Abstract
Lung cancer is the leading cause of cancer death in the world, but the molecular mechanisms for its development have not been well characterized. The suppressors of cytokine signaling (SOCS) are inhibitors of cytokine signaling that function via the Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathway. Eight SOCS proteins with similar structures have been identified so far. SOCS family members, however, have distinct mechanisms of inhibition of JAK/STAT signaling. Abnormalities of the JAK/STAT pathway are associated with cancer. Inhibition of signaling results in growth suppression in various cell types. Recently, the involvement of SOCS-1 in carcinogenesis has been reported. Here, we report identification of frequent hypermethylation in CpG islands of the functional SOCS-3 promoter that correlates with its transcription silencing in cell lines (lung cancer, breast cancer, and mesothelioma) and primary lung cancer tissue samples. Restoration of SOCS-3 in lung cancer cells where SOCS-3 was methylation-silenced resulted in the down-regulation of active STAT3, induction of apoptosis, and growth suppression. Our results suggest that methylation silencing of SOCS-3 is one of the important mechanisms of constitutive activation of the JAK/STAT pathway in cancer pathogenesis. The data also suggest that SOCS-3 therapy may be useful in the treatment of cancer.
The Janus kinase (JAK)/signal transducers and activators of transcription (STAT) signaling plays important roles in a number of important biologic responses, including immune function, cellular growth, differentiation, and hematopoieses (1). Eight proteins, cytokine-inducible SH2-domain-containing protein (CIS) and suppressors of cytokine signaling 1–7 (SOCS-1–7), have been identified in the SOCS family so far. They contain a central SH2 domain, a conserved C terminus (the SOCS box), and a unique N terminus (2–4). Expression of SOCS-1–3 and CIS is induced by cytokine or growth factor stimulation, which directly antagonizes STAT activation as part of a classic feedback loop (4, 5).
SOCS family members have distinct mechanisms of inhibition of JAK/STAT signaling (4). For example, CIS is believed to antagonize signaling by blocking STAT receptor recruitment (6). SOCS-1 is believed to inhibit signaling through direct interaction with JAK activation (7). SOCS-3 can bind both the cytokine receptor and the JAK and is recruited to the tyrosine-phosphorylated receptor, facilitating inhibition of the JAK (8). Another possible mechanism by which SOCS proteins attenuate signaling is to promote protein degradation (9, 10).
It has been reported that abnormalities of the JAK/STAT pathway are associated with cancer (11–15). For example, constitutive activation of the JAK was found in T cell childhood acute lymphoblastic leukemia (16). Transfection of a constitutively activated STAT3 results in tumorigenicity in nude mice (17). Constitutive activation of STAT3 correlates with cell proliferation in breast carcinoma (18) and non-small-cell lung cancer (NSCLC) (19) and also inhibits apoptosis (20–24). On the other hand, inhibition of JAK/STAT signaling results in suppression of cancer cell growth and induces apoptosis in various cancer types (18, 25–29). These results suggest the significance of JAK/STAT activation in oncogenesis.
Aberrant hypermethylation of promoter regions in CpG islands has been shown to be associated with transcriptional silencing of the genes in various cancers (30). Recently, involvement of SOCS-1 in carcinogenesis has also been reported. SOCS-1 was found frequently silenced by hypermethylation in hapatocellular carcinoma (HCC) (31), multiple myeloma (32), and hepatoblastomas (33). SOCS-1 appears to have tumor suppressor activity (34), and restoration of the SOCS-1 gene in HCC cells causes growth suppression and induction of apoptosis (31). However, silencing of SOCS-3 by promoter methylation in human cancer has not been reported. In this study, we demonstrate the correlation of methylation of the SOCS-3 promoter and its transcription silencing as well as growth-suppression activity of SOCS-3 in NSCLC. We also found that this growth suppression was caused by induction of apoptosis in those cancer cells.
Materials
Cell Lines and Tissue Samples. Human NSCLC cell lines (NCI-H1703, -H460, -H838, and -A549), a normal lung cell line (CCL-75, fibroblast), and human breast cancer cell lines (MCF-7, HuL100, BT474, and MDA341) were obtained from the American Type Culture Collection. Human mesothelioma cancer cell lines NCI-H290 and MS-1 were obtained from the National Institutes of Health. These cells, except CCL-75, were cultured in RPMI medium 1640 supplemented with 10% fetal bovine serum, penicillin (100 units/ml), and streptomycin (100 μg/ml). CCL-75 was cultured in MEM with Earle's balanced salt solution containing 2 mM l-glutamine, 1.0 mM sodium pyruvate, 0.1 mM nonessential amino acids, 1.5 g/liter sodium bicarbonate, and 10% FBS. Normal human small airway epithelial cells and bronchial epithelial cells (primary cultures) were obtained from Clonetics (Walkersville, MD) and cultured in the Clonetics small airway epithelial cell growth medium Bullet Kit. All cells were cultured at 37°C in a humid incubator with 5% CO2.
