Summary
The immediate early genes of the α-herpesviruses HSV and VZV are transcriptionally regulated by viral and cellular factors in a complex combinatorial manner. Despite this complexity and the apparent redundancy of activators, the expression of the viral IE genes is critically dependent upon the cellular transcriptional coactivator HCF-1. Although the role of HCF-1 had remained elusive, recent studies have demonstrated that the protein is a component of multiple chromatin modification complexes including the Set1/MLL1 histone H3K4 methyltransferases. Studies using model viral promoter-reporter systems as well as analyses of components recruited to the viral genome during the initiation of infection have elucidated the significance of HCF-1 chromatin modification complexes in contributing to the final state of modified histones assembled on the viral IE promoters. Strikingly, the absence of HCF-1 results in the accumulation of nucleosomes bearing repressive marks on the viral IE promoters and silencing of viral gene expression.
Keywords: HCF-1, herpesvirus, coactivator, Set1, LSD1
Infection by the α-herpesviruses (HSV, herpes simplex virus and VZV, varicella zoster virus) follows a tightly regulated viral gene transcription program that begins with the expression of the viral immediate early (IE) genes and ultimately results in the production of viral progeny and cell lysis. Following a primary infection, these viruses also establish latency in neurons of the host sensory ganglia. Periodic interruption of the latent state can result in recurrent lytic infection, presumably initiated via induced transcription of the viral IE genes. Thus, transcriptional control of the viral IE genes is both a critical regulatory point in the initiation of lytic infection as well as initiation of viral reactivation from latency. Additionally, these genes are transcribed by the host cell RNAPII machinery and have proven to be excellent model systems for the investigation of cellular transcriptional control mechanisms.
Regulatory domains of the α-herpesvirus immediate early genes
Control of α-herpesvirus immediate early gene expression is complex. The enhancer-promoter domains of these genes contain binding sites for members of a number of distinct families of transcription factors. This complexity allows for multiple cooperative regulatory pathways and provides the ability to respond to different cell environments and signals. This multi-path regulation may also reflect the mechanisms involved in the reactivation of these viruses from the latent state.
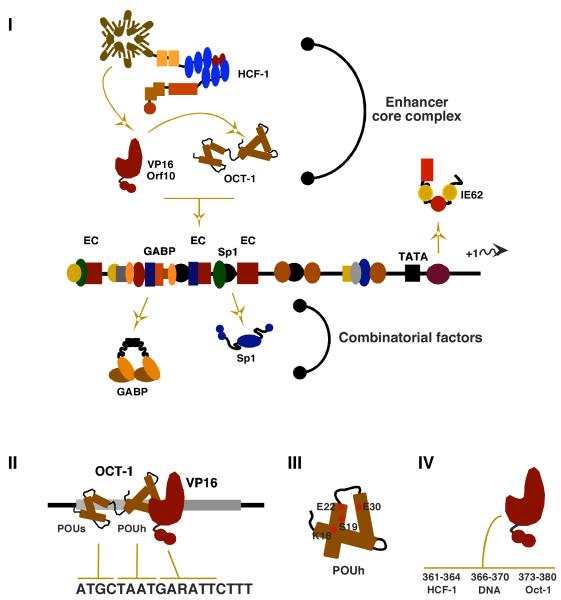
A characteristic IE gene, shown in Figure 1, consists of a basal promoter and an upstream regulatory domain that contains a reiterated enhancer core element (EC; ATGCTAATGARATTCTTT). The EC element nucleates the formation of a multi-protein enhanceosome complex that is the primary mediator of IE gene expression and consists of the cellular POU-homeodomain protein Oct-1, the viral IE activator (VP16 for HSV and ORF10 for VZV), and the cellular transcriptional coactivator HCF-1 rev. in [1, 2].
Figure 1. Components regulating the expression of the α-herpesvirus IE genes.
(I) The IE genes of HSV and VZV enhancer-promoter domains are complex and contain binding sites for multiple factors functioning synergistically or cooperatively. Viral IE activators (VP16 for HSV; ORF10 for VZV) interact with Oct-1 and HCF-1 to form the stable enhanceosome complex. A second VZV IE activator IE62 stimulates expression via its own recognition elements. Cellular factors such as GABP and Sp1 amplify the enhancer core (EC, TAATGARAT) mediated expression of the IE genes but may also function independent of the EC complex to stimulate IE gene expression.
(II) Oct-1 recognizes the EC element via a bipartite DNA binding domain consisting of POU-specific (s) and POU-homeo (h) domains. (III) The viral activator, VP16, recognizes the surface of the Oct-1 POU-homeo domain via specific residues in helix 1 and 2 and provides specificity by recognition of the 3′ sequences of the EC element. (IV) Clustered residues in the carboxyterminal region of VP16 mediate interactions with HCF-1, DNA, and Oct-1.
The assembly of the enhanceosome is dependent upon the recognition of the noncanonical octamer (ATGCTAAT) in the EC element by the Oct-1 bipartite POU-Homeo domain [3-5]. The viral IE activators, which are packaged in the tegument structure of these viruses and released into the cell upon infection, provide specificity by recognition of both the 3′ sequences of the EC element (GARATTCTTT) and the exposed surface of the Oct-1 homeodomain [3, 6-9]. In addition, these IE activators also bind and recruit the cellular coactivator HCF-1 into the EC complex, resulting in a stable enhanceosome assembly [10-12].
In addition to the enhanceosome complex, additional cellular transcription factors such as GABP and Sp1 bind to sites flanking the EC element and contribute to the induced level of IE gene transcription [13-15]. Strikingly, factors like GABP can also provide alternative means of stimulating IE gene expression even in the absence of the enhanceosome nucleating factor Oct-1 [16].
Combinatorial regulation of IE genes is determined at the level of the coactivator HCF-1
Much of what has been learned about HCF-1 to date is derived from investigation of its interactions with both viral and cellular transcription components. HCF-1 was originally identified and purified as a protein required for the stable assembly of the viral IE EC complex [11]. However, the protein is now recognized as an essential cellular coactivator with global impact on gene transcription and cell cycle progression via interactions with multiple cellular transcription factors, coactivators, and chromatin modification components.
