Abstract
Our previous studies showed that establishment of murine cytomegalovirus (MCMV) latency in vivo is associated with repression of immediate-early gene expression, deacetylation of histones bound to the major immediate-early promoter (MIEP), changes in patterns of methylation of histones, and recruitment of cellular repressors of transcription to the MIEP. Here, we have quantitatively analyzed the kinetics of changes in viral RNA expression, DNA copy number, and recruitment of repressors and activators of transcription to viral promoters during the course of infection. Our results show that changes in viral gene expression correlate with changes in recruitment of RNA polymerase and acetylated histones to viral promoters. Binding of the transcriptional repressors histone deacetylase type 2 (HDAC2), HDAC3, YY1, CBF-1/RBP-Jk, Daxx, and CIR to the MIEP and HDACs to other promoters showed a biphasic pattern: some binding was detectable prior to activation of viral gene expression, then decreased with the onset of transcription and increased again as repression of viral gene expression occurred. Potential binding sites for CBF-1/RBP-Jk and YY1 in the MIEP and for YY1 in the M100 promoter (M100P) were identified by in silico analysis. While recruitment of HDACs was not promoter specific, binding of CBF-1/RBP-Jk and YY1 was restricted to promoters with their cognate sites. Our results suggest that sequences within viral promoters may contribute to establishment of latency through recruitment of transcriptional repressors to these genes. The observation that repressors are bound to the MIEP and other promoters immediately upon infection suggests that latency may be established in some cells very early in infection.
Cytomegalovirus (CMV), like other herpesviruses, has the ability to establish a lifelong latent infection. Reactivation from latency, characterized by shedding of virus, occurs frequently and asymptomatically in immunocompetent hosts. In immunocompromised recipients of solid organs and bone marrow transplants, reactivation of CMV is associated with increased risk of acute and chronic allograft rejection, opportunistic infection, graft failure, and patient mortality (54). CMV infection of immunologically immature fetuses in utero is associated with hearing loss, cognitive impairment, cerebral palsy, and visual impairment (16). Thus, CMV is an important opportunistic human pathogen, and an understanding of the mechanisms by which CMV establishes latent infection and reactivates from latency is essential in developing new tools to combat viral infection and its sequelae.
CMV latency has been defined as the absence of infectious virus, despite the presence of viral DNA (56). However, the molecular mechanisms by which latency is established and maintained have not been clear. Immune control of cells expressing viral genes associated with productive infection plays an important role in preventing recurrence of infectious virus. However, transcriptional control of viral gene expression is also thought to be important in controlling viral latency and reactivation. Studies with HCMV have suggested that latency is established through repression of ie-1 and ie-2 gene expression (4, 58). The IE-1 and IE-2 proteins, which are among the first proteins expressed by the virus during productive infection, are transcriptional regulatory proteins that are required for induction of early and late gene expression, viral DNA synthesis, and production of infectious virus (46). Transcription of IE-1 and IE-2 RNAs, which are differentially spliced transcripts initiated at the major immediate-early (ie) promoter (MIEP), is regulated by the MIE enhancer (6, 13). Previous studies have shown that in latently infected cells, the MIEP is bound to deacetylated histones and cellular repressors of transcription, suggesting that latency is established through recruitment of these factors to the MIEP (59). However, studies of HCMV latency in naturally infected cells have been very limited, due to the low frequency of latently infected cells and the inability to manipulate the virus during infection in its host. Thus, it has not been possible to determine the mechanisms by which these factors are recruited to the MIEP.
To circumvent these difficulties, we and others have used murine CMV (MCMV), which is similar to HCMV with respect to genome organization, regulation of transcription, pathogenesis, and ability to establish latent infection and to reactivate, as a model system to study CMV latency in vivo. Studies of HCMV latency have shown that it establishes latent infection in cells of the monocyte lineage and have suggested that, within this lineage, there may be cell-type-specific differences in the ability to establish latency (58). A molecular basis for these differences has not been determined. Because of the lack of availability of other tissues and the low copy number, there have been very few studies that have addressed other sites of HCMV latency. However, in the MCMV model, it is clear that the virus establishes latent infection in multiple organs (5, 31, 44, 51, 64). We previously showed that ie gene expression is repressed during latent infection in mouse kidneys (25, 40). Establishment of latency was also correlated with deacetylation of histones bound to the major immediate-early promoter, with changes in patterns of methylation of histones, and with recruitment of cellular repressors of transcription to the MIEP (40). These changes were specific to viral chromatin, as viral infection had no effect on modifications of histones bound to the promoters of the β-actin and Ant4 cellular genes. These studies support the hypothesis that latency is established through repression of the MIEP but left open the important question of how these repressors are recruited. In order to further investigate this question, we have now quantitatively analyzed viral RNA expression, DNA copy number, changes in histone modifications, and recruitment of transcriptional repressors to the MIEP and other viral promoters at various times postinfection. Our results show that viral promoters in addition to the MIEP are bound to repressors at the very earliest stage of infection. The proportion of genomes bound to these repressors decreased as binding of RNA polymerase and acetylated histones to promoters and activation of viral gene expression occurred, and it subsequently increased when latency was established. Some repressors were recruited in a promoter-specific fashion, suggesting that sequences in the promoters could be involved in the establishment of latency.
MATERIALS AND METHODS
Mice and virus.
Three-week-old, female, specific-pathogen-free BALB/c mice were purchased from Jackson Laboratory, Bar Harbor, ME. Mice were maintained in isolation cages and fed and watered ad libitum. MCMV (Smith strain, ATCC 194-VR) was purchased from the American Type Culture Collection (Rockville, MD). Virus stocks were generated by propagation in salivary glands as previously described (31). Mice were infected by intraperitoneal injection with 105 PFU of MCMV and sacrificed at various times postinfection. This study protocol was reviewed and approved by the Northwestern University Institutional Animal Care and Use Committee.
Nucleic acid extraction.
Kidney DNA was prepared by using a Puregene tissue kit (Gentra, Minneapolis, MN) according to the manufacturer's instruction. Total RNA was isolated and purified with a TRIzol Plus RNA purification kit (Invitrogen, Carlsbad, CA). On-column DNase Ι digestion was performed during RNA purification. DNA and RNA were quantified by A260 with a DU640B spectrophotometer (Beckman Coulter, Fullerton, CA).
Reverse transcription (RT).
Oligo(dT)-primed cDNA was generated from 5 μg of total RNA by using an AffinityScript multiple temperature cDNA synthesis kit (Stratagene, La Jolla, CA) in a 20-μl reaction mixture. Reverse transcriptase was inactivated by heating at 70°C for 15 min. Samples were cooled to 37°C and treated for 30 min with 0.15 U/μl RNaseH (Invitrogen) and 1 μl RNase cocktail (Ambion, Austin, TX). Subsequently, the cDNA was purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA) and quantified using a Quant-iT Oligreen single-stranded DNA (ssDNA) assay kit (Invitrogen).
Real-time PCR analysis.
All real-time PCRs were performed using TaqMan gene expression master mix in an ABI 7500 Fast system using the standard curve (absolute quantitation) assay and regular 7500 mode. Primers and TaqMan minor groove binder (MGB) probes described in Table 1 were designed by ABI or by using Primer Express 3.0 software. Reaction mixtures contained 900 nM primers and 250 nM TaqMan MGB probes. Each sample was analyzed in triplicate. The thermal cycling conditions were 50°C for 2 min and 95°C for 10 min followed by 50 cycles of 95°C for 15 s and 60°C for 1 min.
TABLE 1.
