed the identical DNA affinity purification approach with a DNA fragment lacking RBP-J-binding sites. As expected, neither RBP-J nor KDM5A nor p65 bound to the column (Supplemental Fig. S5A–C). Together, the data suggest that endogenous KDM5A is associated with the endogenous DNA-bound RBP-J corepressor complex in vivo. We further mapped the domain of KDM5A interacting with RBP-J by performing GST pull-down experiments with GST fusion constructs encoding the conserved domains of KDM5A (Fig. 3G; Supplemental Fig. S6). Only the fragments of KDM5A containing the region between PHD2 and PHD3 (“Inter”) as well as the C-terminal part, containing the PHD3, could interact with RBP-J (Fig. 3G). In addition, we identified the RBP-J N-terminal domain (NTD) as the main interacting domain (Fig. 3H). Thus, collectively, these data suggest that KDM5A is a novel component of the RBP-J corepressor complex. Importantly, the RBP-J/KDM5A interaction is evolutionarily conserved from mice to Drosophila (see Fig. 5A–C, below).
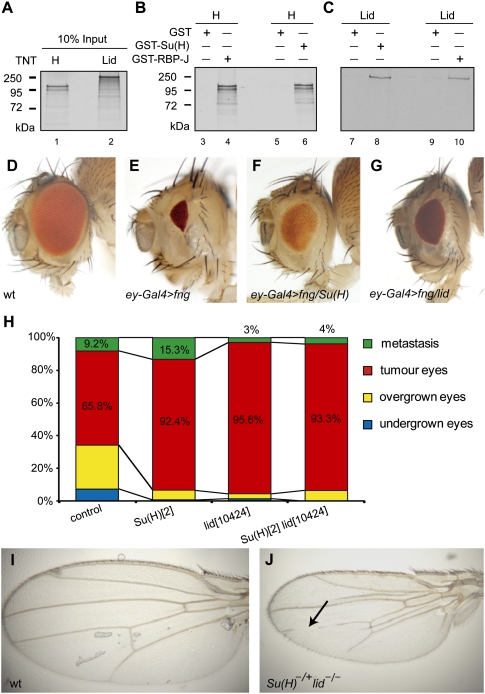
Figure 5.
In vivo interactions of Lid and Su(H) in growth control and tumorigenesis in Drosophila. (A–C) Drosophila RBP-J, called Su(H), and Drosophila KDM5A, called Lid, interact in GST pull-down experiments. (A) Positive control Hairless and Lid were synthesized and 35S-labeled. Hairless (H in B) and Lid (C) bind to GST-Su(H) (lanes 6,8) and to GST-RBP-J (lanes 4,10) immobilized to glutathione-Sepharose beads. (Lanes 3,5,7,9) Lid or Hairless do not interact with GST alone. (D–G) The size of female adult eyes from identically reared animals were scored. (D) Wild type (wt). (E) Overexpression of fringe by ey-Gal4 (abbreviated ey-Gal4 > fng), a Notch pathway modulator, results in a small eye phenotype (∼12% of wild-type eye size). (F,G) Reducing one gene copy of Su(H) (Su[H]del47/+) (F) or lid (lid10424/+) (G) consistently rescued the growth defect caused by reducing Notch pathway activation (60%–70% of wild-type eye size; penetrance 100%). (H) Quantification of adult fly eye tumors and metastasis associated with Notch ligand Delta gain of function and the epigenetic repressors Pipsqueak and Lola (eyeful) control cross and the phenotype in the presence of a mutant copy of lid (+ lid10424/+ +), Su(H), and lid (Su[H]2 lid10424/+ +) or Su(H) mutation alone (Su[H]2 +/+ +). Bars shown represent the mean of total (n > 100) flies scored. Crosses were repeated twice. The overexpression of Delta and pipsqueak and lola genes (eyeful) in the developing eye results in eye overgrowth (25.10% of animals, with >130% increased wild-type eye size) and massive eye tumor overgrowth (65.8% of animals), and metastases in distant tissues within the thorax (9.2% of mutant flies, n > 100). Reducing lid or Su(H) increased the frequency of animals with eye tumor growth and metastases. Reducing lid using a different mutation and RNAi lead to similar enhancement of tumorigenesis (see Supplemental Fig. S9E). (I) Wild-type (wt)wing. (J) Su(H)2 lid10424/+lid10424) wing. Arrow points to vein loss in the male wing when a mutant copy of Su(H) is introduced in a lid mutant background.
RBP-J and KDM5A are found at the Deltex-1 promoter in vivo
We next performed ChIP before and after GSI treatment to identify regions occupied by KDM5A in the Notch target gene Deltex-1. Using α-RBP-J antibodies, we found that RBP-J was bound at −1200 base pairs (bp) upstream of the Deltex-1 TSS (Fig. 3I). RBP-J occupancy at the −1200-bp site was observed before and after GSI treatment, but was absent upon RBP-J knockdown (data not shown). Similarly, using α-KDM5A antibodies, we detected endogenous KDM5A occupancy at the RBP-J-binding site (−1200 bp) of the Deltex-1 promoter (Fig. 3J). Importantly, after GSI treatment, there was an increase in KDM5A occupancy at the RBP-J-binding site (Fig. 3K). The higher enrichment of KDM5A at the Deltex-1 locus upon Notch inactivation supports our hypothesis that RBP-J/KDM5A is directly involved in the gene repression of Notch target genes. In addition, ChIP experiments with the histone acetyltransferase p300, which is part of the RBP-J/Notch/MAML coactivator complex (Oswald et al. 2001), were performed (Supplemental Fig. S7). p300 is localized at the RBP-J-binding site of the Deltex-1 promoter before GSI treatment (Supplemental Fig. S7A), but is removed rapidly upon GSI treatment (Supplemental Fig. S7B). In line with these results, we could observe a decrease of H3K9 acetylation at the RBP-J-binding site at Deltex-1 (Supplemental Fig. S7C).