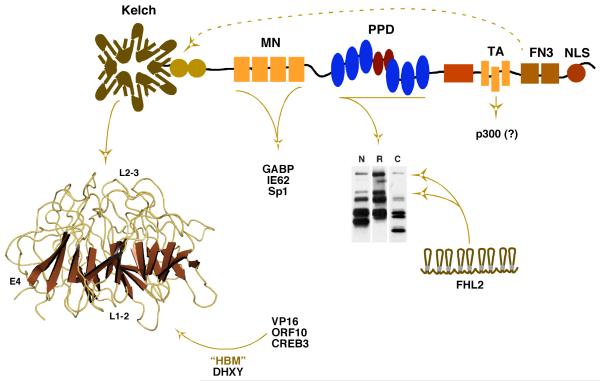
HCF-1 interacts with components that mediate both basal level and viral induced expression of the α-herpesvirus IE genes [17]. These transcriptional activators bind various HCF-1 domains, suggesting that the protein orchestrates a coordinated regulatory process that results in the high level expression of the IE genes upon initial infection (Figure 2). The viral IE activators (VP16 and ORF10) bind the amino-terminal kelch domain [18-20] (Figure 2). Many of the factors that bind this domain, including the viral IE activators, contain a small interaction motif (D/EXHY; referred to as the HBM, HCF binding motif) [18, 21, 22]. However, despite this common motif, mutations in the HCF-1 kelch domain indicate that there are distinct binding determinants for different factors and that multiple HBM proteins may interact with the same HCF-1 molecule [23, 24]. In addition to the viral IE activators, the CREB/ATF family member CREB3/Luman also binds the kelch domain and can induce a representative viral IE gene in an HCF-1 dependent manner [18, 25, 26]. Conversely, Zhangfei, a second bZIP protein that binds this domain can inhibit HCF-1 dependent activation of the HSV IE genes [27, 28] and has been hypothesized to play a role in suppression of lytic infection.
Figure 2. The transcriptional coactivator HCF-1 and interactions regulating the expression of the α-herpesvirus IE genes.
The essential coactivator HCF-1 interacts with numerous factors that impact the expression of the IE genes. An amino-terminal kelch domain is a barrel structure consisting of reiterated units of 4 antiparallel β-sheets connected by flexible loops. The structure presents several protein interaction surfaces composed of: (i) loops connecting sheets 2 and 3 (top surface, L2-3); (ii) loops connecting sheets 1 and 2 (bottom surface, L1-2); and (iii) the 4th β-sheet of each reiterated unit (circumference, E4). Viral and cellular activators (i.e. VP16, ORF10, CREB3/Luman) and coactivators containing an HCF-1 Binding Motif (HBM, D/EHXY) interact with this domain, presumably in a non-exclusive manner. The mid-aminoterminus (MN) binds GABP, IE62, and Sp1. The central proteolytic processing domain (PPD) consists of a series of 20 amino acid reiterations (blue/red ovals) that are the sites of specific proteolytic processing that generates the family of HCF-1 polypeptides shown in the western blot (N, R, C represent antiserum specific to the aminoterminus, repeat, and carboxyterminus, respectively). The coactivator FHL2 interacts with the PPD and this interaction is regulated by HCF-1 processing. A transactivation domain (TA) is required for cooperative stimulation by factors such as VP16 and may function to recruit the coactivator p300/CBP. Two fibronectin type III repeats (FN3) mediate the amino- and carboxyterminal subunit association of HCF-1 as indicated by the dashed arrow. NLS; nuclear localization signal.
The mid-amino terminal (MN) domain of HCF-1 interacts with GA-binding protein (GABP/NRF2) [29], Sp1 [30], and IE62, a second VZV IE activator that is associated with HCF-1 via Sp1 [31]. Sp1 plays a significant role in the basal level expression of the IE genes [13] while GABP plays a co-stimulatory role in the presence of the viral IE activator [15, 17]. As noted above, GABP can also promote the regulated induction of the IE genes even in the absence of the viral EC elements or the EC nucleating factor Oct-1 [16]. Induction by the viral activators through the EC or induction via GABP are two of the multiple mechanisms by which these genes may be regulated. Notably, however, HCF-1 is required for both mechanisms suggesting that the protein mediates some common and essential rate limiting stage.
The central region of HCF-1 (proteolytic processing domain, PPD) interacts with a series of proteins including the transcriptional coactivator FHL2; a protein which functions cooperatively with HCF-1 to stimulate the HSV IE genes [32]. Interestingly, the HCF-1 PPD consists of a series of 20 amino acid reiterations that are the sites of specific proteolytic cleavage. Processing at one or more of these sites generates a family of HCF-1 amino- and carboxy-terminal subunits that do not segregate but rather remain tightly associated [10-12, 33]. As the interaction determinants for FHL2 lie within the PPD, cleavage of HCF-1 at specific reiterations can result in the loss of FHL2 binding. Thus HCF-1 processing may represent a unique mechanism for controlling its interactions and coactivator function. Other studies have suggested that processing of HCF-1 is important in its role in modulation of cell cycle progression although the mechanism involved has not been elucidated [34].
The carboxy-terminal domain of HCF-1 contains several defined functional regions including (i) a transactivation domain that synergistically functions with VP16 and other HCF-1 dependent factors, possibly through cooperative recruitment of p300 [35, 36]; (ii) a series of fibronectin repeats that mediate the amino- and carboxy- HCF-1 subunit association [37]; and (iii) a bi-partite nuclear localization signal (NLS). Interestingly, transfection of an HCF-1 NLS(-) mutant results in cytoplasmic localization of both HCF-1 and VP16, suggesting that HCF-1 functions to cotransport the viral activator upon infection [38]. However, in other studies, depletion of HCF-1 by siRNA did not prevent the nuclear accumulation of VP16, indicating that there are alternative mechanisms by which the viral activator is transported [17].
HCF-1 and the Set1/MLL1 histone methyltransferase family
Given the interactions of HCF-1 with both viral and cellular activators that control the basal and induced levels of the viral IE genes, it is not entirely surprising that the protein is essential for initiation of α-herpesvirus infection [17]. Previously it had been proposed that the protein served as a scaffold to coordinate or couple the various factors (Oct-1, GABP, VP16, Sp1, FHL2) involved in stimulation of IE gene transcription. However, the requirement for HCF-1 to mediate the activation potential of these various factors suggests that it must function at some critical rate limiting stage.