List of primers and probes
| Target region | Sequence | Accession no. (nucleotide no.) |
|---|---|---|
| Mouse β-actin promoter | 5′CGTTCCGAAAGTTGCCTTTTA 3′ (forward) | DD173001 (429-489) |
| 5′GCCGCCGGGTTTTATAGG 3′ (reverse) | ||
| 5′CTCGAGTGGCCGCTG 3′ (probe) | ||
| Mouse Ant4 promoter | 5′CAGGCTAGTGTCTGCACCTG 3′ (forward) | AC146980 (102991-103117) |
| 5′ACCAACCCGGTGATTAACTG 3′ (reverse) | ||
| 5′ATTACAGGCGTGCAGCATCT 3′ (probe) | ||
| Mouse RpL30 gene YY1 | 5′GGCTGGTGTTGGTGAGTGA 3′ (forward) | K02928 (489-599) |
| binding region | 5′ACACAGAGGACAGAAGAGAGGATT 3′ (reverse) | |
| 5′CCAGAGCGTCAAACAC 3′ (probe) | ||
| Mouse HES1 gene RBP-Jk | 5′GGCCTGCGGATCACACA 3′ (forward) | D16464 (188-262) |
| binding region | 5′GGACCAAGGAGAGAGGTAGACA 3′ (reverse) | |
| 5′CACCAGCTCCAGATCC 3′ (probe) | ||
| Mouse Eef2 mRNA | 5′CCACGGGCCTGAAGGT 3′ (forward) | NM_007907.1(1345-1417) |
| 5′GGCTTCAGGTATAGGTCCTCTTTC 3′ (reverse) | ||
| 5′ATGGGCCCCAACTAC 3′ (probe) | ||
| Mouse EeF1α promoter | 5′CGGGTTTGCCGTCAGAAC 3′ (forward) | AC158987 (121163-121239) |
| 5′GCTCGGAGCAGGACCTC 3′ (reverse) | ||
| 5′CCACACCCGCCCCTC 3′ (probe) | ||
| MCMV MIEP | 5′GGTGGTCAGACCGAAGACT 3′ (forward) | U68299 (182873-182944) |
| 5′GCTGAGCTGCGTTCTACGT 3′ (reverse) | ||
| 5′CTGGTCGCGCCTCTTA 3′ (probe) | ||
| MCMV ie-1 mRNA | 5′CAACAGCGGCAGCTTCTTC 3′ (forward) | U68299 (179823-179884) |
| 5′CATGGCGTACTGCCTCTTGA 3′ (reverse) | ||
| 5′ACTGCTCCTCGCCC 3′ (probe) | ||
| MCMV ie-3 mRNA | 5′CAGCCGGAGGCTACGT 3′ (forward) | U68299 (178572-178641) |
| 5′GCTGGGTACTTCCTGGACTTATC 3′ (reverse) | ||
| 5′TTCCCTGCACTTCTTG 3′ (probe) | ||
| IE intron 1 | 5′CCTGTGTTCACACACCAGATATTACA 3′ (forward) | U68299 (182115-182204) |
| 5′GCAGCCTCTGAGTACAATATTCTGT 3′ (reverse) | ||
| 5′CCGCCATGATAAGTCC 3′ (probe) | ||
| MCMV M100 (gM) | 5′GGGTCAAATAGTGAGAGCATCCTT 3′ (forward) | U68299 (145392-145465) |
| promoter | 5′GAAGCGGCGACGTTACC 3′ (reverse) | |
| 5′CTCGGACGGTCTCTCT 3′ (probe) | ||
| MCMV M100 mRNA | 5′TCGCGCGACTCGTAGTG 3′ (forward) | U68299 (144804-144880) |
| 5′GCGTTCAGCAAGTGCATGTAC 3′ (reverse) | ||
| 5′CCTGACGGCCTTCGTG 3′ (probe) | ||
| MCMV M112 (e1) | 5′GCAGACCAAATGCTGATAGTTCCT 3′ (forward) | U68299 (163008-163072) |
| promoter | 5′GGTCGCGACGAGAAAAGTG 3′ (reverse) | |
| 5′TCGCGGTAGATTACG 3′ (probe) | ||
| MCMV M112 mRNA | 5′CGTCTGTAACAAGACCGTCTCTTC 3′ (forward) | U68299 (163423-163533) |
| 5′ACCAGGATGACTTGCAGCAA 3′ (reverse) | ||
| 5′CCGCCACCAGAGTCA 3′ (probe) |
Quantification of MCMV DNA and RNA by real-time PCR.
For absolute quantification of MCMV DNA copy number in kidney tissue, a standard curve was generated by serial dilution of plasmid pIE111 (45), which contains the MIEP and IE genes, in genomic DNA isolated from kidneys of uninfected mice, such that 13.5 μl of the template contained 3 × 106, 3 × 105, 3 × 104, 3 × 103, 3 × 102, or 3 × 101 copies of pIE111 in 800 ng cellular DNA. A standard curve for mouse Eef1α DNA was generated by serial dilution of plasmid pDRIVE-mEF1α (InvivoGene, San Diego, CA), which contains the promoter region of the mouse eukaryotic translation elongation factor 1 alpha (Eef1α) gene, such that 13.5 μl of the template contained 1 × 106, 1 × 105, 1 × 104, 1 × 103, 1 × 102, or 10 copies of pDRIVE-mEF1α in 800 ng sheared salmon sperm DNA. Sample genomic DNA for each reaction (800 ng) was used as a template for PCR in a 30-μl reaction mixture. Genomic DNA from uninfected mouse kidneys (800 ng) was used as a control for detection of nonspecific amplification. The primers and probes used to detect MCMV MIEP and Eef1α promoter DNA are listed in Table 1. The target DNA copy number in each sample was calculated from a standard curve generated in each individual assay. The viral DNA copy number per million cells was determined by dividing the average copy number of MCMV DNA by the Eef1α copy number in the same sample and multiplying by 2 × 106.
Quantification of MCMV IE-1, IE-3, M112, and M100 mRNA was performed using the relative standard curve method as described in the Applied Biosystems support material “Chemistry Guide: Applied Biosystems Real-Time PCR Systems” (http://www.appliedbiosystems.com/support/apptech/, part no. 4348358) with the primers and probes specific to each gene (Table 1). Serially diluted cDNA derived from RNA isolated from kidneys at 5 days postinfection was used to generate the standard curve. Sample cDNA (10 ng) was added to a 20-μl reaction mixture for amplification of virus cDNA; the same amount of cDNA from uninfected mouse kidneys was used as a control for nonspecific amplification. The abundance of the target cDNA in each sample was determined on the basis of a standard curve produced in each run. Mouse eukaryotic Eef2 RNA was employed as an internal control. The relative quantity of MCMV RNA in the sample was determined by dividing the average quantity of MCMV mRNA by that of Eef2, as determined from the standard curve. As a control to detect contamination of the cDNA by genomic DNA, we analyzed cDNAs for the presence of the intronic region between exons 1 and 2 in the immediate-early gene 1/3.
Chromatin immunoprecipitation (ChIP).
Chromatin immunoprecipitation assays were performed essentially as described previously (40). Antibodies against histone H4 (pan), acetyl histone H4 (Lys5, Lys8, Lys12, Lys16), and Daxx were obtained from Millipore Corporation, Billerica, MA. Antibodies for histone deacetylase type 2 (HDAC2) and HDAC3 were purchased from Sigma-Aldrich Inc., St. Louis, MO. Antibodies specifc for RNA polymerase ΙΙ (N-20) and CBF-1/RBP-Jk(H-50) were obtained from Santa Cruz Biotechnology, Santa Cruz, CA. Anti-YY1 and anti-CIR were purchased from Abcam, Cambridge, MA, and Novus Biologicals, Littleton, CO, respectively.
Quantitative analysis of ChIP DNA samples by real-time PCR.
The relative amount of MCMV MIEP DNA in each ChIP sample was determined using the relative standard curve method. Serial dilutions of sheared genomic DNA purified from acutely infected mouse kidneys were used to generate the standard curve. The same set of standards was used for all analyses. For analysis of total histone H4, HDACs, RNA Pol II, YY1, CBF-1/RBP-Jk, Daxx, and CIR, the results are expressed as the percentage of immunoprecipitated chromatin relative to the total input of chromatin in the immunoprecipitation mixture. For analysis of acetylated histone H4, the results are expressed as the ratio of the modified histone to corresponding total histone after normalization to input DNA. ChIP assays were analyzed in triplicate, and the results presented are the averages of results of two to four independent assays plus standard error.
Statistical analysis.
A two-tailed Student's t test was used to determine statistical significance. P values of ≤0.05 were considered significant.
RESULTS
Repression of MCMV ie gene expression in the kidney occurs between day 7 and day 10 postinfection.