KDM5A regulates gene expression at Notch target genes
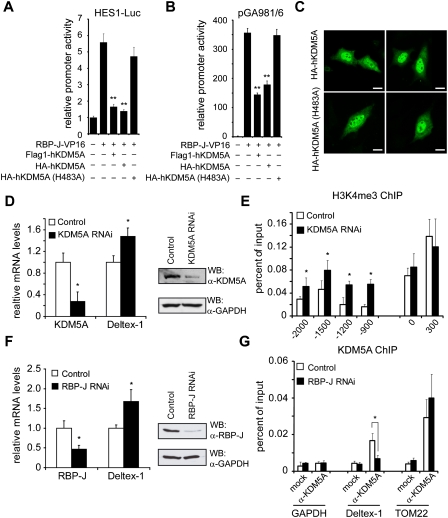
Subsequently, we wanted to investigate the role of KDM5A in repression of Notch target genes using functional assays. Therefore, we used a Hes-1-specific reporter construct in transient transfection assays. Promoter activity stimulated by RBP-J, which was fused to the viral activator domain VP16 (RBP-J-VP16), was reduced more than fourfold by either Flag-tagged or HA-tagged KDM5A (wild type), but not by a demethylase catalytic-dead KDM5A(H483A) mutant (Fig. 4A). In ChIP experiments, we could show that the H3K4 trimethylation is found at the promoter only in the presence of RBP-J-VP16, whereas the mark is absent by simultaneous expression of KDM5A (Supplemental Fig. S8). Similar results were obtained from luciferase assays using the artificial promoter construct pGA981/6 containing several RBP-J-binding sites (Fig. 4B; Oswald et al. 2005; Salat et al. 2008). Correct nuclear localization of both HA-tagged KDM5A (wild type) and KDM5A (H483A) mutant proteins was confirmed by immunofluorescence (Fig. 4C). Thus, the demethylase activity of KDM5A is required to repress the RBP-J-VP16 transcriptional activity.
Figure 4.
KDM5A controls expression of Notch target genes. In transient transfection assays, KDM5A acts as a corepressor of RBP-J. Portions (2 μg) of human Hes-1-specific reporter construct (HES-1-Luc) (A) and reporter construct pGA981/6 (B) were transfected alone or cotransfected with 100 ng of plasmids expressing RBP-J-VP16 together with Flag-KDM5A, HA-KDM5A, or catalytic-inactive HA-KDM5A(H483A). Wild-type but not catalytic-inactive KDM5A was able to repress transcription activity. (Y-axis) Relative promoter activity. Data are mean ± SD of three independent experiments ([**] P < 0.01). (C) Nuclear localization of both KDM5A (wt) (top panel) and the catalytic-dead KDM5A (H483A) (bottom panel) after transient transfection into HeLa cells. HA-tagged KDM5A proteins were analyzed by confocal microscopy 24 h after transfection. Bar, 10 μm. (D) Knockdown of KDM5A leads to an increase in expression of Deltex-1 (mean ± SD, n = 5; [*] P < 0.05, Student's t-test). (E) In KDM5A knockdown cells, H3K4 trimethylation is increased at the RBP-J-binding site (−1.2 kb) of the Deltex-1 promoter, but not at the TSS. The values are mean ± SD for triplicate samples from a representative experiment ([*] P < 0.05, Student's t-test). (F) Knockdown of RBP-J leads to an up-regulation of Deltex-1 expression (mean ± SD, n = 3; [*] P < 0.05, Student's t-test). (G) In ChIP experiments using α-KDM5A antibodies, the recruitment of KDM5A to the Deltex-1 promoter is abrogated in RBP-J knockdown cells. At a Notch-independent gene, mitochondrial TOM22, KDM5A localization remains constant in the presence or absence of RBP-J. The values are mean ± SD for triplicate samples from a representative experiment ([*] P < 0.05, Student's t-test).
In vivo data using KDM5A knockdown cells also support its function in gene repression. A threefold decrease of KDM5A at the mRNA and protein level resulted in a slight increase of Deltex-1 mRNA expression (Fig. 4D). Importantly, H3K4me3 was increased specifically at the RBP-J-binding site, but remained unchanged at the TSS (Fig. 4E).
To confirm that the regulatory interaction of KDM5A at the Deltex-1 promoter depends on RBP-J, RBP-J was knocked down, leading to an increase of Deltex-1 expression (Fig. 4F) similar to the effect of KDM5A knockdown (Fig. 4D). Importantly, KDM5A occupancy at Deltex-1 (position −1200 bp) was reduced down to background levels after RBP-J knockdown, whereas occupancy at TOM22, an RBP-J/Notch-independent KDM5A target gene (Lopez-Bigas et al. 2008), remained constant (Fig. 4G). These data suggest a mechanism by which RBP-J bound to promoters of Notch target genes recruits KDM5A to facilitate histone demethylation.