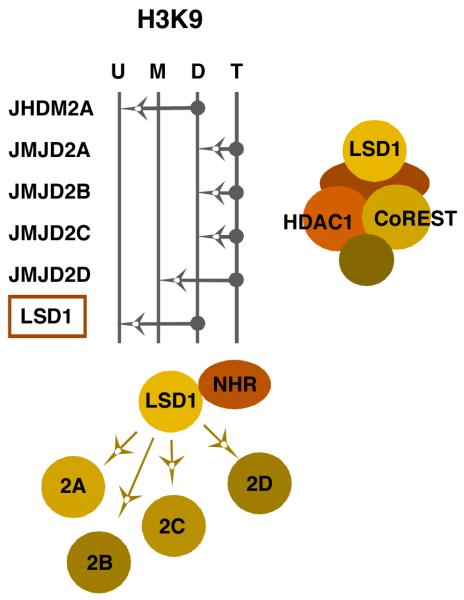
Until recently, the mechanism(s) involved in HCF-1 dependent coactivation remained undefined. A significant clue to its function came from the identification of HCF-1 as a component of the Set1 and MLL1 histone H3-lysine 4 methyltransferase complexes 4 (H3K4 HMT) [24, 39]. These multiprotein enzyme complexes [40] belong to a larger family responsible for the methylation of H3K4 (Figure 3) rev. in [41]; a modification enriched in euchromatin and functionally linked to active transcription. Part of the “histone code”, methylation of H3K4 is concentrated at the promoter and 5′ coding regions of actively transcribed genes and serves as a signal for components that promote efficient gene expression including nucleosome remodeling complexes [42, 43], components of TFIID [44], and the spliceosome [45].
Figure 3. The histone H3-lysine 4 methyltransferase family.
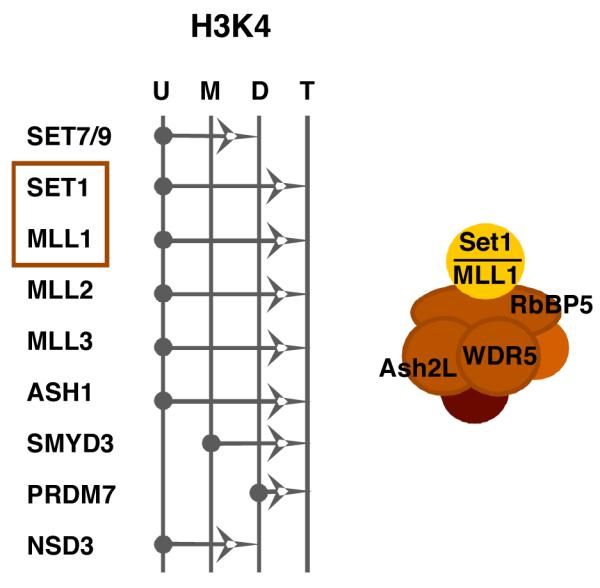
The specificities of the members of the histone H3-lysine 4 (H3K4) family of methyltransferases are shown. The arrows represent the ability of each member to catalyze the addition of a methyl group to an unmodified (U), monomethyl (M), or dimethyl (D) lysine 4 residue. The Set1 and MLL1 methyltransferase complexes contain a set of common core proteins including Ash2L, WDR5, and RbBP5 in addition to the catalytic subunit (Set1 or MLL1). Both hSet1 and MLL1 (boxed) interact with HCF-1 and play a role in HSV and VZV IE gene expression, likely via installation of activating H3K4 methylation at the IE promoters.
The role of chromatin in the regulation of the expression of the α-herpesvirus genes has been controversial. The viral genome is not packaged with histones in the capsid [46, 47] and it enters the cell nucleus devoid of nucleosomes. Historically, the model for the induction of the viral IE genes was derived from the role of the viral activators (VP16 and IE62) in recruitment of RNAPII initiation factors (i.e.; TFIIB, TBP, mediator components) [1, 48-52] rev. in [1]. However, recent data have suggested that chromatin does play a significant role in IE gene expression including: (i) VP16 dependent recruitment of HAT (p300 and CBP) and nucleosome remodeling components (BRG1 and hBRM) to viral IE promoters [53]; (ii) infection with an HSV mutant virus lacking the VP16 transactivation domain resulted in accumulation of histones on the IE promoters, suggesting that the protein was important for reducing stable nucleosome occupancy [53]; (iii) histone modifications associated with transcriptional activation were found at viral promoters [54, 55]; and (iv) HIRA dependent assembly of the histone variant H3.3 on viral IE promoters was important for IE transcription [56]. The accumulating data and the consideration that the virus must utilize cellular transcription components whose activity and fidelity is regulated by chromatin is the basis for a more complex model of IE gene transcription.
HCF-1 mediated chromatin control of IE genes
Using a model IE reporter assay system, Narayanan et al. demonstrated that HCF-1 plays a central role in the regulation of chromatin as part of the mechanism by which this essential protein stimulates viral IE gene transcription [31]. In this study: (i) HCF-1 was directly recruited to the model promoter in a viral IE activator dependent manner; (ii) HCF-1 had no impact on the assembly of the RNAPII initiation complex even though the protein was essential for transcriptional activation; (iii) recruitment of both Set1 and MLL1 to the promoter was concomitant with and dependent upon HCF-1; and (iv) recruitment of HCF-1 resulted in loss of repressive (histone H3-lysine 9 methylation) and gain of activating (histone H3-lysine 4 methylation) chromatin marks. HCF-1 dependent recruitment of Set1 and MLL1, was also observed at the viral genomic IE promoters during the initial stages of infection. These results demonstrated that the viral activators recruit components of the RNAPII initiation complex but require the HCF-1 dependent recruitment of the Set1/MLL1 HMTs to install positive chromatin modifications to promote efficient transcription initiation.