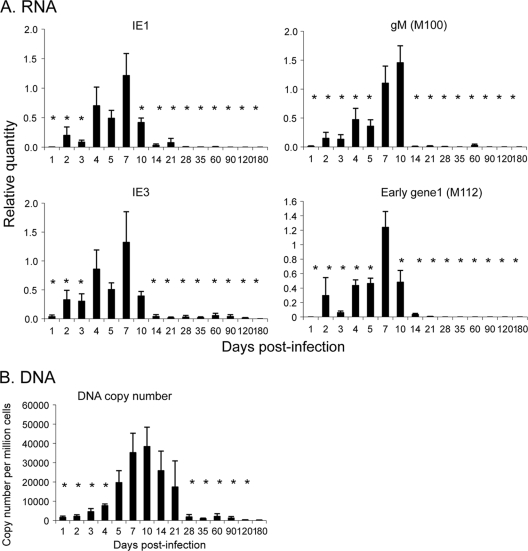
In order to understand the mechanism of establishment of latency, it is important to know when repression of viral gene expression occurs during the course of infection. We therefore examined the kinetics of expression of IE-1 and IE-3 RNAs and accumulation of viral DNA during acute infection in the kidney. IE-1 and IE-3 RNAs were analyzed by real-time PCR analysis of purified cDNA using the relative standard curve method as described in Materials and Methods. The intron region between exons 1 and 2 was analyzed as a control for genomic DNA contamination of the cDNA. DNA and RNA from uninfected mice were used as controls for nonspecific amplification. These controls were negative (data not shown). Our results show that expression of IE-1 RNA was detectable at day 2 postinfection (p.i.), peaked at day 7, and dropped significantly at day 10 relative to the peak at day 7 (Fig. 1A). Expression of IE-3, which is transcribed from the same promoter as IE-1, followed the same trend as expression of IE-1.
FIG. 1.
Kinetics of viral gene expression (A) and DNA copy number (B) in kidneys of MCMV-infected mice. RNA and DNA were isolated from each mouse at various times postinfection (n = 6 to 10/time point). (A) Relative quantification of MCMV RNA was performed as described in Materials and Methods. Results are shown as the normalized average quantity plus standard error. *, P ≤ 0.05 relative to the peak of expression (day 7 for IE-1, IE-3, and M112; day 10 for M100). (B) Absolute quantification of MCMV DNA copy number, determined as described in Materials and Methods. The data are expressed as the average number of copies per million cells plus standard error. *, P ≤ 0.05 relative to the result on day 10.
MCMV DNA was detectable in the kidney within 1 day p.i. but did not accumulate significantly above this level until day 4. In contrast to IE RNA expression, which peaked at day 7, MCMV DNA copy number peaked at day 10 and fell gradually until day 28, where it plateaued to levels seen in latently infected mice (Fig. 1B). The decrease in DNA copy number likely reflects clearance of infected cells by the host immune response. However, our results show that loss of ie gene expression precedes the decrease in DNA copy number and is therefore not due simply to clearance of infected cells. These data demonstrate that repression of IE RNA expression in the kidneys of infected mice occurs between day 7 and day 10.
CMV gene expression during productive infection occurs in a cascade fashion typical of other herpesviruses, in which the immediate-early genes induce expression of the early genes, followed by DNA replication and expression of the late genes. We therefore analyzed expression of M112 (e1) and M100 (gM), which are representative of the early and late genes, respectively. The kinetics of expression of M112 RNA was indistinguishable from that of the immediate-early genes during acute infection in vivo (Fig. 1A). In contrast, relatively high levels of M100 RNA were still observed at day 10 p.i., when expression of IE RNA had fallen, but DNA copy number was still high. Interestingly, expression of this RNA was rapidly lost after day 10, despite continued presence of the DNA (Fig. 1A and B). With the exception of results for one mouse at day 60, expression of M112 and M100 RNA was undetectable after day 28. Taken together, our results suggest that latency is established in the kidney by day 28 p.i.
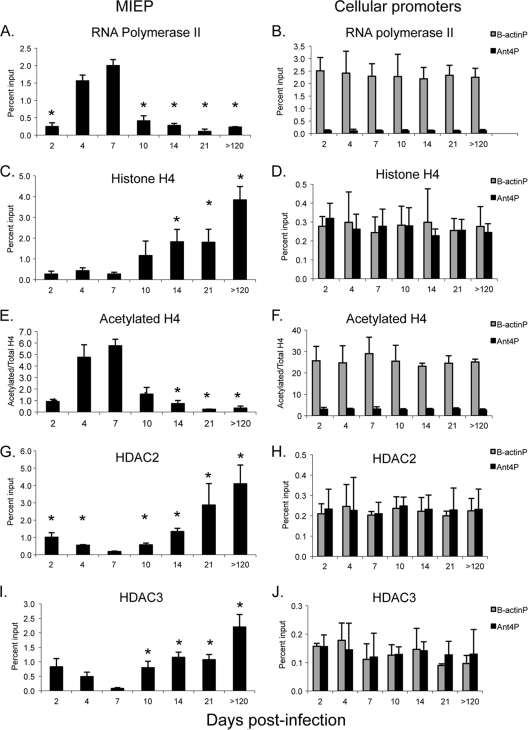
Transcriptional activation of ie gene expression correlates with recruitment of RNA polymerase and acetylated H4 to the MIEP.
We then examined binding of RNA polymerase II to the MIEP during acute infection (Fig. 2A). As expected, binding of RNA polymerase paralleled ie gene expression, with binding detectable at day 2, peaking at day 7, and falling significantly at day 10. In addition, we analyzed binding of RNA polymerase II to the β-actin and Ant4 cellular promoters as controls for transcriptionally active and inactive genes, respectively (Fig. 2B). β-Actin is widely expressed in all organs; The Ant4 gene is a developmentally regulated gene which is silenced through DNA methylation in adult mouse tissues (61). Binding of RNA polymerase to the β-actin promoter was observed at all time points at comparable levels. Thus, the changes in binding of RNA polymerase to the MIEP during the course of infection are not due to differences in the chromatin preparations. At the peak of infection, the percentage of MIEP DNA bound to RNA polymerase was similar to that of β-actin, indicating that the proportion of viral genomes being actively transcribed was similar to that of a highly active cellular gene and, thus, is biologically relevant. In contrast, very little binding of RNA polymerase to the transcriptionally inactive Ant4 promoter was observed, indicating the specificity of the binding to the MIEP that we observed.
FIG. 2.
Kinetics of recruitment of histones, HDACs, and RNA polymerase to the MIEP in kidneys of mice infected with MCMV. Chromatin from kidneys of infected mice was isolated at various times after infection and immunoprecipitated with the indicated antibodies. Immunoprecipitated DNA was amplified with MIEP-specific primers and probe (A, C, E, G, and I) or with primers and probe specific for the β-actin or Ant 4 promoters (B, D, F, H, and J), and the percentage of input DNA bound to antibody was calculated as described in Materials in Methods. n = 3 to 4/time point, except for day 21, where n = 2. Each sample was analyzed in triplicate. Results for panels A to D and G to H are shown are the mean percentage of input DNA bound to antibody plus standard error. Results for panels E and F are shown as the ratio of acetylated histone H4 to total H4, calculated from the percent input determined with antibodies specific to acetylated and total H4. The ratio is greater than 1 due to differences in the efficiencies of the antibodies. The horizontal axis indicates the days postinfection. *, P < 0.05 relative to the result on day 7.
We previously demonstrated that MCMV DNA is bound to histones during infection and that the percentage of genomes associated with histones rises dramatically in latent infection (40). Here, we have examined the kinetics of histone deposition to the MIEP (Fig. 2C). Our previous studies showed that binding of histone H4 to the MIEP was detectable during acute infection; here, we show that binding is detectable as early as day 2. Statistically significant accumulation of histones above this level was observed starting at day 14, when repression of ie gene expression was apparent (Fig. 1). At the peak of ie gene expression at day 7, binding of H4 to the MIEP was comparable to that of cellular genes (Fig. 2C and D). In contrast to cellular promoters, whose association with H4 did not change during the course of infection, binding of H4 to the MIEP increased more than 14-fold in latently infected mice compared to that in mice at 7 days postinfection.
Transcriptional activity of cellular genes is controlled by posttranslational modifications of histones bound to promoter regions (32). Acetylation of histone H4 is associated with active transcription, while deacetylation is associated with transcriptional repression. Because the percentage of genomes associated with histones changes during the course of infection, we analyzed the ratio of acetylated histones to total histones bound to the MIEP. Our results show that binding of acetylated histone H4 to the MIEP paralleled that of RNA polymerase, with a peak at day 7 and a significant drop at day 10 (Fig. 2E). In contrast, binding of acetylated H4 to the β-actin promoter, which was analyzed as a positive control, remained high throughout the course of infection (Fig. 2F). Very little binding of acetylated H4 to the Ant4 promoter, which was analyzed as a negative control, was observed at any time point.