Fresh lung cancer tissue and adjacent normal lung tissue from patients undergoing resection of early stage lung cancers were collected at the time of surgery and immediately snap-frozen in liquid nitrogen. These tissue samples were kept at –170°C in a liquid nitrogen freezer before use.
Northern Blotting and Semiquantitative RT-PCR. Total RNA from lung cancer cell lines, fresh lung cancer, and paired adjacent normal tissue were isolated by using TRIzol reagent (Life Technologies, Carlsbad, CA). Poly(A) RNA of those samples was isolated further from the total RNA by using Oligotex mRNA Kit (Qiagen, Valencia, CA). A SOCS-3 cDNA insert from the cDNA construct in pCDNA3 vector was used as a probe for the Northern blot. Northern blotting was carried out as described (31). The same membrane was then reprobed with a specific probe of L19 ribosomal protein as a standard. RT-PCR was performed in the GeneAmp PCR system 9700 by using the One-Step RT-PCR Kit from Life Technologies, according to the manufacturer's protocol. Primers for RT-PCR were obtained from Operon Technologies (Alameda, CA). Primer sequences for a 579-bp fragment of the human SOCS-3 cDNA were: 5′-GTCACCCACAGCAAGTTTCC-3′ (forward) and 5′-CCGACAGAGATGCTGAAGAG-3′ (reverse). A 395-bp fragment of a gene encoding the L19 ribosomal protein was used as an internal control, and their primer sequences were: 5′-GAAATCGCCAATGCCAACT-3′ (forward) and 5′-TCTTAGACCTGCGAGCCTCA-3′ (reverse). 5-Aza-2′-deoxycytidine (Sigma) treatment was performed as described (32).
Western Blotting. Standard protocol was used. Antiphospho-Stat3 (Tyr-705) rabbit polyclonal antibody and anti-β-actin mouse monoclonal antibody were obtained from Cell Signaling Technology (Beverly, MA). Anti-human SOCS-3 mouse monoclonal antibody was obtained from IBL (Gunma, Japan).
Sequencing Analysis. Genomic DNA of the cell lines and fresh tissue samples were extracted by using DNA STAT-60 reagent (Tel-Test, Friendswood, TX), according to the manufacturer's protocol. Bisulfite modification of genomic DNA was carried out as described (35). Bisulfite-treated genomic DNA was amplified by using primers [5′-GTGTAGAGTAGTGATTAAATA-3′ (forward) and 5′-TCCTTAAAACTAAACCCCCTC-3′ (reverse)] designed to amplify nucleotides –1084 to –671 of the SOCS-3 promoter region (the start codon ATG of SOCS-3 is defined as +1). The PCR products were cloned into TOPO-TA pCR2.1 vector (Life Technologies), and multiple randomly picked clones from each sample were sequenced at the DNA-sequencing Core Facility of the University of California at San Francisco Cancer Center.
Methylation-Specific PCR. Bisulfite-treated genomic DNA was amplified by using either a methylation- or unmethylation-specific primer set. HotStarTaq DNA polymerase (Qiagen, Chatsworth, CA) was used in the experiments. Sequences of the methylation-specific primers were: 5′-TATATATTCGCGAGCGCGGTTT-3′ (forward) and 5′-CGCTGCGCCCAGATGTT-3′ (reverse), corresponding to the SOCS-3 promoter region sequences –1005 to –983 and –754 to –737, respectively. Sequences of the unmethylation-specific primers were 5′-TGTGGTGGTTGTTTATATATTTGTGAGTGTGGTT-3′ (forward) and 5′-CAACCAACAATAACCCACACTACACCCA-3′ (reverse), corresponding to the SOCS-3 promoter region sequences –1018 to –984 and –748 to –720, respectively.