HCF-1 coupled demethylase and methyl-transferase activities: circumventing the accumulation of repressive chromatin
Two observations suggested that modulation of chromatin to promote transcription of the viral IE genes is more complex than the simple installation of activating marks on newly assembled nucleosomes. In cells depleted of HCF-1, chromatin bearing repressive histone H3K9 methylation accumulated on the viral IE promoters [31, 57]. Even in the presence of HCF-1, repressive chromatin could be detected at early times post-infection and the levels of these repressive marks decreased as HCF-1 was recruited to the IE promoters. Interestingly this accumulation of repressive chromatin likely reflects an initial cellular response to infection (or naked DNA) and suggested that a histone H3K9 demethylase would be required to circumvent the accumulation of these marks during induction of the viral IE gene expression. In support of this, Liang et al. subsequently demonstrated that Lysine Specific Demethylase 1 (LSD1) was also required for expression of the α-herpesvirus IE genes.
Like the histone methyltransferases, LSD1 is a member of a histone demethylase (HDM) family with distinct specificities for the removal of mono-, di-, or tri-methylation of histone H3K9 (Figure 4) [58-60]. LSD1 was the first demethylase to be identified [61] and it is unique as it can function to remove either positive histone H3K4 methylation or repressive histone H3K9 methylation, depending upon the protein complex it is incorporated into [62]. In the repressive CoREST complex, LSD1 removes activating H3K4 methylation [63, 64]. In contrast, LSD1 can remove repressive H3K9 methylation when bound to a ligand-activated nuclear hormone receptor [65-69]. Although the biochemical mechanism by which the specificity of the enzyme is determined is unknown, a recent study of the yeast LSD1 homolog (SWIRM1/SWIRM2) suggested that additional histone modifications might be a determining factor [70]. While these mechanistic questions remain to be answered, it is clear that LSD1 is associated with a large percentage of actively transcribed genes and plays a significant role in transcriptional activation in addition to its well characterized role in the repressive CoREST complex.
Figure 4. The histone H3-lysine 9 demethylase family: positive and negative LSD1 complexes.
The specificities of the members of the histone H3-lysine 9 (H3K9) family of demethylases are shown. The arrows represent the ability of each member to catalyze the removal of a methyl group from a trimethyl (T), dimethyl (D), or monomethyl (M) lysine 9 residue. LSD1 has a dual specificity; removing positive H3K4 methylation when in the CoREST complex or removing repressive H3K9 methylation when complexed with other components such as nuclear hormone receptors (NHR). Removal of repressive H3K9 trimethylation requires the cooperative activity of one of the other members of the family with the complimenting activity (JMJD2A, B, C, or D). LSD1 is highlighted (boxed) to emphasize its role in viral IE gene expression.
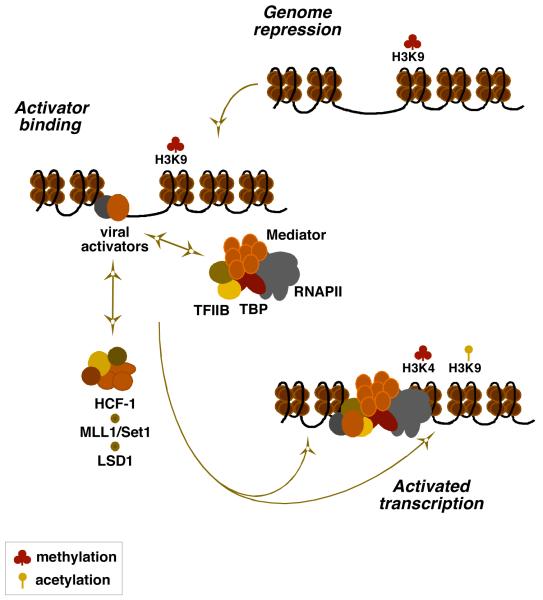
In infection with HSV or VZV, the depletion of LSD1 or the inhibition of its activity results in accumulation of repressive chromatin on the viral genome and ablation of viral IE gene expression. Similar to the Set1/MLL1 HMTs, recruitment of LSD1 is also HCF-1 dependent and the protein is associated with the HCF-1-HMT complex. The data supports the model, shown in Figure 5, whereby infection results in the rapid cell-mediated deposition of repressive chromatin on the viral genome. Viral IE activators, packaged in the tegument structure of the viral capsid, are released and transported to the nucleus where they function, in part, to recruit components of the cellular RNAPII initiation complex. Importantly, recruitment of the essential transcriptional coactivator HCF-1 is required to couple HDM and HMT activities to circumvent the accumulation of repressive chromatin and promote the installation of positive chromatin for initiation of IE gene transcription.
Figure 5. An HCF-1 complex couples methyltransferase and demethylase activities in the initiation of IE gene expression.
A model for the role of the HCF-1 coactivator complex in the regulation of the α-herpesvirus IE genes is shown. Upon infection, the viral genome is subject to the accumulation of chromatin bearing repressive marks (H3K9-methylation). The viral IE activators (VP16 or ORF10 and IE62) bind to the IE regulatory domains and recruit components of the RNAPII complex to the promoter. Transcriptional activation requires the recruitment of HCF-1 dependent chromatin modification activities (H3K4 methyltransferase Set1 or MLL1 and H3K9 demethylase LSD1) that ultimately results in a decrease in repressive marks and an increase in positive marks to promote IE gene transcription. Coordinated H3K4 methylation and H3K9 acetylation is proposed although the specific acetyltransferase complexes involved have not yet been determined.
The concept of coupling
An HCF-1 dependent complex containing both histone methyltransferase and demethylase activities is essential for the expression of the IE genes of HSV and VZV [29, 56]. As the “histone code” becomes more developed and complex, it is clear that mechanisms must exist to regulate and coordinate these signals. Numerous examples of modification interdependency and cross-talk have been described rev. in [71, 72]. However, direct coupling of various modification components is also a mechanism by which these modifications may be regulated to efficiently activate or repress gene expression. In addition to the HCF-1 complex, other coupled modification enzymes include: (i) the histone H4K16 acetyltransferase MOF and the H3K4 methyltransferase MLL1 [73]; (ii) LSD1 and HDAC1/2 in the CoREST complex [63, 74]; and (iii) the histone H3K27 demethylase UTX and the H3K4 methyltransferase MLL3/4 [75].