Lid, the Drosophila homolog of KDM5A, and Suppressor of Hairless [Su(H)]/RBP-J physically interact and limit Notch-induced growth and tumorigenesis in vivo
Consistent with the interaction between RBP-J and KDM5A in mammalian cells, we found that Su(H) and Lid, the Drosophila homologs of RBP-J and KDM5A, respectively, interact physically (Fig. 5A–C) and provoke similar responses on several Notch loss-of-function and gain-of-function contexts in vivo (Fig. 5D–J). Su(H) has been shown to interact physically and genetically with corepressor Hairless (H) (for review, see Bray 2006; Maier 2006). In GST pull-down experiments, GST-Su(H) interacted strongly not only with Hairless (Fig. 5B), but also with Lid (Fig. 5C). Drosophila Lid also interacted with GST-RBP-J (Fig. 5C, lane 10). The same holds true for Drosophila GST-Su(H) interaction with mouse KDM5A (data not shown). Furthermore, we could map the Lid/Su(H) to the C-terminal part of Lid (data not shown), analogous to the interaction of KDM5A/RBP-J (Fig. 3D). This suggests that the interaction between KDM5A and RBP-J is evolutionarily conserved, and that Drosophila Su(H)/Lid might also interact in vivo. To dissect the biological relevance of the Su(H)/Lid interaction, we studied their mutants in genetic assays that are sensitive to the dose of Notch pathway genes. For this purpose, the developing eye-antennal imaginal disc of Drosophila larvae (the precursors of the adult fly eye) provides an excellent experimental model system (Dominguez and Casares 2005).
The Notch signaling response in growth control can be determined in changes of fly eye size by using precise temporal and spatial gene manipulation via the eye-specific driver ey-Gal4 (Hazelett et al. 1998) and UAS transgenes of Notch pathway components (Artavanis-Tsakonas et al. 1999; Bray et al. 2005), combined with gene dosage-sensitive assays. In the first assay, we used eye imaginal discs misexpressing the Notch modulator fringe by the ey-Gal4 driver (hereafter ey-Gal4 > fng), which reduces Notch signaling at the conserved eye growth organizer, resulting in a specific and reproducible eye defect (Fig. 5D,E; Dominguez and de Celis 1998; Dominguez et al. 2004). Either mutation in Su(H) (Fig. 5F) or lid (Fig. 5G) partially reverted the small eye phenotype, similarly to the effect of the corepressor of Su(H), Hairless (data not shown). In a second assay, we used eye imaginal discs coexpressing an oncogenic combination of Notch ligand Delta with two neighboring epigenetic repressors, Pipsqueak and Lola (collectively called eyeful) (Ferres-Marco et al. 2006), under the same eye-specific promoter (hereafter ey-Gal4 > Dl > eyeful), which results in invasive eye tumors (Ferres-Marco et al. 2006). Consistent with the above results, dose reduction of Su(H)/RBP-J enhanced eye tumor formation (Fig. 5H; Supplemental Fig. S9). Similarly, lowering the level of lid using two independently generated P-element insertions, lid10424 and lidk06801, also enhanced tumorigenesis (Fig. 5H; Supplemental Fig. S9), and, moreover, the effect of lid mutation was epistatic to Su(H) (Fig. 5H, right column). We confirmed that reducing endogenous lid, and not an additional mutation in the background, is responsible for the enhancement of tumorigenesis by Notch signaling by using two lid RNAi transgenes from fly stocks of National Institute of Genetics (NIG-FLY) stock center. Again, reduction of lid enhanced the incidence of tumor growth (Supplemental Fig. S9E). Together, these observations are most compatible with the view that Lid is required for endogenous Su(H) repressor function in vivo.
As predicted by our model, and as a further confirmation of the functional link between lid/KDM5A and Su(H)/RBP-J, we found that reduction in the amount of Su(H) gene dosage in a homozygous lid background leads to phenotypes of bristle loss and vein loss characteristic of Notch gain of function (Fig. 5I,J; Supplemental Tables 2, 3). Loss of function of Hairless also causes interruption of wing vein similar to the Su(H) lid/lid animals (Furriols and Bray 2001). For this experiment, we generated double-mutant strains carrying a Su(H) mutation and a lid mutation on the same chromosome and assayed different mutations (Supplemental Tables 2, 3). Taken together, these results unveil a specific contribution of the histone lysine demethylase to gene silencing by the Notch nuclear effector RBP-J that is evolutionarily conserved.
Discussion
Histone lysine demethylases reversibly remove methyl marks, thus facilitating changes in chromatin formation and transcriptional regulation. Histone demethylases have therefore been proposed as promising therapeutic targets of human diseases, including cancer, that are often associated with aberrant histone methylation. In this study, we identified KDM5A as an enzyme responsible for the removal of H3K4me3 at Notch target genes, and showed that KDM5A interacts directly with RBP-J via a domain located between PHD2 and PHD3 domain and its C-terminal PHD3 domain. Interestingly, the PHD3 domain was shown recently to bind to H3K4me3 (Wang et al. 2009). Although it has been suggested that the Arid domain of KDM5A can bind to a short DNA sequence, CCGCCC (Tu et al. 2008), the importance of this finding is challenged by the fact that this sequence is very common in CGIs, yet KDM5A is found only at a small number of genes in ChIP-on-chip experiments (Lopez-Bigas et al. 2008; Pasini et al. 2008). Morover, 11 of these putative KDM5A DNA-binding sites can be found at the CGI of Deltex-1, but only one is present in the Deltex-1 enhancer. Therefore, no correlation exists between the position with a high density of putative KDM5A-binding sites and the dynamically regulated H3K4 trimethylation site at the Deltex-1 gene. Furthermore, in EMSA assays we could not corroborate binding of KDM5A at the proposed sites (data not shown). Thus, we hypothesize that the PHD3 domain of KDM5A binds to H3K4m3 at active promoters, and once the H3K4me3 substrate is demethylated, KDM5A is released.