Biphasic recruitment of repressors to the MIEP.
Deacetylation of histones is catalyzed by histone deacetylases (HDACs). Relative to the situation on day 7, binding of HDAC2 to the MIEP began to increase at day 10 and continued to increase as latency was established (Fig. 2G). Thus, binding of HDAC2 to the MIEP correlated inversely with binding of RNA polymerase and acetylated histone H4. Interestingly, binding of HDAC2 to the MIEP was initially observed at day 2 and day 4 and then fell at day 7 before increasing again at day 10. Binding of HDAC3 to the MIEP followed the same trend (Fig. 2I). As controls, we analyzed binding of HDACs to the β-actin and Ant4 promoters (Fig. 2H and J). As expected, little binding of HDACs to the transcriptionally active β-actin promoter was observed. We also observed little binding of HDACs to the transcriptionally silent Ant4 promoter. As we noted previously, this gene is silenced by DNA methylation, and its expression is apparently not regulated by histone deacetylation (40, 61). No changes in the levels of HDACs bound to cellular promoters were observed at different time points, indicating that differences in binding of HDACs to the MIEP were not due to differences in chromatin preparations.
Repression of ie gene expression is likely to occur through interaction of repressive factors in addition to HDACs with the MIEP. Recruitment of these factors may occur directly through recognition of specific DNA sequences in the promoter/enhancer region or indirectly through interaction with other factors. We used MatInspector (release 8.01; Genomatix) (9) to identify potential binding sites for repressive transcription factors in the MCMV enhancer. This analysis identifies sites using position weight matrices, in which every nucleotide in the sequence is assigned a score based on its conservation with other residues at that position in the database and, thus, accounts for the fact that some positions are more highly conserved than others (73). The algorithm defines a short, highly conserved core sequence common to all sequences in the database and calculates a conservation score (Ci value) at each position of the matrix. This is used to identify sequences in the gene of interest with perfect matches for the core sequence and calculates the similarity of the sequence to the matrix. A matrix score of 1 indicates that the test sequence corresponds to the most conserved nucleotide at each position of the matrix. Many of the sites in the MIEP identified by MatInspector are sequences recognized by positively acting transcription factors, such as NF-κB, AP-1, Sp1, and CREB, which may have a role in driving IE gene expression during productive infection or during reactivation from latency.
Of particular interest to this study, we identified 10 sites with high matrix similarity to known CBF-1/RBP-Jk sites and 12 sites with high matrix similarity to known YY1 sites (Table 2). YY1 is a DNA-binding transcription factor (26, 30) that acts as a repressor of some promoters and an activator of others (17, 66). Previous studies have shown that YY1 activates the ribosomal protein L30 (RpL30) promoter and represses the HCMV enhancer (19, 39). We previously demonstrated that YY1 is bound to the MIEP in kidneys of latently infected mice (40). CBF1/RBP-Jk is a component of the Notch signaling pathway (reviewed in references 7 and 28). It is constitutively bound to its cognate sites and, in the absence of Notch ligands (Jagged and Delta-like), acts as a repressor of transcription through recruitment of HDACs and other corepressors, including the CBF-1/RBP-Jk-associated corepressor CIR. CBF-1/RBP-Jk binds constitutively to the promoter of the developmental regulatory gene Hes1 (28). CBF-1/RBP-Jk and CIR have not been previously associated with regulation of CMV IE gene expression. Previous studies have also suggested that Daxx may have a role in silencing ie gene expression (29, 43, 62, 63, 78).
TABLE 2.
Potential CBF1/RBP-Jk and YY1 sites in the MIEP
| Site | Positiona | Strand | Sequenceb | Matrix similarity |
|---|---|---|---|---|
| CBF1/RBP-Jk | 183049-183063 | − | ttaaTGGGaaagtac | 0.959 |
| CBF1/RBP-Jk | 183067-183081 | + | gctaTGGGaaagtac | 0.961 |
| CBF1/RBP-Jk | 183128-183142 | + | atggTGGGaaagtac | 0.982 |
| CBF1/RBP-Jk | 183211-183225 | + | ataaTGGGaaaaacc | 0.946 |
| CBF1/RBP-Jk | 183396-183410 | + | gtaaTGGGaaaaacc | 0.946 |
| CBF1/RBP-Jk | 183588-183602 | − | tcaaTGGGaaaaacc | 0.947 |
| CBF1/RBP-Jk | 183838-183852 | − | ccaaTGGGaaaaacc | 0.947 |
| CBF1/RBP-Jk | 183926-183940 | + | gtaaTGGGaaaaacc | 0.946 |
| CBF1/RBP-Jk | 184002-184016 | + | atgcTGGGaaatggt | 0.944 |
| CBF1/RBP-Jk | 184111-184125 | + | cgcgTGGGaaattgg | 1.000 |
| YY1 | 182955-182973 | − | aaattCCATattggcacgc | 0.951 |
| YY1 | 183052-183072 | + | ctttcCCATtaatcagctatg | 0.894 |
| YY1 | 183202-183222 | − | ttttcCCATtattggcacgta | 0.851 |
| YY1 | 183220-183239 | + | aaaacCCATtgactcacccc | 0.842 |
| YY1 | 183387-183407 | − | ttttcCCATtactggcacgta | 0.852 |
| YY1 | 183405-183425 | + | aaaacCCATtggcttacctcc | 0.820 |
| YY1 | 183841-183861 | + | ttttcCCATtggctcacctcc | 0.841 |
| YY1 | 183856-183876 | + | acctcCCATtgacccaatgta | 0.842 |
| YY1 | 183935-183955 | + | aaaacCCATtggcttacctcc | 0.820 |
| YY1 | 183950-183970 | + | acctcCCATtgacccaatgta | 0.842 |
| YY1 | 183978-183998 | + | aaaggCCATtgagtcaccacc | 0.884 |
| YY1 | 184046-184066 | + | ggccgCCATtagagtgcatga | 0.923 |
MCMV genomic coordinates (53).
Capital letters indicate core sequence. Bold letters indicate Ci values of >60.
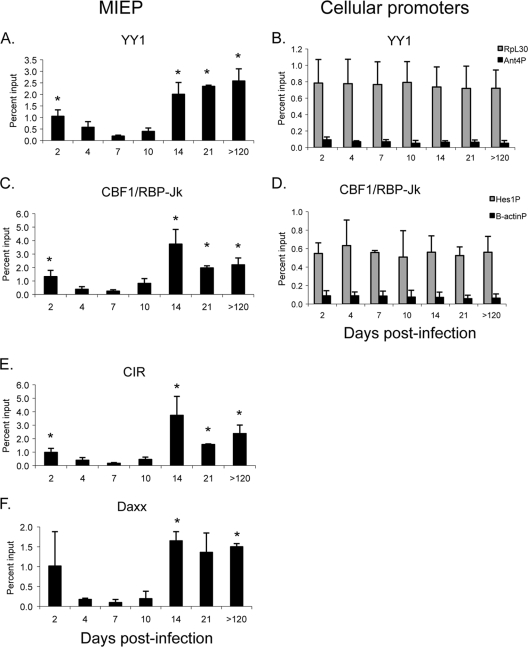
We therefore examined interaction of YY1, CBF-1/RBP-Jk, CIR, and Daxx with the MIEP as a function of time of infection (Fig. 3A, C, E, and F). Our results show that all of these repressors bind to the MIEP in latently infected mice with a pattern similar to that of HDACs. Recruitment of repressors to the MIEP was detectable as early as day 2 postinfection, prior to the activation of ie gene expression (Fig. 1). This was followed by a loss of binding as activation of ie gene expression occurred at days 4 to 7, with a nadir of binding at day 7, when transcription of the IE genes reached its peak. Relative to the findings for day 7, there was a statistically significant increase in the percentage of genomes bound to these factors at day 14, which did not increase further at later times. As positive controls, we analyzed binding of YY1 and CBF-1/RBP-Jk to the promoter regions of RpL30 and Hes1, respectively (Fig. 3B and D). In contrast to the MIEP, binding of these factors did not change during the course of infection, indicating that differences in binding of YY1 and CBF-1/RBP-Jk to the MIEP were not due to variability in the chromatin preparations. Furthermore, the level of binding to the MIEP in latently infected mice was similar to or higher than that observed with cellular promoters, indicating that the binding activity is likely to be biologically relevant. Very little binding to the negative controls (the Ant4 promoter for YY1 and the β-actin promoter for CBF-1/RBP-Jk) was observed, indicating the specificity of the interaction with the MIEP. Changes in the levels of binding of these factors to the MIEP are unlikely to be due to differences in the levels of expression, since real-time PCR analysis of YY1 and CBF-1 RNA levels and Western blot analysis of YY1 protein levels showed no change in expression of these genes over the course of infection (data not shown). Binding of CIR and Daxx to the MIEP followed the same biphasic pattern observed for YY1 and CBF-1/RBP-Jk (Fig. 3E and F). Collectively, our results demonstrate that (i) cellular repressors are recruited to the MIEP during infection and (ii) there are dramatic changes in the proportion of MIEP molecules bound to these repressors as the infection progresses from the very early stages of infection prior to activation of IE gene expression to acute infection and then to latency.