Transient Transfection and Colony Formation Assay. For transient transfection experiments, cells (2 × 105) were plated in six-well plates 24 h before transfection. Lipofectamine 2000 (Life Technologies) was used to mediate transfection by using 5.0 μg of SOCS-3 cDNA construct in pCDNA3 vector (kindly provided by Atsuo Sasaki, Medical Institute of Bioregulation, Kyushu University, Higashi-ku, Fukuoka-shi, Japan) or 5.0 μg of empty pCDNA3 vector as control, according to the manufacturer's protocol. Transfected cells were striped and plated on 10-cm cell culture dishes 48 h after transfection. The cells were then selected by G418 (400 μg/ml). Colonies were stained by using 0.5% methylene blue and counted 4 wk after transfection.
Apoptosis Analysis. One week after transfection (as described above), the cells were harvested by trypsinization and stained by using an Annexin V FITC Apoptosis Detection Kit (Oncogene Science), according to the manufacturer's protocol. Stained cells were then immediately analyzed by flow cytometry (FACScan, Becton Dickinson). Early apoptotic cells with exposed phosphatidylserine but intact cell membranes bound to annexin V-FITC but excluded propidium iodide. Cells in necrotic or late apoptotic stages were labeled with both annexin V-FITC and propidium iodide.
Statistical Analysis. Data shown represent mean values (±SD). Student's t test was used for comparing activities of different constructs and treatments.
Results
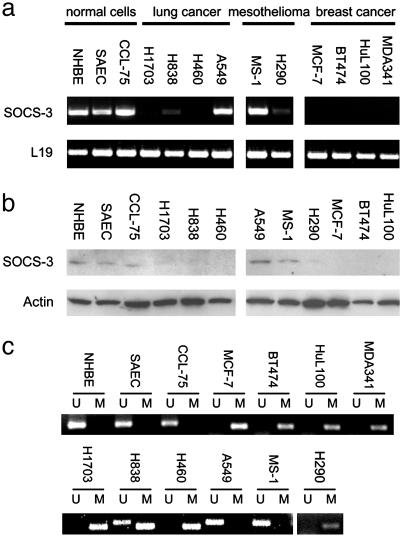
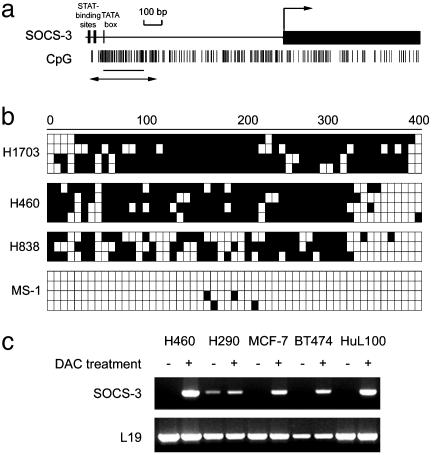
Correlation of Promoter Hypermethylation with Silencing of SOCS-3 in Cell Lines. We first examined SOCS-3 expression in cell lines (Fig. 1a). We found that the SOCS-3 transcript was missing or dramatically underexpressed in three of four NSCLC cell lines, one of two mesothelioma cell lines, and all four breast cancer cell lines tested. In contrast, SOCS-3 expression was detectable in all three normal controls, including two normal primary cell cultures (bronchial epithelial cells and small airway epithelial cells). We also detected SOCS-3 protein in those three normal cells, but not in those cancer cell lines missing or underexpressing the SOCS-3 transcript (Fig. 1b). To identify an underlying mechanism for loss of SOCS-3 expression in cancer cells, we then analyzed by methylation the status of CpG islands in the SOCS-3 promoter region in these cell lines. All cancer cell lines tested (NCI-H1703, -H460, -H838, and -H290; MCF-7; BT474; HuL100; and MDA341) lacking SOCS-3 expression were found to be hypermethylated by using methylation-specific PCR (MSP) (Fig. 1c). In contrast, no hypermethylation was seen in all three normal controls that expressed SOCS-3 (Fig. 1c). We also used bisulfite sequencing to analyze details of the methylation status of 55 CpG sites in the 413 bp of the SOCS-3 promoter region in several cell lines (Fig. 2a). This region contains both functional STAT-binding sites and a TATA box that are highly conserved in both mouse and rat SOCS-3 promoters and can function as a minimal promoter (19). Consistent with MSP results, we found that these CpG islands in all three NSCLC cell lines tested were densely methylated. The mesothelioma cell line, MS-1, which has SOCS-3 expression showed no dense methylation in these CpG islands (Fig. 2b). In addition, we found that the SOCS-3 expression was restored after demethylating agent 5-aza-2′-deoxycytidine treatment in those cell lines lacking SOCS-3 expression (Fig. 2c). These results suggest that the status of SOCS-3 expression in NSCLC, breast cancer, and mesothelioma cell lines correlates with dense methylation of the functional and conserved SOCS-3 promoter region.