The Polycomb group (PcG) proteins play important roles in maintaining gene silencing during development and adult tissue homeostasis, and recent studies have shown that histone demethylase KDM5A is part of a Polycomb complex (Pasini et al. 2008). Thus, a KDM5A/Polycomb complex, recruited to Notch target genes, could facilitate the removal of active mark H3K4me3 and the subsequent addition of the repressive H3K27me3 marks. Paradoxically, original genetic analysis of Drosophila lid mutations classified Lid as a member of the Trithorax group of genes. Biochemical experiments suggest that Drosophila Lid affects the HDAC activity of Rpd3 (Lee et al. 2009), and molecular and genetic data show that Lid facilitates activation of dMYC target genes in a demethylase-independent manner (Secombe et al. 2007), explaining in part the original classification of lid as a positive transcriptional regulator. However, more recent data and this study support a key role for KDM5A demethylase in the dynamics of gene silencing.
Human KDM5A-containing and Drosophila KDM5A/Lid-containing complexes have been analyzed by several groups (Hayakawa et al. 2007; Pasini et al. 2008; Lee et al. 2009; Moshkin et al. 2009). KDM5A is found to be part of an MRG15-containing complex comprising multiple subunits, including Sin3B, HDAC1/2, and RbAp46, and two histone acetyltrasferases, TRRAP and Tip60 (Hayakawa et al. 2007). Interestingly, similar to our data, these studies unveiled effects of KDM5A/RBP2 on H3K4 trimethylation away from the TSSs in intergenic regions. Moreover, our findings on dynamic removal of H3K4me3 (Fig. 2A) associated with changes in acetylated H3K9 (Supplemental Fig. S7C) also point to a combined action of KDM5A and histone deacetylases. Importantly, Drosophila KDM5A/Lid complexes also contain histone deacetylase activity along with histone chaperones ASF1 and NAP1 (Lee et al. 2009; Moshkin et al. 2009). In agreement with our data, several of these Drosophila corepressors affect Notch target gene expression (Moshkin et al. 2009). However, although all of these data clearly point to a repressive role of KDM5A/Lid, how these enzymes silence specific genes was unknown.
KDM5A is a member of the KDM5 family, which consists of four proteins (KDM5A–D) in mammals. Particularly, KDM5A is highly expressed in the hematopoietic system (Nottke et al. 2009). KDM5A (RBP2)-deficient mice appear grossly normal but display a mild hematopoietic phenotype, especially in the myeloid compartment (Klose et al. 2007). The relatively mild phenotype of KDM5A mouse knockout might be explained in part by some redundancy between the KDM5 paralogs. Yet, one of the up-regulated genes in the KDM5A knockout microarray is Ifi2004 (Klose et al. 2007), a Notch target gene (Weng et al. 2006), indicating that KDM5 paralogs might play redundant as well as specific roles. Loss of KDM5 orthologs in organisms that encode a single KDM5 gene show more severe phenotypes, underscoring the important role of KDM5 demethylases in development. Thus, mutations in Drosophila KDM5A homolog lid often result in lethality before hatching, some animals show a small optic brain lobe and small imaginal discs (Gildea et al. 2000), and functional inactivation of the C. elegans KDM5A ortholog, Rbr-2, results in undeveloped vulvas or a multivulval phenotype (Christensen et al. 2007).
Although the discovery of histone demethylases implicates a reversible state of epigenetic gene silencing, it was unanticipated that these chromatin-modifying enzymes exert pathway-specific effects on gene regulation. The dynamic switch-off (and back on) system for Notch target gene expression used in this study allowed us to reveal dynamic changes of histone modifications at Notch target genes. We found that the H3K4me3 is removed with a half-time of about 4 h after inhibition of the Notch pathway by the GSI. These 4 h cannot be explained by out-dilution through cell cycling; the cell division time here is 24 h. Modulation of H3K4 methylation has also been observed in other biological systems, such as the circadian variation of the transcription of the albumin D-element-binding protein gene in the mouse liver (Ripperger and Schibler 2006) or the X inactivation in early embryonic development, where loss of H3K4me3 is one of the earliest and most characteristic features of chromosome-wide silencing (Heard et al. 2004).