FIG. 3.
Biphasic recruitment of cellular repressors to the MIEP in kidneys of mice infected with MCMV. Chromatin from kidneys of infected mice was isolated at various times after infection and immunoprecipitated with the indicated antibodies. Immunoprecipitated DNA was amplified with primers and probe specific for the MIEP (A, C, E, and F), the RpL30 or Ant 4 promoters (B), or the Hes1 or β-actin promoters (D) and the percentage of input DNA bound to antibody was calculated as described in Materials and Methods. n = 2 to 4/time point. Each sample was analyzed in triplicate. Results shown are the mean percentage of input DNA bound to antibody plus standard error. The horizontal axis indicates the days postinfection. *, P < 0.05 relative to the result on day 7.
YY1 and CBF-1/RBP-Jk are specifically recruited to the MIEP in latent infection.
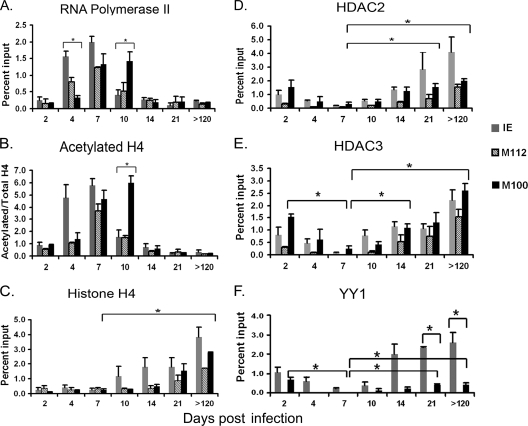
The observations that the MIEP contains sequences similar to binding sites for YY1 and CBF-1/RBP-Jk and that these factors are bound to the MIEP suggested that these factors might be recruited to the MIEP through direct recognition of sequences in the MIEP. To test this hypothesis further, we examined interaction of YY1 and CBF-1/RBP-Jk with the promoters for the early gene M112 and the late gene M100. In addition, we analyzed binding of RNA polymerase, histone H4, acetylated H4, HDAC2, and HDAC3 to these promoters. These data are shown in Fig. 4, with the results of ChIP analysis of the MIEP in Fig. 2 and 3 included for comparison. Our results show that at day 4, when viral gene expression is increasing, binding of RNA polymerase to the MIEP was significantly higher than binding to the M100 promoter (M100P), while at day 10, the converse was true (Fig. 4A). These changes in recruitment of RNA polymerase to the promoter correlate with changes in RNA expression (Fig. 1). Binding of acetylated histone H4 to these promoters followed the same trend (Fig. 4B). These results are consistent with the temporal regulation of gene expression typical of herpesviruses. Binding of H4 to the MIEP appeared to be higher than binding to other promoters at some time points, but these differences were not significant (Fig. 4C). In latently infected mice, binding of H4 to all viral promoters was approximately 10-fold higher than binding to cellular promoters (2 to 4% of input versus ∼0.3% of input [compare Fig. 2D and Fig. 4C]). These results suggest that, like that of HCMV (11, 18, 49), the entire MCMV genome is likely to be chromatinized.
FIG. 4.
Kinetics of recruitment of transcriptional regulatory factors to viral promoters. Chromatin from kidneys of infected mice was isolated at various times after infection and immunoprecipitated with the indicated antibodies. Immunoprecipitated DNA was amplified with primers and probe specific for the M112P or M100P as described in Materials and Methods, and the percentage of input DNA bound to antibody was calculated as described in Materials and Methods. The data for ChIP analysis of the MIEP in Fig. 2 and 3 are shown with analysis of M112P and M100P for comparison. n = 2 to 4/time point. Each sample was analyzed in triplicate. Results for panels A, C, D, E, and F shown are the mean percentage of input DNA bound to antibody plus standard error. Results for panel B are shown as the ratio of acetylated H4 to total H4. The horizontal axis indicates the days postinfection. *, P < 0.05. For the sake of clarity, not all statistically significant differences are indicated: significant differences in binding to the MIEP are shown in Fig. 2 and 3; significant differences in binding of RNA polymerase and acetylated H4 to the M100P and M112P and in binding of H4, HDAC2, and HDAC3 to M112P at different times are not shown.
Recruitment of HDAC2 and HDAC3 to the M100P was also observed, and the percentages of M100 DNA bound to HDACs at all time points followed the same trend observed with the MIEP (Fig. 4D and E). Binding of HDACs to the M112 promoter (M112P) appeared to be lower than binding to the MIEP and M100P at early times, but these differences were not significant. HDACs were bound at similar levels to all promoters analyzed in latently infected mice. Thus, in latently infected mice, deacetylated histones are likely to be distributed throughout the genome.
In contrast to HDACs, which do not bind to specific DNA sequences, we observed promoter-specific binding of YY1 and CBF-1/RBP-Jk. Using Genomatix analysis, we identified a potential YY1 binding site in the M100P, in addition to that in the MIEP. No YY1 sites were identified in the M112P. ChIP analysis of chromatin isolated from infected mice at various times showed that binding of YY1 to the M100P was detectable and that binding to this promoter followed the biphasic pattern observed with binding of YY1 to the MIEP, with initial binding very early in infection, followed by a statistically significant decrease at day 7, and an increased level in latently infected mice (Fig. 4F). However, the percentage of MIEP molecules bound to YY1 was significantly higher than the percentage bound to M100 during latent infection. In contrast, binding to M112P, which lacks potential YY1 sites, was negligible at all time points. We also examined binding of CBF-1/RBP-Jk to the M112 and M100 promoters, which lack CBF-1/RBP-Jk binding sites. In contrast to the results for the MIEP (Fig. 3C), very little interaction with these promoters was observed at any time point (data not shown). These observations support the hypothesis that binding of YY1 and CBF-1/RBP-Jk to the MIEP may be mediated by direct interaction with specific sequences within the promoter, and they suggest that these factors may play a role in recruitment of corepressor molecules and, thus, in repression of IE gene expression during latency.
DISCUSSION
In this study, we have analyzed the kinetics of viral RNA expression, DNA amplification, and recruitment of transcriptional repressors to viral promoters during the course of infection in vivo, as the transition is made from acute to latent infection. Strong activation of ie-1 and ie-3 gene expression was observed starting at day 4 and peaking at day 7 (Fig. 1). This was accompanied by binding of acetylated histones and RNA polymerase to the MIEP (Fig. 2). The pattern of expression of the early gene e1 (M112) was similar to that of the IE genes, as was binding of RNA polymerase and acetylated H4 to the M112 promoter (Fig. 4). In contrast, expression of the late gene M100 peaked at day 10, when DNA replication was maximal (Fig. 1). Changes in expression of M100 RNA also correlated with binding of RNA polymerase and acetylated H4 to the promoter (Fig. 1 and 4). Thus, our results are consistent with the regulatory cascade of viral gene expression, in which immediate-early genes activate expression of genes required for DNA replication, which allows expression of late genes. As expected, activation of gene expression correlated with recruitment of transcriptional activators to the promoter regions.
Activation of gene expression was followed by repression, and there was some variability in the kinetics of repression among different classes of genes. Following the peak of ie and e1 gene expression at day 7, RNA abundance dropped at day 10, while the DNA copy number remained high. Thus, repression of ie and e1 gene expression occurred in the kidney between day 7 and day 10 postinfection, while repression of M100 was not observed until day 14. Repression of gene expression correlated with loss of RNA polymerase and acetylated H4 to promoters.