Multiple HCF-1 chromatin modulation complexes
HCF-1 couples LSD1 and the Set/MLL1 modification activities. However, HCF-1 has also been found in association with other chromatin modulation activities including the ATAC/STAGA (GCN5/PCAF) acetyltransferase complex [76, 77], sin3a/HDAC complex [24], and other members of the MLL family. The association of HCF-1 with these activities suggests that the protein plays a key role in the coordination of multiple modifications. Current investigations are based upon the anticipation that other HCF-1 chromatin modification/modulation activities may also be critical for the regulation of the viral IE genes.
A role for HCF-1 in reactivation of α-herpesviruses from latency
In addition to the lytic replication cycle, α-herpesviruses also establish latent infections in the neurons of sensory ganglia. Periodically, these latent infections may be interrupted, presumably via stimulation of the expression of the viral lytic IE genes in the absence of the viral IE activators. Importantly, despite the numerous transcription factors that can contribute to the induced expression of the viral IE genes, HCF-1 is essential. Even in the absence of the Oct-1/VP16 enhancer core complex, the IE genes can be induced in an HCF-1 dependent manner; suggesting that multiple alternative pathways exist to regulate these genes.
Strikingly, although HCF-1 is ubiquitously expressed and localized in the nucleus of most cell types, the protein is uniquely sequestered in the cytoplasm of sensory neurons. Stimuli that results in viral reactivation in a mouse model system results in rapid transport of HCF-1 to the nucleus[78, 79] and recruitment to latent viral IE promoters[80], indicating that HCF-1 functions as part of the signaling mechanism involved in the initiation of viral reactivation (Figure 6).
Figure 6. Specific sequestering and transport of HCF-1 in sensory neurons: a role for HCF-1 in regulation of α-herpesvirus reactivation from latency.
(I) HCF-1 is ubiquitously expressed and localized in the nucleus of most cell types (Liver and Kidney are examples shown) but is specifically sequestered in the cytoplasm of unstimulated sensory neurons (Trigeminal Ganglia).
(II) Stimuli that result in viral reactivation cause rapid transport and accumulation of HCF-1 in the nucleus of sensory neurons in a mouse trigeminal ganglia explant model system. The percentage of neurons exhibiting HCF-1 nuclear localization post explant in both mock infected and HSV latently infected trigeminal ganglia is graphically represented as a function of time.
With respect to the potential role of HCF-1 in this reactivation process, latent viral genomes are nucleosomal [81] and recent studies have suggested that the appropriate positive and negative chromatin modifications correlate with the states of latency and reactivation [82-90] rev. in [91]. The data suggests that chromatin modification components play a role in regulating this switch. Strikingly, inhibition of the histone demethylase LSD1; a component of the HCF-1 modification complex that functions during viral lytic infection, results in suppression of HSV reactivation from latency [57]. It is, therefore, tempting to propose that similar to its role in the lytic replication cycle, HCF-1 plays a controlling role in this process by modulating the state of chromatin at the viral IE promoters during reactivation.
Regulation of cellular transcription by HCF-1
HCF-1 was originally identified as a required component of the HSV enhancer core complex. However, the critical role of the protein in cellular transcriptional processes has been shown in a number of studies including (i) the identification of transcription factors of the krupple, Ets, THAP, and E2F families that interact with HCF-1 and/or for which the protein functions as a required transcriptional coactivator [25, 29, 30, 36, 92-95]; (ii) synergistic or cooperative interaction with coactivators including PGC, PRC, and FHL2 [32, 96, 97]; (iii) the requirement for HCF-1 for several stages of cell cycle progression [34, 98-100]; (iv) the role of HCF-1 in promoting efficient mRNA splicing [101]; and (v) alteration in the expression of cellular genes upon inhibition of HCF-1 nuclear accumulation [102].
Cell cycle control by HCF-1 is a particularly revealing example of the multi-point impact of HCF-1 on cellular transcription. HCF-1 was originally determined to be an important component of cell cycle control by the isolation of a cell line carrying an HCF-1 ts-mutant that arrested in G0 at the nonpermissive temperature [98]. Subsequently it was demonstrated that the protein is required for both G0-G1 progression and proper cytokinesis, with distinct domains of HCF-1 mediating distinct stages [34, 99]. Investigations of the mechanisms involved in HCF-1 dependent cell cycle progression have revealed that HCF-1: (i) regulates the expression of pRb through the Ets factor GABP [93]; (iii) regulates the expression of the histone H4K20 methyl-transferase PR-Set7 (disregulation results in upregulation of H4K20 methylation and defects in cytokinesis) [103]; (iv) interacts with and mediates the activity of members of the E2F family of cell cycle transcription factors [94, 95]; (v) interferes with the recruitment of p300 to the Myc-interacting protein Miz-1, thereby preventing Miz-1 mediated cell cycle arrest [104]; and (vi) regulates the level of mitotic histone H3S10 phosphorylation [105]. These studies clearly indicate that HCF-1 is essential to regulate components that play key roles in various stages of cell-cycle progression and illustrate the significance of this coactivator for cellular transcriptional programs in addition to its central role in α-herpesvirus infection.
Going forward
Many questions regarding the role of chromatin and the regulation of chromatin modulating components in both α-herpesvirus lytic and latency-reactivation cycles remain to be addressed. Importantly, the state of viral chromatin in terms of the localization and density of nucleosomes has not yet been determined. With respect to the regulation of the viral IE genes upon infection, the identification of the enzymatic complexes, mechanisms of recruitment, and the regulation of these activities involved are active areas of focus. Importantly, histone modifications serve as signals for the recruitment of components that repress or promote transcription, yet the roles of these modifications in IE gene transcription have not been determined. Finally, histone modification complexes have also been shown to modify other targets [106-108] and may be involved in modification of non-histone components that impact the regulation of the viral IE genes.
In a more general sense, investigation of the role of chromatin and chromatin modulation components in these viral systems may provide insights into the linkage between chromatin assembly and modification. Additionally, these studies should illuminate the mechanisms by which these viruses have evolved to control and utilize this process to circumvent repressive impacts but utilize positive components to promote the expression of their genes.