Fig. 1.
Correlation of methylation in the promoter region with silencing of the SOCS-3 gene in cell lines. (a) RT-PCR results from four NSCLC cells, two mesothelioma cells, four breast cancer cell lines, and three normal cells. The fragment of human SOCS-3 cDNA amplified is 579 bp. A 395-bp fragment of the L19 ribosomal protein gene is used as a positive control for RNA quality and loading. (b) Western analysis of SOCS-3 protein in cancer cell lines and normal controls. β-Actin protein was used as loading control. (c) MSP analysis of cell lines. Bands (298 bp) in lanes labeled “U” are unmethylated DNA product amplified with unmethylation-specific primers. Bands (268 bp) in lanes labeled “M” are methylated DNA product amplified with methylation-specific primers.
Fig. 2.
(a) A scheme of the 5′ SOCS-3 promoter region and SOCS-3 gene. The large black bar represents the ORF of SOCS-3 gene with an arrow on top indicating the translation start site. The location of two STAT-binding sites and one TATA box in the SOCS-3 promoter region is indicated. Vertical bars represent CpG islands. The line and double arrow below the CpG islands represent regions analyzed by MSP and bisulfite sequencing, respectively. (b) Bisulfite-sequencing analysis of cell lines. Open and filled squares represent unmethylated and methylated CpG islands, respectively. We sequenced four clones of PCR products amplified from bisulfite-treated genomic DNA for each cell line. (c) Reactivation of SOCS-3 expression by 5-aza-2′-deoxycytidine (DAC) treatment in cell lines. RT-PCR was performed after DAC treatment (1.0 μM for 96 h). A 395-bp fragment of the L19 ribosomal protein gene was used as a control for RNA quality and loading.
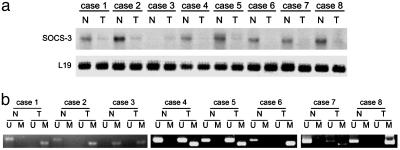
Correlation of Promoter Hypermethylation with Silencing of SOCS-3 in Primary NSCLC Tissue Samples. We next analyzed the SOCS-3 expression and methylation status in the SOCS-3 promoter region in primary NSCLC tissue samples. Among eight matched pairs of surgically resected early-stage lung cancers we examined, seven cancer samples (87.5%) were found to have no SOCS-3 mRNA or less than their matched normal samples (Fig. 3a). Consistent with lack of or diminished SOCS-3 expression, we found aberrant methylation in all seven tumor samples but not in their matched normal samples by MSP (Fig. 3b). In case 3, we found methylation in both cancer and its matched normal samples. The methylation observed in normal tissue may be due to unavoidable contamination of cancer cells in the noncancer specimen or premalignant changes of peritumoral normal tissue. Partial methylation observed in the tumor samples can also be interpreted by unavoidable contamination of normal cells in the cancer specimens. These data indicate that silencing of SOCS-3 is correlated with hypermethylation of CpG islands in its promoter in primary NSCLC tissue samples.
Fig. 3.
Correlation of methylation in the promoter region with silencing of the SOCS-3 gene in primary NSCLC tissue samples. (a) Northern blot of eight matched pairs of normal and lung cancer tissue probed with SOCS-3 cDNA. A specific probe of L19 ribosomal protein was used as a standard. (b) MSP analysis of eight matched pairs of normal (N) and lung cancer (T) tissue. Bands in lanes U and M are as indicated in Fig. 1c.
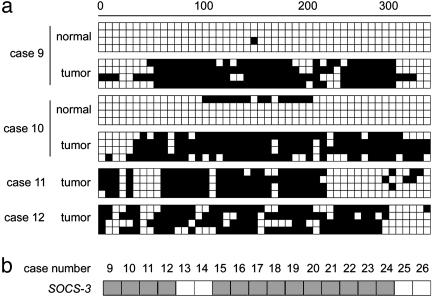
We also examined methylation of CpG islands in the SOCS-3 promoter in 18 additional surgically resected NSCLC primary tissue samples by either bisulfite sequencing or MSP. Of the 18 tumor samples, two had matched normal tissue. Six tissue samples, including the two matched pairs, were selected for bisulfite sequencing. We detected dense methylation in those CpG sites in four of the six tumor samples, including both tumor samples of the two matched pairs. In contrast, only one of four clones sequenced from one paired normal sample demonstrated minimal regional methylation (Fig. 4a). MSP analysis also detected methylation in 10 of the remaining 12 tumor samples (data not shown). In summary, methylation in the SOCS-3 promoter region was detected in 14 of the 18 (77.8%) NSCLC primary cancer tissue samples that we examined (Fig. 4b).