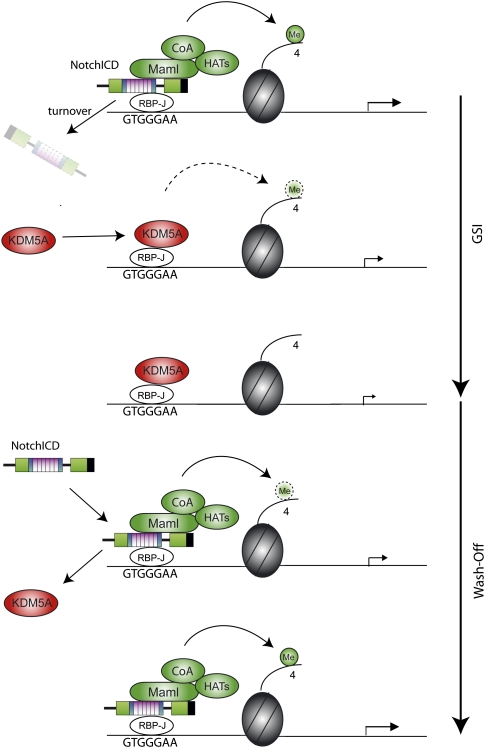
Disruption of Notch signaling results in a reduction of H3K4me3 at RBP-J sites, while reactivation re-establishes H3K4me3 levels (Fig. 2A; model in Fig. 6). This suggests that switch-off and switch-on of Notch target genes depends on a tightly controlled balance of histone H3 methylation and demethylation. Our study further identifies the histone demethylase KDM5A as a fundamental element in the switch-off process. For re-establishing H3K4me3 levels, Notch-IC could recruit an H3K4me3 methyltransferase to RBP-J sites of Notch target genes (Fig. 6). In contrast to Drosophila genes, ∼70% of mammalian genes possess CGIs (Saxonov et al. 2006), including many Notch target genes like Deltex-1, Hes-1, Hes-5, Nrarp, and Hey-1. Genome-wide studies proposed that H3K4 trimethylation remains very constant at CGI-containing promoters in different cell types (Barrera et al. 2008; Heintzman et al. 2009). Our study showed that H3K4me3 stays stable at the CGIs of Deltex-1 and Hes-1 after switching off Notch, but is regulated at the RBP-J-binding site. This finding shows for the first time that H3K4me3 does not necessarily have to be removed from the entire promoter to facilitate gene silencing. Instead, modulation of H3K4me3 at specific regulator elements could be sufficient to regulate gene expression. It will be of interest if the dynamic versus constant H3K4me3 is a more common feature of CGI-containing promoters. Recent technical advances in analyzing histone modifications genome-wide might help to address this question.
Figure 6.
Model for dynamic H3K4 trimethylation at Notch target genes: When Notch signaling is active, NotchICD migrates into the nucleus, associates with transcription factor RBP-J, and recruits Mastermind (MAML) and other coactivators (CoA) including a histone acetyltransferase (HAT). Upon GSI treatment, NotchICD gets degraded and RBP-J recruits KDM5A, which demethylates H3K4me3. When GSI is washed off, NotchICD replaces KDM5A and recruits a coactivator complex that re-establishes H3K4 trimethylation and, subsequently, transcriptional activity.
In summary, this study unveils that histone methylation is dynamically regulated by Notch signaling: Inhibition of Notch leads to a reduction of H3K4me3 levels at regulatory RBP-J sites, while reactivation of signaling re-establishes high levels of H3K4me3. Our biochemical and in vivo data support a role for the histone H3K4me3 demethylase KDM5A/Lid in facilitating the switch from activation to repression state via Su(H)/RBP-J in both Drosophila and mammals (Fig. 6). Thus, the histone lysine demethylase KDM5A/Lid is a crucial factor in the silencing process. With the in vivo evidence of Drosophila lid/KDM5A in Notch-induced tumorigenesis, this study suggests a pathway-specific tumor suppressor role of KDM5A in cancer, and provides the basis for studies in novel strategies to manipulate Notch-mediated carcinogenesis.
Materials and methods
Reagents, antibodies, and cell culture
For ChIP experiments, the following antibodies were used: H3K4me3 (Abcam, ab12209; Diagenode, pAb-003-050), H3 (Abcam, ab1791), KDM5A (Abcam, ab65769), RBP-J (Millipore, AB5790), p300 (Santa Cruz Biotechnologies, sc-585), H3K9Ac (Abcam, ab4441). For Western blotting, the following antibodies were used: RBP-J (Cosmo Bio Co., Clone T6709; secondary: anti-rat IgG [Dianova]), KDM5A (Bethyl Laboratories, A330-897A), anti-p65 (Santa Cruz Biotechnologies, sc372-G), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Abcam, ab8245).
Beko mouse T cells were grown in Iscove's modified Dulbecco medium (Gibco) with 2% fetal calf serum (FCS), nonessential amino acids, 0.3 mg/L Primatone, and 5 mg/L insulin. The cell line was maintained at 37°C under 5% CO2. The cells were treated with GSI (10 μg/mL DAPT; Calbiochem, catalog no. 565770) or DMSO only. GSI was removed by washing the cells twice with phosphate-buffered saline (PBS). For tamoxifen induction, the cells were treated with 0.75 μg/mL 4-OHT.
ChIP
H3K4me3 and acetylated H3K9 ChIP experiments were performed via native ChIP (NChIP) using the Epigenome Network of Excellence (NoE) protocol “Chromatin immunoprecipitation on native chromatin from cells and tissues” (PROT22) with the following modifications: Four million cells were used per ChIP. The chromatin was digested with MNase to mono- and dinucleosomes. After the digestion, the chromatin was centrifuged at 10,000 rpm. The pellet was resuspended with 500 μL of dialysis buffer and incubated overnight at 4°C. After centrifugation at 13,000 rpm, the supernatants (S2 fraction) were used further as described in the protocol. The S1 and P fractions were discarded.