Repression of viral gene expression is likely due to recruitment of chromatin remodeling complexes that render the promoters inaccessible to the transcription apparatus. As previously noted (40), histone H4 and HDACs were bound to the MIEP at significantly higher levels in latently infected mice than in acutely infected mice. Here, we show that H4 and HDACs are also bound to other viral promoters at higher levels in latently infected mice, suggesting that the entire genome may be in a highly chromatinized, repressive state in latency. In addition to HDACs, we have shown that a number of cellular repressors, including YY1, CBF-1/RBP-Jk, CIR, and Daxx, are bound to the MIEP during latent infection.
Surprisingly, recruitment of repressors to viral promoters was biphasic: binding was detectable at very early times postinfection, prior to activation of ie gene expression, then decreased with the onset of ie gene expression and increased again at day 10 to 14, when repression of gene expression occurred. Loss of these repressors correlated with increased binding of RNA polymerase and acetylated histones and activation of immediate-early and early gene expression between day 4 and day 7 p.i., followed by amplification of viral DNA and activation of late gene expression. These results are similar to recent studies of HCMV infection of permissive cells, which showed that the genome becomes rapidly associated with histones bearing markers of repressed chromatin, which are then lost as the viral program of transcription becomes activated (18).
Previous studies with HCMV have shown that during infection of fibroblasts in vitro, the viral genome becomes rapidly associated with ND10 bodies in the nucleus (2, 43). These multiprotein complexes are dynamic structures comprised of transcriptional repressors, including PML, Daxx, and ATRX, which are thought to act as a form of host intrinsic defense against infection through repression of ie gene expression (62, 63, 75, 78). Two viral proteins that activate ie gene expression, the tegument protein pp71 and the immediate-early protein IE-1, antagonize Daxx and HDACs and disrupt formation of ND10 bodies (1, 2, 35, 42, 48, 62, 63). Like HCMV IE-1, MCMV IE-1 binds to host cellular repressors and blocks HDAC activity (74). It is tempting to speculate that cellular repressors are recruited to MCMV DNA when the genome enters the nucleus and that the decrease in binding of these proteins to the MIEP observed from day 2 to 7 is due to disruption of these interactions mediated by viral tegument and IE-1 proteins. However, there may be other explanations for the loss of binding of repressors to the enhancer. Activation of cellular signaling pathways triggered by viral entry (27, 82) may help to derepress viral promoters. Alternatively, the apparent loss of repressors bound to the MIEP could be due to new DNA being synthesized that lacks repressors, rather than loss of repressors from viral DNA that was present at day 2.
Following activation of gene expression and loss of repressors, a second phase of binding of repressors to viral promoters was observed from day 10 to day 21, which correlated with repression of viral gene expression. During the course of infection in vivo, the adaptive immune response eliminates productively infected cells, leaving behind latently infected cells, which are presumably invisible to the host immune response due to transcriptional silencing of gene expression. Thus, the apparent increase in binding of repressive factors to the viral promoters observed during the second phase of binding could be due largely to a change in the ratio of productively versus latently infected cells as productively infected cells are cleared by the immune response.
While infected cells are abundant and readily detectable during acute infection, the frequency of latently infected cells is very low, suggesting that establishment of latency is a rare event (5, 31, 50, 55, 64, 71). At the peak of IE RNA expression, binding of repressors to the MIEP fell significantly but was still detectable. These observations suggest that while most genomes are free of cellular repressors between day 4 and day 7, allowing for expression of the ie genes and productive infection, some viral genomes may remain in a repressive state with these factors bound to the MIEP. These cells may form a reservoir of latently infected cells which never transition to productive infection in the days immediately following infection. Thus, latency and productive infection may be mutually exclusive pathways which are chosen at the outset of infection. The observation that only 10% of infecting HCMV genomes are actively transcribed in permissive cells is consistent with this model (18). However, our results do not exclude the possibility that latency may be established in some cells as a result of abortive infection subsequent to activation of viral gene expression.
Recruitment of some cellular repressors to viral promoters is likely to be mediated by recognition of specific DNA sequences in the promoter, while others may be recruited indirectly, as a result of interactions with these sequence-specific DNA-binding factors. The MCMV IE-3 protein, like HCMV IE-2, is a repressor of the MIEP as well as an activator of early gene expression (45). Studies with HCMV IE-2 have shown that it represses the MIEP through binding to a cis repression sequence immediately upstream of the transcription start site and recruitment of chromatin remodeling complexes (11, 57, 72). Thus, recruitment of repressors to the MIEP and repression of ie gene expression observed between day 7 and day 10 may be due in part to autorepression. However, repressors must be recruited to viral promoters through alternative means prior to the activation of ie gene expression during acute infection and following the establishment of latency, when the IE genes are not expressed in most latently infected cells (56).
Our studies of latently infected mice show that while HDACs bind to all viral promoters tested, YY1 and CBF-1/RBP-Jk bind specifically to promoters containing their cognate recognition sequences: CBF-1/RBP-Jk bound only to the MIEP, while YY1 bound to both the MIEP and M100P. These results support the hypothesis that these factors are recruited to these promoters though direct interaction with DNA sequences in the promoter region. Since these repressors are part of large corepressor complexes (7, 17), binding of these factors to the DNA may recruit additional repressors to viral promoters. Interestingly, although YY1 bound to both the MIEP and the M100P at comparable levels at day 2, binding of YY1 to the M100P was significantly lower than binding to the MIEP in latent infection. The significance of this observation will require further analysis.
YY1 is a ubiquitously expressed (3, 14) member of the polycomb group of proteins, which form chromatin-modifying complexes that mediate transcriptional silencing (70). YY1 has fundamental roles in embryogenesis, differentiation, replication, cellular proliferation, and inflammation and has been shown to regulate expression of many genes (17). YY1 was so named because it can function as either an activator or repressor of transcription, depending on a variety of factors, including its binding position relative to the transcription start site, intracellular concentration, and posttranslational modifications, such as acetylation (8, 67, 80). These dual functions are mediated by different domains of the protein (66) which interact with repressors, including HDACs and SAP30 (23, 79), transcription factors, including Sp1 and c-myc (65, 68), or coactivators such as CBP, pCAF, and p300 (3, 8, 80). YY1 binds directly to DNA through recognition of specific sequences (26, 30) and thus can serve as an adaptor to recruit either activating or repressive complexes (37) to promoter regions. YY1 has been shown to be a repressor of the HCMV enhancer, which has multiple YY1 binding sites (39).
Intracellular signaling events can modulate YY1 activity. For example, under resting conditions, YY1 constitutively occupies a single, high-affinity proximal site in the beta-interferon promoter, leading to recruitment of HDACs and transcriptional repression. In response to viral infection, however, YY1 occupies a second, lower-affinity distal site in the promoter, leading to recruitment of CBP and histone acetyltransferases (HATs), acetylation of histones bound to the promoter, and activation of transcription (47, 77). In other cases, YY1 may act solely as a repressor whose activity can be abrogated through competition with other transcription factors, such as serum response factor, AP-1, or NF-κB, that recognize overlapping sites in promoter regions (36, 41, 81). AP-1 and NF-κB are activated in response to many inflammatory signaling pathways and control expression of many genes that mediate innate and adaptive immunity. Thus, inflammation can alter the activity of YY1, either through conversion of YY1 from a repressor to an activator or through competition with activating factors for DNA binding sites.
Like YY1, CBF-1 is a DNA-binding protein that recruits corepressors or coactivators and, thus, can act as a repressor or an activator of expression, depending on the cellular signaling environment. CBF-1 is the major downstream effector of the Notch signaling pathway. In the absence of Notch signaling, CBF-1 binds to promoters of target genes and recruits corepressor complexes (7, 28). Interaction of transmembrane forms of Notch with its ligands, Jagged and Delta-like, results in cleavage of Notch and translocation of the intracellular domain into the nucleus, where it binds to CBF-1, displaces transcriptional repressors, and recruits activators that drive expression of target genes. The Notch signaling system is conserved from Drosophila to vertebrates and regulates the expression of genes involved in embryonic development (7, 28). In the immune system, Notch signaling is involved in the development and function of T cells, macrophages, NK cells, and dendritic cells (21). Recent studies indicate that CBF1 mediates Toll-like receptor-induced expression of some inflammatory cytokines, including TNF, IL-6, and IL-12, independent of Notch signaling (22). Thus, like YY1, CBF-1 is an important mediator of inflammation as well as embryonic development.