Acknowledgements
We thank T. Pierson for critical reading of this manuscript. Studies done in the Kristie Laboratory were supported by the Intramural Research Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations
- HCF-1
Host Cell Factor-1
- HSV
herpes simplex virus
- VZV
varicella zoster virus
- HMT
histone methyltransferase
- HDM
histone demethylase
- MLL1
mixed-lineage leukemia
- LSD1
Lysine-specific demethylase 1
- H3K4
histone H3-lysine 4
- H3K9
histone H3-lysine 9
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Kristie TM. Early events pre-initiation of viral gene expression. In: Arvin A, Campadielli-Fiume G, Roizman B, Whitley R, Yamanishi K, editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge University Press; Cambridge: 2006. pp. 395–452. [Google Scholar]
- [2].Roizman B, Sears AE. Herpes simplex viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fundamental Virology. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 1043–1107. [Google Scholar]
- [3].Kristie TM, Sharp PA. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV alpha-trans-activator protein. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- [4].Sturm RA, Herr W. The POU domain is a bipartite DNA-binding structure. Nature. 1988;336:601–604. doi: 10.1038/336601a0. [DOI] [PubMed] [Google Scholar]
- [5].Verrijzer CP, Kal AJ, van der Vliet PC. The oct-1 homeo domain contacts only part of the octamer sequence and full oct-1 DNA-binding activity requires the POU-specific domain. Genes Dev. 1990;4:1964–1974. doi: 10.1101/gad.4.11.1964. [DOI] [PubMed] [Google Scholar]
- [6].Pomerantz JL, Kristie TM, Sharp PA. Recognition of the surface of a homeo domain protein. Genes Dev. 1992;6:2047–2057. doi: 10.1101/gad.6.11.2047. [DOI] [PubMed] [Google Scholar]
- [7].Babb R, Huang CC, Aufiero DJ, Herr W. DNA recognition by the herpes simplex virus transactivator VP16: a novel DNA-binding structure. Mol Cell Biol. 2001;21:4700–4712. doi: 10.1128/MCB.21.14.4700-4712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu Y, Gong W, Huang CC, Herr W, Cheng X. Crystal structure of the conserved core of the herpes simplex virus transcriptional regulatory protein VP16. Genes Dev. 1999;13:1692–1703. doi: 10.1101/gad.13.13.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lai JS, Cleary MA, Herr W. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeo domain. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- [10].Kristie TM, Pomerantz JL, Twomey TC, Parent SA, Sharp PA. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- [11].Kristie TM, Sharp PA. Purification of the cellular C1 factor required for the stable recognition of the Oct-1 homeodomain by the herpes simplex virus alpha-trans-induction factor (VP16) J Biol Chem. 1993;268:6525–6534. [PubMed] [Google Scholar]
- [12].Wilson AC, LaMarco K, Peterson MG, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- [13].Jones KA, Tjian R. Sp1 binds to promoter sequences and activates herpes simplex virus ‘immediate-early’ gene transcription in vitro. Nature. 1985;317:179–182. doi: 10.1038/317179a0. [DOI] [PubMed] [Google Scholar]
- [14].LaMarco KL, McKnight SL. Purification of a set of cellular polypeptides that bind to the purine-rich cis-regulatory element of herpes simplex virus immediate early genes. Genes Dev. 1989;3:1372–1383. doi: 10.1101/gad.3.9.1372. [DOI] [PubMed] [Google Scholar]
- [15].Triezenberg SJ, LaMarco KL, McKnight SL. Evidence of DNA: protein interactions that mediate HSV-1 immediate early gene activation by VP16. Genes Dev. 1988;2:730–742. doi: 10.1101/gad.2.6.730. [DOI] [PubMed] [Google Scholar]
- [16].Nogueira ML, Wang VEH, Tantin D, Sharp PA, Kristie TM. Herpes simplex virus infections are arrested in Oct-1-deficient cells. PNAS. 2004;101:1473–1478. doi: 10.1073/pnas.0307300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Narayanan A, Nogueira ML, Ruyechan WT, Kristie TM. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J Biol Chem. 2005;280:1369–1375. doi: 10.1074/jbc.M410178200. [DOI] [PubMed] [Google Scholar]
- [18].Freiman RN, Herr W. Viral mimicry: common mode of association with HCF by VP16 and the cellular protein LZIP. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hughes TA, La Boissiere S, O’Hare P. Analysis of functional domains of the host cell factor involved in VP16 complex formation. J Biol Chem. 1999;274:16437–16443. doi: 10.1074/jbc.274.23.16437. [DOI] [PubMed] [Google Scholar]
- [20].Wilson AC, Freiman RN, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lu R, Yang P, Padmakumar S, Misra V. The herpesvirus transactivator VP16 mimics a human basic domain leucine zipper protein, luman, in its interaction with HCF. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Simmen KA, Newell A, Robinson M, Mills JS, Canning G, Handa R, Parkes K, Borkakoti N, Jupp R. Protein interactions in the herpes simplex virus type 1 VP16-induced complex: VP16 peptide inhibition and mutational analysis of host cell factor requirements. J Virol. 1997;71:3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mahajan SS, Wilson AC. Mutations in host cell factor 1 separate its role in cell proliferation from recruitment of VP16 and LZIP. Mol Cell Biol. 2000;20:919–928. doi: 10.1128/mcb.20.3.919-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lu R, Yang P, O’Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lu R, Misra V. Potential role for luman, the cellular homologue of herpes simplex virus VP16 (alpha gene trans-inducing factor), in herpesvirus latency. J Virol. 2000;74:934–943. doi: 10.1128/jvi.74.2.934-943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Akhova O, Bainbridge M, Misra V. The neuronal host cell factor-binding protein Zhangfei inhibits herpes simplex virus replication. J Virol. 2005;79:14708–14718. doi: 10.1128/JVI.79.23.14708-14718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lu R, Misra V. Zhangfei: a second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 2000;28:2446–2454. doi: 10.1093/nar/28.12.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vogel JL, Kristie TM. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. Embo J. 2000;19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gunther M, Laithier M, Brison O. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol Cell Biochem. 2000;210:131–142. doi: 10.1023/a:1007177623283. [DOI] [PubMed] [Google Scholar]