Fig. 4.
Methylation analysis of additional NSCLC primary tissue samples. (a) Bisulfite-sequencing examples of the SOCS-3 promoter region in NSCLC primary tissue samples. Open and filled squares represent unmethylated and methylated CpG islands, respectively. We sequenced four clones of PCR products amplified from bisulfite-treated genomic DNA for each sample. The region analyzed is as indicated in Fig. 2a. (b) Summary of methylation status of the SOCS-3 promoter region in 18 NSCLC primary tissue samples. The gene name is indicated on the left, and case numbers are indicated at the top. Each gray (methylation) or open (no methylation) square represents a primary tumor.
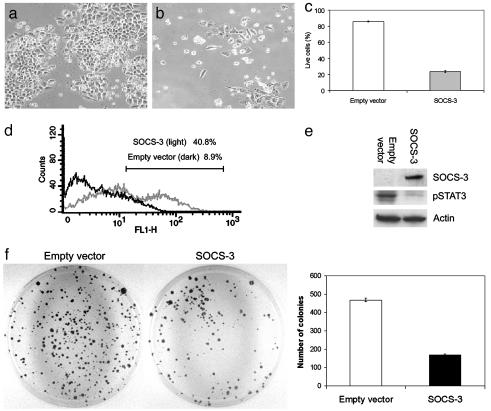
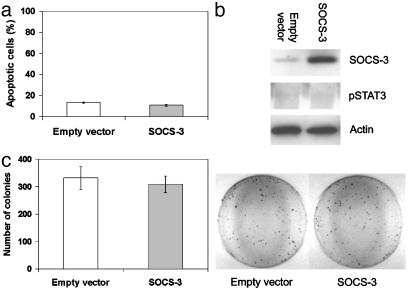
Restoration of SOCS-3 Causes Cell Growth Suppression. We investigated whether restoration of SOCS-3 would result in growth suppression in the lung cancer cell lines where SOCS-3 was silenced by promoter hypermethylation. One week after transfection and subsequent drug selection, we found significant decreases in live cell numbers in H460 cells transfected with SOCS-3 (≈24% cells are alive) compared with the empty vector-transfected control transfectants (≈86% cells are alive) (P < 0.005) (Fig. 5 a–c). Flow cytometry analysis showed a significantly higher level of apoptosis induction in SOCS-3-transfected H460 cells than empty vector-transfected H460 cells (≈41% and 9% at 1 wk after transfection, respectively) (P < 0.005) (Fig. 5d). These results indicate that restoration of SOCS-3 expression induces apoptosis and can suppress cell growth. Also of importance was the observation that restoration of SOCS-3 decreased the level of constitutive STAT3 phosphorylation in the H460 cells tested (Fig. 5e). In addition, after selection of drug-resistant colonies for 4 wk, we found that the colony numbers of SOCS-3 transfected cells decreased compared with that of empty vector-transfected cells (P < 0.005) (Fig. 5f). In contrast, when we transfected a SOCS-3 construct into mesothelioma cell line MS-1, where SOCS-3 is not silenced and the promoter is not methylated, no apoptosis was observed (Fig. 6a). Furthermore, phosphorylated STAT3 was not detected in MS-1 cells, consistent with the presence of SOCS-3 in these cells (Fig. 6b). After selection of drug-resistant colonies, no significant difference in colony numbers between SOCS-3 and empty vector-transfected MS-1 cells was seen (P = 0.4) (Fig. 6c).
Fig. 5.