CBF-1 and/or YY1 has been implicated in the control of latency and reactivation of human immunodeficiency virus (HIV), human papillomavirus (HPV), and Kaposi's sarcoma-associated herpesvirus (KSHV) (20, 33, 34, 38, 76). Previous studies have suggested that an inflammatory immune response can lead to reactivation of latent CMV through activation of the ie enhancer (10, 12, 15, 24, 25, 52, 60, 69). The observations that (i) YY1 and CBF-1 are bound to the MIEP in MCMV latently infected mice and (ii) the activity of these factors can be modulated by cellular signaling pathways, and particularly by inflammatory stimuli, suggest that they could also have a role in regulating the switch from CMV latency to reactivation.
In summary, we have demonstrated in this study that several cellular repressors are bound to the MIEP and other viral promoters in latently infected mice. While these data are correlative, they provide strong circumstantial evidence in support of the hypothesis that (i) transcriptional silencing of viral gene expression through recruitment of cellular repressors leads to the establishment of latency; (ii) some of these repressors may be recruited directly to promoters through recognition of specific sequences, leading to binding of additional corepressors through protein-protein interactions; and (iii) latency may be established in some cells at very early times after infection. Investigation of the requirement for the putative YY1 and CBF-1/RBP-Jk binding sites in the MIEP for establishment of latency should provide further insight into the mechanisms by which MCMV establishes latent infection.
Acknowledgments
This work was supported by Public Health Service grant R21 AI76771 from the NIH.
Footnotes
Published ahead of print on 27 January 2010.
REFERENCES
- 1.Ahn, J. H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Austen, M., B. Luscher, and J. M. Luscher-Firzlaff. 1997. Characterization of the transcriptional regulator YY1. The bipartite transactivation domain is independent of interaction with the TATA box-binding protein, transcription factor IIB, TAFII55, or cAMP-responsive element-binding protein (CPB)-binding protein. J. Biol. Chem. 272:1709-1717. [DOI] [PubMed] [Google Scholar]
- 4.Bain, M., M. Reeves, and J. Sinclair. 2006. Regulation of human cytomegalovirus gene expression by chromatin remodeling, p. 167-183. In M. Reddehase (ed.), Cytomegaloviruses: molecular biology and immunology. Caister Academic Press, Norfolk, UK.
- 5.Balthesen, M., M. Messerle, and M. J. Reddehase. 1993. Lungs are a major organ site of cytomegalovirus latency and recurrence. J. Virol. 67:5360-5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshart, M., F. Weber, G. Jahn, K. Dorsch-Hasler, B. Fleckenstein, and W. Schaffner. 1985. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 7.Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678-689. [DOI] [PubMed] [Google Scholar]
- 8.Bushmeyer, S., K. Park, and M. L. Atchison. 1995. Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem. 270:30213-30220. [DOI] [PubMed] [Google Scholar]
- 9.Cartharius, K., K. Frech, K. Grote, B. Klocke, M. Haltmeier, A. Klingenhoff, M. Frisch, M. Bayerlein, and T. Werner. 2005. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21:2933-2942. [DOI] [PubMed] [Google Scholar]
- 10.Cook, C. H., J. Trgovcich, P. D. Zimmerman, Y. Zhang, and D. D. Sedmak. 2006. Lipopolysaccharide, tumor necrosis factor alpha, or interleukin-1β triggers reactivation of latent cytomegalovirus in immunocompetent mice. J. Virol. 80:9151-9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuevas-Bennett, C., and T. Shenk. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82:9525-9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Docke, W. D., S. Prosch, E. Fietze, V. Kimel, H. Zuckermann, C. Klug, U. Syrbe, D. H. Kruger, R. von Baehr, and H. D. Volk. 1994. Cytomegalovirus reactivation and tumour necrosis factor. Lancet 343:268-269. [DOI] [PubMed] [Google Scholar]
- 13.Dorsch-Hasler, K., G. M. Keil, F. Weber, M. Jasin, W. Schaffner, and U. H. Koszinowski. 1985. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate early mRNA in murine cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 82:8325-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drews, D., M. Klar, C. Dame, and A. U. Brauer. 2009. Developmental expression profile of the YY2 gene in mice. BMC Dev. Biol. 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fietze, E., S. Prosch, P. Reinke, J. Stein, W. D. Docke, G. Staffa, S. Loning, S. Devaux, F. Emmrich, and R. von Baehr. 1994. Cytomegalovirus infection in transplant recipients. The role of tumor necrosis factor. Transplantation 58:675-680. [PubMed] [Google Scholar]
- 16.Gaytant, M. A., E. A. Steegers, B. A. Semmekrot, H. M. Merkus, and J. M. Galama. 2002. Congenital cytomegalovirus infection: review of the epidemiology and outcome. Obstet. Gynecol. Surv. 57:245-256. [DOI] [PubMed] [Google Scholar]
- 17.Gordon, S., G. Akopyan, H. Garban, and B. Bonavida. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25:1125-1142. [DOI] [PubMed] [Google Scholar]
- 18.Groves, I. J., M. B. Reeves, and J. H. Sinclair. 2009. Lytic infection of permissive cells with human cytomegalovirus is regulated by an intrinsic ‘pre-immediate-early’ repression of viral gene expression mediated by histone post-translational modification. J. Gen. Virol. 90:2364-2374. [DOI] [PubMed] [Google Scholar]
- 19.Hariharan, N., D. E. Kelley, and R. P. Perry. 1989. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes Dev. 3:1789-1800. [DOI] [PubMed] [Google Scholar]
- 20.He, G., and D. M. Margolis. 2002. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol. Cell. Biol. 22:2965-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hertzog, P. 2008. A notch in the toll belt.[Comment]. Immunity 29:663-665. [DOI] [PubMed] [Google Scholar]
- 22.Hu, X., A. Y. Chung, I. Wu, J. Foldi, J. Chen, J. D. Ji, T. Tateya, Y. J. Kang, J. Han, M. Gessler, R. Kageyama, and L. B. Ivashkiv. 2008. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity 29:691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, N. E., C. H. Lin, Y. S. Lin, and W. C. Yu. 2003. Modulation of YY1 activity by SAP30. Biochem. Biophys. Res. Commun. 306:267-275. [DOI] [PubMed] [Google Scholar]
- 24.Hummel, M., and M. I. Abecassis. 2002. A model for reactivation of CMV from latency. J. Clin. Virol. 25:S123-S136. [DOI] [PubMed] [Google Scholar]
- 25.Hummel, M., Z. Zhang, S. Yan, I. DePlaen, P. Golia, T. Varghese, G. Thomas, and M. I. Abecassis. 2001. Allogeneic transplantation induces expression of cytomegalovirus immediate-early genes in vivo: a model for reactivation from latency. J. Virol. 75:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyde-DeRuyscher, R. P., E. Jennings, and T. Shenk. 1995. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 23:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isaacson, M. K., L. K. Juckem, and T. Compton. 2008. Virus entry and innate immune activation. Curr. Top. Microbiol. Immunol. 325:85-100. [DOI] [PubMed] [Google Scholar]
- 28.Kageyama, R., T. Ohtsuka, and T. Kobayashi. 2007. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 134:1243-1251. [DOI] [PubMed] [Google Scholar]
- 29.Kalejta, R. F. 2008. Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Curr. Top. Microbiol. Immunol. 325:101-115. [DOI] [PubMed] [Google Scholar]
- 30.Kim, J. 2009. YY1's longer DNA-binding motifs. Genomics 93:152-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koffron, A. J., M. Hummel, B. K. Patterson, S. Yan, D. B. Kaufman, J. P. Fryer, F. P. Stuart, and M. I. Abecassis. 1998. Cellular localization of latent murine cytomegalovirus. J. Virol. 72:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 33.Lace, M. J., Y. Yamakawa, M. Ushikai, J. R. Anson, T. H. Haugen, and L. P. Turek. 2009. Cellular factor YY1 downregulates the human papillomavirus 16 E6/E7 promoter, P97, in vivo and in vitro from a negative element overlapping the transcription-initiation site. J. Gen. Virol. 90:2402-2412. [DOI] [PubMed] [Google Scholar]