KDM5A, p300, and H3 ChIPs were performed via cross-linking ChIP. Beko cells were fixed with 1% formaldehyde for 10 min (20 min for KDM5A) at room temperature. The cross-linking reaction was stopped with 125 mM glycine. Nuclear extracts were prepared by using a cell lysis buffer (50 mM Tris-HCl at pH 8.0, 2 mM EDTA, 0.1% NP-40, 10% glycerol, 2 mm dithiothreitol). The nuclei were resuspended in 600 μL of resuspension buffer (50 mM Tris-HCl at pH 8.0, 1% SDS, 5 mM EDTA, 2 mM DTT) and sonicated. Chromatin was 10-fold diluted with dilution buffer (50 mM Tris-HCl at pH 8.0, 200 mM NaCl, 5 mM EDTA, 0.5% NP-40) and precleared with presaturated protein A beads. The precleared chromatin extract was incubated overnight with the appropriate antibody. The immunoprecipitates were immobilized on protein A-Sepharose beads for 2 h. After washing the beads with 500 mM NaCl- and 500 mM LiCl-containing buffers, chromatin was eluted from the beads with elution buffer (Tris-EDTA, 2% SDS) at room temperature. The cross-linking of chromatin was reversed for 6 h at 65°C in the presence of 300 mM NaCl and RNase A. Chromatin was precipitated with ethanol, dissolved in Tris-EDTA-PK buffer (10 mM Tris-HCl at pH 7.5, 5 mM EDTA, 0.25% SDS), and treated with proteinase K for 2 h at 45°C. After purification by phenol-chloroform extraction, the chromatin was precipitated overnight at 20°C in the presence of 300 mM NaCl, tRNA, glycogen, and ethanol.
For ChIP at the luciferase construct, ∼5 million HeLa cells were transfected with 0.5 μg of pGL2-HES1-Luc reporter construct together with 5 μg of RBP-VP16 and 5 μg of HA-hKDM5A expression vectors. After 24 h, the cells were trypsinized and washed, and H3K4me3 ChIP was performed via NChIP (see above).
The enriched chromatin was analyzed by quantitative real-time PCR. The values were calculated as percent of the input.
MNase digestion
Chromatin was prepared like in NChIP and then split in two parts. One part was digested with MNase in excess for 30 min at 37° to mononucleosomes. Afterward, the DNA from the digested and undigested chromatin was extracted with phenol/chloroform extraction. The ratio between digested versus undigested DNA was measured by real-time PCR.
Beko cells with constitutively expressing dn-MAML-ER and Notch-IC-ER
Beko cells were infected with the retroviral vectors MIGR dn-MAML-ER IRES GFP or MIGR Notch-IC-ER IRES GFP. GFP-positive cells were sorted.
Knockdown in pre-T-cell line Beko
For knockdown of KDM5A, Beko cells were transfected with pSIR-siRNA vector and were selected for puromycin resistance, which expresses either the specific hairpin against KDM5A (target sequence: TGTCAAAGTGAGAGCACA) or a scrambled hairpin (TGAAATTAATGGCTATGAGTT) as control.
For the knockdown of RBP-J, the miRNA environment for microRNA (miRNA) 155 was cloned from the vector pRTS (provided by Dr. M. Hölzel) (Holzel et al. 2007) into the retroviral vector MIGR (provided by Dr. W. Pear) containing an IRES Tomato as fluorescence marker. The miRNAs were designed using the BLOCK-iT RNAi Designer of Invitrogen. The miRNAs were cloned in the miRNA environment of the retroviral vector via BbsI cutting sites. In order to check knockdown efficiency by FACS, a control vector containing GFP, a stop codon, and an RBP-J-cDNA fragment containing the hairpin region (GFP-stop-RBP-J) were cloned and introduced into Beko cells. These GFP-stop-RBP-J control cells were infected with retroviral constructs containing miRNA directed against RBP-J (GGTGTCCTAACTACCGTCTTT). Tomato-positive and GFP-low (indicating a successful knockdown) were sorted and analyzed. As negative control for the experimentsm a miRNA directed against GFP was used (ACAAGCTGGAGTACAACTACA).
RNA extraction, RT–PCR, and quantitative PCR
Total RNA was isolated from Beko cells using Trizol reagents (Thermo Scientific, no. AB-0305). For cDNA synthesis, 1 μg of RNAs was synthesized using random hexamers and reverse transcriptase according to the manufacturer's protocol (Invitrogen). Real-time PCR reactions were performed using a 7300 ABI PRISM sequence detector system (Applied Biosystems) according to the manufacturer's recommendations. Quantitative PCRs were performed using ABsolute QPCR ROX mix (Thermo Scientific, no. AB-1139), gene-specific oligonucleotides, and double-dye probes (see Supplemental Table 1) under the following conditions: 2 min at 50°C and 15 min at 95°C, and then 45 cycles of 15 sec at 95°C and 1 min at 60°C. The results were normalized to endogenous GUSB (glucuronidase β) or GAPDH mRNA expression levels.
For miniarrays, the PCR array PAMM-059 from SABiosciences was used.
Western blotting
The proteins resolved in SDS–polyacrylamide gels (10%) were transferred electrophoretically for 1 h at room temperature to polyvinylidene difluoride membranes (Millipore) at 50 mA by using a Tris-glycine buffer system. The membranes were preblocked for 1 h in a solution of 2% or 3% milk powder in 0.1% Tween20 in PBS (PBS-T) before the antibodies were added.