Growth suppression in NSCLC cells by restoration of the SOCS-3 gene. (a and b) Morphology of H460 cells (SOCS-3 methylation-silenced) after transfection of empty vector or SOCS-3 expression vector and subsequent selection with G418 for 1 wk, respectively. (c) Percentage of live H460 cells after transfection of empty vector or SOCS-3 expression vector and subsequent selection with G418 for 1 wk. Results are means with error bars (SD). (d) Example of apoptotic analysis using flow cytometry on H460 cells 1 wk after transfection of empty vector or SOCS-3 expression vector. FL1-H represents annexin V-FITC staining. (e) Western analysis of endogenous phosphorylated STAT3 (active form) and SOCS-3 proteins in H460 cells after transfection of empty vector or SOCS-3 expression vector. β-Actin was used as loading control. Whole-cell extracts were prepared after transfection and selection with G418 for 4 wk. (f) Colony formation assay using H460 cells. The cells were transfected with empty vector or SOCS-3 expression vector and selected with G418 for 4 wk. The bar graph shows the average of colony numbers in triplicate experiments. Error bars are SD.
Fig. 6.
Transfection of SOCS-3 construct into cell line MS-1 where the SOCS-3 promoter is not methylated. (a) Graph of apoptotic analysis using flow cytometry on MS-1 cells 1 wk after transfection of empty vector or SOCS-3 expression vector. (b) Western analysis of endogenous phosphorylated STAT3 and SOCS-3 proteins in MS-1 cells after transfection of empty vector or SOCS-3 expression vector. β-Actin was used as loading control. Whole-cell extracts were prepared after transfection and selection with G418 for 3 wk. (c) Colony formation assay using MS-1 cells. The bar graph shows the average of colony numbers in triplicate experiments. Error bars are SD. The cells were transfected with empty vector or SOCS-3 expression vector and selected with G418 for 3 wk.
Discussion
Constitutive activation of STAT proteins is frequently found in tumor cells (11–24), suggesting that aberrant JAK/STAT signaling plays important roles in malignant progression by regulating cell growth and survival. SOCS gene family proteins function as negative regulators of the JAK/STAT signaling pathway (4, 5). Ectopic expression of the SOCS-1 gene has been shown to be able to block the transforming activity of oncogenic forms of JAK, in addition to its physiology role in inhibiting cytokine signaling (10). Aberrant hypermethylation of promoter regions that silences transcription of the genes has been recognized as a mechanism for inactivating tumor suppressor genes in cancer (36, 37), such as VHL (38), MGMT (39), MLH1 (40), DAPK1 (41), and SFRPs (42). Recently hypermethylation of the SOCS-1 gene was also reported to be associated with its silencing in a few tumor types (31–33). SOCS-3 inhibits STAT3 phosphorylation by binding to JAK-proximal sites on cytokine receptors to inhibit JAK activity (43). SOCS-3 was also shown to play a negative regulatory role in STAT3 activation and intestinal inflammation (44, 45). However, a mechanism for involvement of SOCS-3 in human cancers has not been reported. In the present study, we found that frequent dense hypermethylation of the functional SOCS-3 promoter region was correlated with silencing of the SOCS-3 gene in NSCLC samples and other cancer cell lines we tested. We also found that NSCLC cell lines where SOCS-3 was silenced had higher levels of STAT3 phosphorylation (19). In contrast, the normal cells that expressed SOCS-3 showed little or no phosphorylation of STAT3 (19). When we restored SOCS-3 expression in those cancer cells where SOCS-3 was silenced by promoter methylation, cell growth was suppressed due to induction of apoptosis. In addition, we found a decreased level of phosphorylated STAT3 after restoration of SOCS-3 in those cancer cells. Taken together, our results argue that SOCS-3 silencing as a result of promoter methylation may be an important cause of constitutive activation of the JAK/STAT pathway in NSCLC.
Our finding of SOCS-3 silencing by promoter methylation reveals an important epigenetic event during the development of NSCLC. SOCS-3 is normally activated by JAK/STAT signaling in normal cells, which in turn inhibits JAK activity and subsequent STAT3 activation. During development of NSCLC, however, once SOCS-3 is silenced by promoter hypermethylation, this important negative regulatory machinery is suppressed. This may result in cells becoming more sensitive to aberrant growth-stimulating signals that function through the JAK/STAT pathway, leading to cell growth and survival. In addition, our experiments showed hypermethylation of the SOCS-3 promoter region also correlates with silencing of SOCS-3 in breast cancer and mesothelioma cell lines. Therefore, the phenomenon of SOCS-3 silencing as a result of promoter methylation may be a common event during oncogenesis. SOCS-3 itself may function as an important tumor suppressor gene. Our results also suggest potential uses of SOCS-3 as therapies for treatment of NSCLC and other cancer types. The high prevalence of hypermethylation in the SOCS-3 promoter also supports targeted therapies using the JAK/STAT pathway. Reexpression of SOCS-3 can functionally restore the endogenous negative regulatory machinery of the cytokine signaling cascade, thus restoring the apoptotic machinery in these cancer cells.