- 34.Lan, K., D. A. Kuppers, and E. S. Robertson. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jkappa, the major downstream effector of the Notch signaling pathway. J. Virol. 79:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, H. R., D. J. Kim, J. M. Lee, C. Y. Choi, B. Y. Ahn, G. S. Hayward, and J. H. Ahn. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, T. C., Y. Shi, and R. J. Schwartz. 1992. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal alpha-actin transcription in embryonic myoblasts. Proc. Natl. Acad. Sci. U. S. A. 89:9814-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le May, N., Z. Mansuroglu, P. Leger, T. Josse, G. Blot, A. Billecocq, R. Flick, Y. Jacob, E. Bonnefoy, and M. Bouloy. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang, Y., and D. Ganem. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 100:8490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, R., J. Baillie, J. G. Sissons, and J. H. Sinclair. 1994. The transcription factor YY1 binds to negative regulatory elements in the human cytomegalovirus major immediate early enhancer/promoter and mediates repression in non-permissive cells. Nucleic Acids Res. 22:2453-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, X. F., S. Yan, M. Abecassis, and M. Hummel. 2008. Establishment of murine cytomegalovirus latency in vivo is associated with changes in histone modifications and recruitment of transcriptional repressors to the major immediate-early promoter. J. Virol. 82:10922-10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu, S. Y., M. Rodriguez, and W. S. Liao. 1994. YY1 represses rat serum amyloid A1 gene transcription and is antagonized by NF-kappa B during acute-phase response. Mol. Cell. Biol. 14:6253-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lukashchuk, V., S. McFarlane, R. D. Everett, and C. M. Preston. 2008. Human cytomegalovirus protein pp71 displaces the chromatin-associated factor ATRX from nuclear domain 10 at early stages of infection. J. Virol. 82:12543-12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maul, G. G. 2008. Initiation of cytomegalovirus infection at ND10. Curr. Top. Microbiol. Immunol. 325:117-132. [DOI] [PubMed] [Google Scholar]
- 44.Mercer, J. A., C. A. Wiley, and D. H. Spector. 1988. Pathogenesis of murine cytomegalovirus infection: identification of infected cells in the spleen during acute and latent infections. J. Virol. 62:987-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Messerle, M., B. Buhler, G. M. Keil, and U. H. Koszinowski. 1992. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J. Virol. 66:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2702-2772. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 47.Mokrani, H., O. Sharaf el Dein, Z. Mansuroglu, and E. Bonnefoy. 2006. Binding of YY1 to the proximal region of the murine beta interferon promoter is essential to allow CBP recruitment and K8H4/K14H3 acetylation on the promoter region after virus infection. Mol. Cell. Biol. 26:8551-8561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. U. S. A. 101:17234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nitzsche, A., C. Paulus, and M. Nevels. 2008. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol. 82:11167-11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollock, J. L., R. M. Presti, S. Paetzold, and H. W. Virgin IV. 1997. Latent murine cytomegalovirus infection in macrophages. Virology 227:168-179. [DOI] [PubMed] [Google Scholar]
- 51.Pollock, J. L., and H. W. Virgin IV. 1995. Latency, without persistence, of murine cytomegalovirus in the spleen and kidney. J. Virol. 69:1762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prosch, S., K. Staak, J. Stein, C. Liebenthal, T. Stamminger, H.-D. Volk, and D. Kruger. 1995. Stimulation of the human cytomegalovirus IE enhancer/promoter in HL-60 cells by TNF alpha is mediated via induction of NF-kB. Virology 208:197-206. [DOI] [PubMed] [Google Scholar]
- 53.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Razonable, R. R., and C. V. Paya. 2002. Beta-herpesviruses in transplantation. Rev. Med Microbiol. 13:163-176. [Google Scholar]
- 55.Reddehase, M. J., M. Balthesen, M. Rapp, S. Jonjic, I. Pavic, and U. H. Koszinowski. 1994. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J. Exp. Med. 179:185-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddehase, M. J., C. O. Simon, C. K. Seckert, N. Lemmermann, and N. K. Grzimek. 2008. Murine model of cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325:315-331. [DOI] [PubMed] [Google Scholar]
- 57.Reeves, M., J. Murphy, R. Greaves, J. Fairley, A. Brehm, and J. Sinclair. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 80:9998-10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reeves, M., and J. Sinclair. 2008. Aspects of human cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325:297-313. [DOI] [PubMed] [Google Scholar]
- 59.Reeves, M. B., P. A. MacAry, P. J. Lehner, J. G. Sissons, and J. H. Sinclair. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. U. S. A. 102:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reinke, P., S. Prosch, F. Kern, and H. D. Volk. 1999. Mechanisms of human cytomegalovirus (HCMV) (re)activation and its impact on organ transplant patients. Transpl. Infect. Dis. 1:157-164. [DOI] [PubMed] [Google Scholar]
- 61.Rodic, N., M. Oka, T. Hamazaki, M. R. Murawski, M. Jorgensen, D. M. Maatouk, J. L. Resnick, E. Li, and N. Terada. 2005. DNA methylation is required for silencing of ant4, an adenine nucleotide translocase selectively expressed in mouse embryonic stem cells and germ cells. Stem Cells 23:1314-1323. [DOI] [PubMed] [Google Scholar]
- 62.Saffert, R. T., and R. F. Kalejta. 2007. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 81:9109-9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seckert, C. K., A. Renzaho, H. M. Tervo, C. Krause, P. Deegen, B. Kuhnapfel, M. J. Reddehase, and N. K. Grzimek. 2009. Liver sinusoidal endothelial cells are a site of murine cytomegalovirus latency and reactivation. J. Virol. 83:8869-8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seto, E., B. Lewis, and T. Shenk. 1993. Interaction between transcription factors Sp1 and YY1. Nature 365:462-464. [DOI] [PubMed] [Google Scholar]
- 66.Shi, Y., J.-S. Lee, and K. Galvin. 1997. Everything you ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed] [Google Scholar]
- 67.Shi, Y., E. Seto, L. S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 68.Shrivastava, A., S. Saleque, G. V. Kalpana, S. Artandi, S. P. Goff, and K. Calame. 1993. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science 262:1889-1892. [DOI] [PubMed] [Google Scholar]
- 69.Simon, C. O., C. K. Seckert, D. Dreis, M. J. Reddehase, and N. K. Grzimek. 2005. Role for tumor necrosis factor alpha in murine cytomegalovirus transcriptional reactivation in latently infected lungs. J. Virol. 79:326-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simon, J. A., and R. E. Kingston. 2009. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10:697-708. [DOI] [PubMed] [Google Scholar]
- 71.Slobedman, B., and E. S. Mocarski. 1999. Quantitative analysis of latent human cytomegalovirus. J. Virol. 73:4806-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stinski, M. F., and D. T. Petrik. 2008. Functional roles of the human cytomegalovirus essential IE86 protein. Curr. Top. Microbiol. Immunol. 325:133-152. [DOI] [PubMed] [Google Scholar]
- 73.Stormo, G. D. 2000. DNA binding sites: representation and discovery. Bioinformatics 16:16-23. [DOI] [PubMed] [Google Scholar]
- 74.Tang, Q., and G. G. Maul. 2003. Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J. Virol. 77:1357-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tyagi, M., and J. Karn. 2007. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weill, L., E. Shestakova, and E. Bonnefoy. 2003. Transcription factor YY1 binds to the murine beta interferon promoter and regulates its transcriptional capacity with a dual activator/repressor role. J. Virol. 77:2903-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woodhall, D. L., I. J. Groves, M. B. Reeves, G. Wilkinson, and J. H. Sinclair. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652-37660. [DOI] [PubMed] [Google Scholar]
- 79.Yang, W. M., C. Inouye, Y. Zeng, D. Bearss, and E. Seto. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. U. S. A. 93:12845-12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao, Y. L., W. M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye, J., M. Cippitelli, L. Dorman, J. R. Ortaldo, and H. A. Young. 1996. The nuclear factor YY1 suppresses the human gamma interferon promoter through two mechanisms: inhibition of AP1 binding and activation of a silencer element. Mol. Cell. Biol. 16:4744-4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yurochko, A. D. 2008. Human cytomegalovirus modulation of signal transduction. Curr. Top. Microbiol. Immunol. 325:205-220. [DOI] [PubMed] [Google Scholar]