- [31].Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci U S A. 2007;104:10835–10840. doi: 10.1073/pnas.0704351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Vogel JL, Kristie TM. Site-specific proteolysis of the transcriptional coactivator HCF-1 can regulate its interaction with protein cofactors. Proc Natl Acad Sci U S A. 2006;103:6817–6822. doi: 10.1073/pnas.0602109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wilson AC, Peterson MG, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- [34].Julien E, Herr W. Proteolytic processing is necessary to separate and ensure proper cell growth and cytokinesis functions of HCF-1. Embo J. 2003;22:2360–2369. doi: 10.1093/emboj/cdg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Luciano RL, Wilson AC. An activation domain in the C-terminal subunit of HCF-1 is important for transactivation by VP16 and LZIP. Proc Natl Acad Sci U S A. 2002;99:13403–13408. doi: 10.1073/pnas.202200399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Luciano RL, Wilson AC. HCF-1 functions as a coactivator for the zinc finger protein Krox20. J Biol Chem. 2003;278:51116–51124. doi: 10.1074/jbc.M303470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wilson AC, Boutros M, Johnson KM, Herr W. HCF-1 amino- and carboxy-terminal subunit association through two separate sets of interaction modules: involvement of fibronectin type 3 repeats. Mol Cell Biol. 2000;20:6721–6730. doi: 10.1128/mcb.20.18.6721-6730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].La Boissiere S, Hughes T, O’Hare P. HCF-dependent nuclear import of VP16. Embo J. 1999;18:480–489. doi: 10.1093/emboj/18.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- [41].Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- [43].Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- [45].Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Oh J, Fraser NW. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J Virol. 2008;82:3530–3537. doi: 10.1128/JVI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pignatti PF, Cassai E. Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. J Virol. 1980;36:816–828. doi: 10.1128/jvi.36.3.816-828.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Arnosti DN, Preston CM, Hagmann M, Schaffner W, Hope RG, Laughlan G, Luisi BF. Specific transcriptional activation in vitro by the herpes simplex virus protein VP16. Nucleic Acids Res. 1993;21:5570–5576. doi: 10.1093/nar/21.24.5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Uhlmann T, Boeing S, Lehmbacher M, Meisterernst M. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J Biol Chem. 2007;282:2163–2173. doi: 10.1074/jbc.M608451200. [DOI] [PubMed] [Google Scholar]
- [50].Yang F, DeBeaumont R, Zhou S, Naar AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Yang M, Hay J, Ruyechan WT. Varicella-zoster virus IE62 protein utilizes the human mediator complex in promoter activation. J Virol. 2008;82:12154–12163. doi: 10.1128/JVI.01693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Perera LP. The TATA motif specifies the differential activation of minimal promoters by varicella zoster virus immediate-early regulatory protein IE62. J Biol Chem. 2000;275:487–496. doi: 10.1074/jbc.275.1.487. [DOI] [PubMed] [Google Scholar]
- [53].Herrera FJ, Triezenberg SJ. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J Virol. 2004;78:9689–9696. doi: 10.1128/JVI.78.18.9689-9696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Huang J, Kent JR, Placek B, Whelan KA, Hollow CM, Zeng PY, Fraser NW, Berger SL. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J Virol. 2006;80:5740–5746. doi: 10.1128/JVI.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kent JR, Zeng PY, Atanasiu D, Gardner J, Fraser NW, Berger SL. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J Virol. 2004;78:10178–10186. doi: 10.1128/JVI.78.18.10178-10186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Placek BJ, Huang J, Kent JR, Dorsey J, Rice L, Fraser NW, Berger SL. The histone variant H3.3 regulates gene expression during lytic infection by Herpes Simplex Virus, HSV-1. J Virol. 2008 doi: 10.1128/JVI.01276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks α-herpesvirus lytic replication and reactivation from latency. Manuscript in review. 2009 doi: 10.1038/nm.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Agger K, Christensen J, Cloos PA, Helin K. The emerging functions of histone demethylases. Curr Opin Genet Dev. 2008 doi: 10.1016/j.gde.2007.12.003. [DOI] [PubMed] [Google Scholar]
- [59].Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- [60].Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- [61].Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- [62].Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, Liu F, Taylor H, Lozach J, Jayes FL, Korach KS, Glass CK, Fu XD, Rosenfeld MG. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- [63].Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- [64].Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- [65].Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju BG, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu XD, Rosenfeld MG. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Metzger E, Wissmann M, Schule R. Histone demethylation and androgen-dependent transcription. Curr Opin Genet Dev. 2006;16:513–517. doi: 10.1016/j.gde.2006.08.013. [DOI] [PubMed] [Google Scholar]
- [67].Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- [68].Wissmann M, Yin N, Muller JM, Greschik H, Fodor BD, Jenuwein T, Vogler C, Schneider R, Gunther T, Buettner R, Metzger E, Schule R. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- [69].Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- [70].Opel M, Lando D, Bonilla C, Trewick SC, Boukaba A, Walfridsson J, Cauwood J, Werler PJ, Carr AM, Kouzarides T, Murzina NV, Allshire RC, Ekwall K, Laue ED. Genome-wide studies of histone demethylation catalysed by the fission yeast homologues of mammalian LSD1. PLoS ONE. 2007;2:e386. doi: 10.1371/journal.pone.0000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135:604–607. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- [72].Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- [74].Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Guelman S, Suganuma T, Florens L, Swanson SK, Kiesecker CL, Kusch T, Anderson S, Yates JR, 3rd, Washburn MP, Abmayr SM, Workman JL. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol Cell Biol. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- [78].Kolb G, Kristie TM. Association of the cellular coactivator HCF-1 with the Golgi apparatus in sensory neurons. J Virol. 