The mechanisms that subvert normal highly regulated JAK/STAT signaling are still not fully defined. Understanding how oncogene products alter JAK/STAT signaling should provide further insights into the role of abnormal JAK/STAT activation during oncogenesis and may suggest a mechanistic basis for circumventing the oncogenic process and targeted therapies for the treatment of cancer.
Acknowledgments
We thank Dr. Atsuo Sasaki for kindly providing the SOCS-3 cDNA construct. This work was supported by the Larry Hall Memorial Trust and The Kazan McClain Edises Abrams Fernandez Lyons & Farrise Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: JAK, Janus kinase; STAT, signal transducers and activators of transcription; SOCS, suppressors of cytokine signaling; MSP, methylation-specific PCR; NSCLC, non-small-cell lung cancer.
References
- 1.Cooney, R. N. (2002) Shock 17, 83–90. [DOI] [PubMed] [Google Scholar]
- 2.Masuhara, M., Sakamoto, H., Matsumoto, A., Suzuki, R., Yasukawa, H., Mitsui, K., Wakioka, T., Tanimura, S., Sasaki, A., Misawa, H., et al. (1997) Biochem. Biophys. Res. Commun. 239, 439–446. [DOI] [PubMed] [Google Scholar]
- 3.Hilton, D. J., Richardson, R. T., Alexander, W. S., Viney, E. M., Willson, T. A., Sprigg, N. S., Starr, R., Nicholson, S. E., Metcalf, D. & Nicola, N. A. (1998) Proc. Natl. Acad. Sci. USA 95, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Shea, J. J., Gadina, M. & Schreiber, R. D. (2002) Cell 109, Suppl., S121–S131. [DOI] [PubMed] [Google Scholar]
- 5.Aaronson, D. S. & Horvath, C. M. (2002) Science 296, 1653–1655. [DOI] [PubMed] [Google Scholar]
- 6.Yoshimura, A., Ohkubo, T., Kiguchi, T., Jenkins, N. A., Gilbert, D. J., Copeland, N. G., Hara, T. & Miyajima, A. (1995) EMBO J. 14, 2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasukawa, H., Misawa, H., Sakamoto, H., Masuhara, M., Sasaki, A., Wakioka, T., Ohtsuka, S., Imaizumi, T., Matsuda, T., Ihle, J. N., et al. (1999) EMBO J. 18, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholson, S. E., De Souza, D., Fabri, L. J., Corbin, J., Willson, T. A., Zhang, J. G., Silva, A., Asimakis, M., Farley, A., Nash, A. D., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 6493–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamizono, S., Hanada, T., Yasukawa, H., Minoguchi, S., Kato, R., Minoguchi, M., Hattori, K., Hatakeyama, S., Yada, M., Morita, S., et al. (2001) J. Biol. Chem. 276, 12530–12538. [DOI] [PubMed] [Google Scholar]
- 10.Frantsve, J., Schwaller, J., Sternberg, D. W., Kutok, J. & Gilliland, D. G. (2001) Mol. Cell. Biol. 21, 3547–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia, R., Yu, C. L., Hudnall, A., Catlett, R., Nelson, K. L., Smithgall, T., Fujita, D. J., Ethier, S. P. & Jove, R. (1997) Cell Growth Differ. 8, 1267–1276. [PubMed] [Google Scholar]
- 12.Ihle, J. N. (1995) Nature 377, 591–594. [DOI] [PubMed] [Google Scholar]
- 13.Boudny, V. & Kovarik, J. (2002) Neoplasma 49, 349–355. [PubMed] [Google Scholar]
- 14.Li, L. & Shaw, P. E. (2002) J. Biol. Chem. 277, 17397–17405. [DOI] [PubMed] [Google Scholar]
- 15.Kaur, N., Wohlhueter, A. L. & Halvorsen, S. W. (2002) Cell. Signalling 14, 419–429. [DOI] [PubMed] [Google Scholar]