In vitro protein translation
Proteins were translated in vitro in the presence of [35S]methionine using the reticulocyte lysate-coupled transcription/translation system (Promega). Translation and labeling quality were monitored by SDS-PAGE.
GST pull-down assay
The GST fusion proteins were expressed in Escherichia coli strain BL21 (Stratagene) and stored as whole bacterial lysates at −80°C. Approximately 1 μg of GST protein and GST fusion protein were immobilized with glutathione-Sepharose beads (GE Healthcare, no. 17-5132-01) and incubated together with the in vitro translated proteins in buffer A (40 mM HEPES at pH 7.5, 5 mM MgCl2, 0.2 mM EDTA, 0.5% Nonidet P40 [NP-40], 100 mM KCl) under rotation for 1 h at 4°C. Beads were washed three times with 600 μL of buffer A and three times with 600 μL of buffer B (equivalent to buffer A, but containing 300 mM KCl). After the washing steps, the beads were resuspended in SDS-PAGE loading buffer, and the proteins were separated by SDS-PAGE. The gels were dried and exposed to X-ray films.
Preparation of cell extracts
Cells were washed three times in PBS and pelleted by centrifugation at 300g. The pellet was resuspended in 5 vol of ice-cold 3-[(cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) lysis buffer consisting of 10 mM CHAPS, 50 mM Tris/HCl (pH 7.9), 150 mM NaCl, 2 mM EDTA, 5 mM NaF, 10 μg/mL Aprotinin, 10 μg/mL Leupeptin, 1 mM Dithiothreitol (DTT), and 0.5 mM phenylmethylsulfonyl-fluoride (PMSF), and was incubated for 40 min on ice. The lysate was cleared at 80,000g for 30 min. Protein concentrations were determined by the Bradford method (Bio-Rad) and extracts were used for DNA affinity purification, in EMSA, for immunoprecipitation, and for Western blotting.
Coimmunoprecipitation
Immunoprecipitation experiments were carried out using whole-cell extracts from HEK293 cells 24 h after cotransfection with Flag-RBP-J and expression plasmids for Flag/HA-KDM5A. Cells were lysed with 700 μL of CHAPS lysis buffer. The extracts were incubated with 4 μL of anti-HA antibody (HA11 monoclonal AB; Covance, no. MMS-101P) overnight at 4°C. The reaction was then incubated with protein A-Sepharose beads (4 Fast-fFow, GE Healthcare, no.17-0618-01) for 2 h at 4°C. The beads were washed six times with CHAPS lysis buffer and were resuspended in SDS–polyacrylamide gel loading buffer.
Purification of RBP-J DNA-binding complexes
DNA affinity purification using Beko pre-T cells and EMSA experiments were performed as described previously (Oswald et al. 2005).
EMSAs
Cellular extract (5–10 μg) or eluted fractions were used for EMSAs in a binding buffer consisting of 10 mM Tris/HCl (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, 0.5 mM dithiothreitol (DTT), and 4% glycerol. For binding reaction, 2 μg of poly-(dI–dC) (Pharmacia) and ∼0.5 ng of 32P-labeled oligonucleotides were added. The sequence of the double-stranded oligonucleotide SL233 (5′-CCTGGAACTATTTTCCCACGGTGCCCTTCCGCCCATTTTCCCACGAGTCG-3′) corresponds to the two RBP-J-binding sites within the EBV TP-1 promoter. For supershift experiments, 0.1 μg of antibody, directed against RBP-J (Cosmo Bio Co., Clone K0043), was added to the reaction mixture (Hamaguchi et al. 1992). The reaction products were separated using 5% polyacrylamide gels with 1× Tris-glycine-EDTA (TGE) at room temperature. Gels were dried and exposed to X-ray films.
Drosophila husbandry
The mutant stocks and genotypes of animals and larvae used for genetic interactions and mosaic analyses used are all described at http://flybase.org. Mutant stocks and transgenes used were Su(H)Δ47; Su(H)1; Su(H)2; lid10424; lidk06801 (from Bloomington); UAS-fng, UAS-Dl; GS88A8 (eyeful) (Ferres-Marco et al. 2006); UAS-GFPnls; UAS-Su(H)VP16; UAS-RNAi lid (two transgenes were used, lid9088R-1 and lid9088R-2, from NIG-FLY, Kyoto); ey-Gal4; hsp70-FLP; Actin 5c > FRT > y+ > FRT > Gal4 (AyGal4). Two different recombinants of Su(H) and lid were generated in the study: Su(H)2 lid10424/CyO twi-Gal4 UAS-GFP and y w; FRT40A Su(H)Δ47 lid10424/CyO twi-Gal4 UAS-GFP.
Statistical analyses
Data are presented as mean ± SD; statistical analyses were performed using unpaired Student's t-test. P < 0.05 was considered significant.