2008;82:9555–9563. doi: 10.1128/JVI.01174-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kristie TM, Vogel JL, Sears AE. Nuclear localization of the C1 factor (host cell factor) in sensory neurons correlates with reactivation of herpes simplex virus from latency. Proc Natl Acad Sci U S A. 1999;96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Whitlow Z, Kristie TM. Recruitment of the transcriptional coactivator HCF-1 to viral immediate early promoters during initiation of HSV-1 reactivation from latency. J Virol. 2009 doi: 10.1128/JVI.01115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Deshmane SL, Fraser NW. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J Virol. 1989;63:943–947. doi: 10.1128/jvi.63.2.943-947.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Amelio AL, Giordani NV, Kubat NJ, O’Neil J E, Bloom DC. Deacetylation of the herpes simplex virus type 1 latency-associated transcript (LAT) enhancer and a decrease in LAT abundance precede an increase in ICP0 transcriptional permissiveness at early times postexplant. J Virol. 2006;80:2063–2068. doi: 10.1128/JVI.80.4.2063-2068.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Amelio AL, McAnany PK, Bloom DC. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol. 2006;80:2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Chen Q, Lin L, Smith S, Huang J, Berger SL, Zhou J. CTCF-dependent chromatin boundary element between the latency-associated transcript and ICP0 promoters in the herpes simplex virus type 1 genome. J Virol. 2007;81:5192–5201. doi: 10.1128/JVI.02447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Coleman HM, Connor V, Cheng ZS, Grey F, Preston CM, Efstathiou S. Histone modifications associated with herpes simplex virus type 1 genomes during quiescence and following ICP0-mediated de-repression. J Gen Virol. 2008;89:68–77. doi: 10.1099/vir.0.83272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Giordani NV, Neumann DM, Kwiatkowski DL, Bhattacharjee PS, McAnany PK, Hill JM, Bloom DC. During herpes simplex virus type 1 infection of rabbits, the ability to express the latency-associated transcript increases latent-phase transcription of lytic genes. J Virol. 2008;82:6056–6060. doi: 10.1128/JVI.02661-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kubat NJ, Amelio AL, Giordani NV, Bloom DC. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J Virol. 2004;78:12508–12518. doi: 10.1128/JVI.78.22.12508-12518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kubat NJ, Tran RK, McAnany P, Bloom DC. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J Virol. 2004;78:1139–1149. doi: 10.1128/JVI.78.3.1139-1149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Neumann DM, Bhattacharjee PS, Giordani NV, Bloom DC, Hill JM. In vivo changes in the patterns of chromatin structure associated with the latent herpes simplex virus type 1 genome in mouse trigeminal ganglia can be detected at early times after butyrate treatment. J Virol. 2007;81:13248–13253. doi: 10.1128/JVI.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wang QY, Zhou C, Johnson KE, Colgrove RC, Coen DM, Knipe DM. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc Natl Acad Sci U S A. 2005;102:16055–16059. doi: 10.1073/pnas.0505850102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nat Rev Microbiol. 2008;6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- [92].Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, Songyang Z, Thomson JA, Zwaka TP. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–1174. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Delehouzee S, Yoshikawa T, Sawa C, Sawada J, Ito T, Omori M, Wada T, Yamaguchi Y, Kabe Y, Handa H. GABP, HCF-1 and YY1 are involved in Rb gene expression during myogenesis. Genes Cells. 2005;10:717–731. doi: 10.1111/j.1365-2443.2005.00873.x. [DOI] [PubMed] [Google Scholar]
- [94].Knez J, Piluso D, Bilan P, Capone JP. Host cell factor-1 and E2F4 interact via multiple determinants in each protein. Mol Cell Biochem. 2006;288:79–90. doi: 10.1007/s11010-006-9122-x. [DOI] [PubMed] [Google Scholar]
- [95].Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–119. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- [96].Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- [97].Vercauteren K, Gleyzer N, Scarpulla RC. PGC-1-related coactivator complexes with HCF-1 and NRF-2beta in mediating NRF-2(GABP)-dependent respiratory gene expression. J Biol Chem. 2008;283:12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- [99].Reilly PT, Herr W. Spontaneous reversion of tsBN67 cell proliferation and cytokinesis defects in the absence of HCF-1 function. Exp Cell Res. 2002;277:119–130. doi: 10.1006/excr.2002.5551. [DOI] [PubMed] [Google Scholar]
- [100].Reilly PT, Wysocka J, Herr W. Inactivation of the retinoblastoma protein family can bypass the HCF-1 defect in tsBN67 cell proliferation and cytokinesis. Mol Cell Biol. 2002;22:6767–6778. doi: 10.1128/MCB.22.19.6767-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Ajuh P, Chusainow J, Ryder U, Lamond AI. A novel function for human factor C1 (HCF-1), a host protein required for herpes simplex virus infection, in pre-mRNA splicing. Embo J. 2002;21:6590–6602. doi: 10.1093/emboj/cdf652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Khurana B, Kristie TM. A protein sequestering system reveals control of cellular programs by the transcriptional coactivator HCF-1. J Biol Chem. 2004;279:33673–33683. doi: 10.1074/jbc.M401255200. [DOI] [PubMed] [Google Scholar]
- [103].Julien E, Herr W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell. 2004;14:713–725. doi: 10.1016/j.molcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- [104].Piluso D, Bilan P, Capone JP. Host cell factor-1 interacts with and antagonizes transactivation by the cell cycle regulatory factor Miz-1. J Biol Chem. 2002;277:46799–46808. doi: 10.1074/jbc.M206226200. [DOI] [PubMed] [Google Scholar]
- [105].Lee S, Horn V, Julien E, Liu Y, Wysocka J, Bowerman B, Hengartner MO, Herr W. Epigenetic Regulation of Histone H3 Serine 10 Phosphorylation Status by HCF-1 Proteins in C. elegans and Mammalian Cells. PLoS ONE. 2007;2:e1213. doi: 10.1371/journal.pone.0001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, Berger SL. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- [107].Pless O, Kowenz-Leutz E, Knoblich M, Lausen J, Beyermann M, Walsh MJ, Leutz A. G9a-mediated lysine methylation alters the function of CCAAT/enhancer-binding protein-beta. J Biol Chem. 2008;283:26357–26363. doi: 10.1074/jbc.M802132200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Rathert P, Dhayalan A, Murakami M, Zhang X, Tamas R, Jurkowska R, Komatsu Y, Shinkai Y, Cheng X, Jeltsch A. Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol. 2008;4:344–346. doi: 10.1038/nchembio.88. [DOI] [PMC free article] [PubMed] [Google Scholar]