Acknowledgments
We thank Drs. M. Hölzel, D. van Essen, Y. Shi, W. Pear, U. Zimber-Strobl, and K. Helin for providing us with plasmids, and the NIG-FLY from Japan for RNAi flies. We thank Dr. J. Kisielow for providing us with the Beko preT-cell line. We thank S. Schirmer, R. Rittelmann, M. Krötschel, and K. Grubisic for excellent technical assistance. We thank Drs. Grosschedl, S. Saccani, D. van Essen, and P. Heun for critical reading of the manuscript. Research in the T.B. laboratory is funded by the DFG (BO-1639 and Collaborative Research Center grant SFB592/C3) and the Max-Planck Society. Research in the F.O. laboratory is supported by the DFG (Collaborative Research Center grants SFB518/A18 and SFB497/B9). Work in the laboratory of M.D. is funded by the Ministerio de Ciencia e Innovación (BFU2006-05150 and MEC-CONSOLIDER CSD2007-00023), Asociación Española Contra el Cáncer (AECC), Generalitat Valenciana (PROMETEO 2008/134), Foundation Marcelino Botin, and a European Union Research grant (UE-HEALH-F2-2008-201666).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.563210.
Supplemental material is available at http://www.genesdev.org.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Barrera LO, Li Z, Smith AD, Arden KC, Cavenee WK, Zhang MQ, Green RD, Ren B. Genome-wide mapping and analysis of active promoters in mouse embryonic stem cells and adult organs. Genome Res. 2008;18:46–59. doi: 10.1101/gr.6654808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: A simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Bray S, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev Cell. 2005;8:279–286. doi: 10.1016/j.devcel.2004.11.020. [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Casares F. Organ specification-growth control connection: New in-sights from the Drosophila eye–antennal disc. Dev Dyn. 2005;232:673–684. doi: 10.1002/dvdy.20311. [DOI] [PubMed] [Google Scholar]
- Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–278. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36:31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat Struct Mol Biol. 2007;14:344–346. doi: 10.1038/nsmb1217. [DOI] [PubMed] [Google Scholar]
- Fattaey AR, Helin K, Dembski MS, Dyson N, Harlow E, Vuocolo GA, Hanobik MG, Haskell KM, Oliff A, Defeo-Jones D, et al. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- Ferres-Marco D, Gutierrez-Garcia I, Vallejo DM, Bolivar J, Gutierrez-Avino FJ, Dominguez M. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature. 2006;439:430–436. doi: 10.1038/nature04376. [DOI] [PubMed] [Google Scholar]
- Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Lopez R, Shearn A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi Y, Yamamoto Y, Iwanari H, Maruyama S, Furukawa T, Matsunami N, Honjo T. Biochemical and immunological characterization of the DNA binding protein (RBP-J κ) to mouse J κ recombination signal sequence. J Biochem. 1992;112:314–320. doi: 10.1093/oxfordjournals.jbchem.a123898. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, Nakayama J. RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells. 2007;12:811–826. doi: 10.1111/j.1365-2443.2007.01089.x. [DOI] [PubMed] [Google Scholar]
- Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development. 1998;125:3741–3751. doi: 10.1242/dev.125.18.3741. [DOI] [PubMed] [Google Scholar]
- Heard E, Chaumeil J, Masui O, Okamoto I. Mammalian X-chromosome inactivation: An epigenetics paradigm. Cold Spring Harb Symp Quant Biol. 2004;69:89–102. doi: 10.1101/sqb.2004.69.89. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Rohrmoser M, Orban M, Homig C, Harasim T, Malamoussi A, Gruber-Eber A, Heissmeyer V, Bornkamm G, Eick D. Rapid conditional knock-down–knock-in system for mammalian cells. Nucleic Acids Res. 2007;35:e17. doi: 10.1093/nar/gkl1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan MX, Kopan R. SnapShot: Notch signaling pathway. Cell. 2007;128:1246e-1–1246e-2. doi: 10.1016/j.cell.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathrein KL, Chari S, Winandy S. Ikaros directly represses the notch target gene Hes1 in a leukemia T cell line: Implications for CD4 regulation. J Biol Chem. 2008;283:10476–10484. doi: 10.1074/jbc.M709643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Koch U, Radtke F. Notch and cancer: A double-edged sword. Cell Mol Life Sci. 2007;64:2746–2762. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes & Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The H3K4 demethylase lid associates with and inhibits histone deacetylase Rpd3. Mol Cell Biol. 2009;29:1401–1410. doi: 10.1128/MCB.01643-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D. Hairless: The ignored antagonist of the Notch signalling pathway. Hereditas. 2006;143:212–221. doi: 10.1111/j.2007.0018-0661.01971.x. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–793. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol. 2001;21:7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knochel W, et al. SHARP is a novel component of the Notch/RBP-Jκ signalling pathway. EMBO J. 2002;21:5417–5426. doi: 10.1093/emboj/cdf549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-Jκ/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK–BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Salat D, Liefke R, Wiedenmann J, Borggrefe T, Oswald F. ETO, but not leukemogenic fusion protein AML1/ETO, augments RBP-Jκ/SHARP-mediated repression of notch target genes. Mol Cell Biol. 2008;28:3502–3512. doi: 10.1128/MCB.01966-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes & Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Teng YC, Yuan C, Wu YT, Chan MY, Cheng AN, Lin PH, Juan LJ, Tsai MD. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat Struct Mol Biol. 2008;15:419–421. doi: 10.1038/nsmb.1400. [DOI] [PubMed] [Google Scholar]
- van Oevelen C, Wang J, Asp P, Yan Q, Kaelin WG, Jr, Kluger Y, Dynlacht BD. A role for mammalian Sin3 in permanent gene silencing. Mol Cell. 2008;32:359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg AE, Pedersen K, Lendahl U, Roeder RG. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol. 2002;22:7812–7819. doi: 10.1128/MCB.22.22.7812-7819.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes & Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]