- 16.Lacronique, V., Boureux, A., Valle, V. D., Poirel, H., Quang, C. T., Mauchauffe, M., Berthou, C., Lessard, M., Berger, R., Ghysdael, J., et al. (1997) Science 278, 1309–1312. [DOI] [PubMed] [Google Scholar]
- 17.Bromberg, J. F., Wrzeszczynska, M. H., Devgan, G., Zhao, Y., Pestell, R. G., Albanese, C. & Darnell, J. E., Jr. (1999) Cell 98, 295–303. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, F., Li, C., Halfter, H. & Liu, J. (2003) Oncogene 22, 894–905. [DOI] [PubMed] [Google Scholar]
- 19.He, B., You, L., Uematsu, K., Matsangou, M., Xu, Z., He, M., McCormick, F. & Jablons, D. M. (2003) Biochem. Biophys. Res. Commun. 301, 386–391. [DOI] [PubMed] [Google Scholar]
- 20.Catlett-Falcone, R., Landowski, T. H., Oshiro, M. M., Turkson, J., Levitzki, A., Savino, R., Ciliberto, G., Moscinski, L., Fernandez-Luna, J. L., Nunez, G., et al. (1999) Immunity 10, 105–115. [DOI] [PubMed] [Google Scholar]
- 21.Rebbaa, A., Chou, P. M. & Mirkin, B. L. (2001) Mol. Med. 7, 393–400. [PMC free article] [PubMed] [Google Scholar]
- 22.Shen, Y., Devgan, G., Darnell, J. E., Jr., & Bromberg, J. F. (2001) Proc. Natl. Acad. Sci. USA 98, 1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zamo, A., Chiarle, R., Piva, R., Howes, J., Fan, Y., Chilosi, M., Levy, D. E. & Inghirami, G. (2002) Oncogene 21, 1038–1047. [DOI] [PubMed] [Google Scholar]
- 24.Grandis, J. R., Drenning, S. D., Zeng, Q., Watkins, S. C., Melhem, M. F., Endo, S., Johnson, D. E., Huang, L., He, Y. & Kim, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 4227–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blaskovich, M. A., Sun, J., Cantor, A., Turkson, J., Jove, R. & Sebti, S. M. (2003) Cancer Res. 63, 1270–1279. [PubMed] [Google Scholar]
- 26.Yamashita, H., Iwase, H., Toyama, T. & Fujii, Y. (2003) Oncogene 22, 1638–1652. [DOI] [PubMed] [Google Scholar]
- 27.Kanai, M., Konda, Y., Nakajima, T., Izumi, Y., Kanda, N., Nanakin, A., Kubohara, Y. & Chiba, T. (2003) Oncogene 22, 548–554. [DOI] [PubMed] [Google Scholar]
- 28.Mora, L. B., Buettner, R., Seigne, J., Diaz, J., Ahmad, N., Garcia, R., Bowman, T., Falcone, R., Fairclough, R., Cantor, A., et al. (2002) Cancer Res. 62, 6659–6666. [PubMed] [Google Scholar]
- 29.Buettner, R., Mora, L. B. & Jove, R. (2002) Clin. Cancer Res. 8, 945–954. [PubMed] [Google Scholar]
- 30.Laird, P. W. (2003) Nat. Rev. Cancer 3, 253–266. [DOI] [PubMed] [Google Scholar]
- 31.Yoshikawa, H., Matsubara, K., Qian, G. S., Jackson, P., Groopman, J. D., Manning, J. E., Harris, C. C. & Herman, J. G. (2001) Nat. Genet. 28, 29–35. [DOI] [PubMed] [Google Scholar]
- 32.Galm, O., Yoshikawa, H., Esteller, M., Osieka, R. & Herman, J. G. (2003) Blood 101, 2784–2788. [DOI] [PubMed] [Google Scholar]
- 33.Nagai, H., Naka, T., Terada, Y., Komazaki, T., Yabe, A., Jin, E., Kawanami, O., Kishimoto, T., Konishi, N., Nakamura, M., et al. (2003) J. Hum. Genet. 48, 65–69. [DOI] [PubMed] [Google Scholar]
- 34.Rottapel, R., Ilangumaran, S., Neale, C., La Rose, J., Ho, J. M., Nguyen, M. H., Barber, D., Dubreuil, P. & de Sepulveda, P. (2002) Oncogene 21, 4351–4362. [DOI] [PubMed] [Google Scholar]
- 35.Herman, J. G., Graff, J. R., Myohanen, S., Nelkin, B. D. & Baylin, S. B. (1996) Proc. Natl. Acad. Sci. USA 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaenisch, R. & Bird, A. (2003) Nat. Genet. 33, Suppl., 245–254. [DOI] [PubMed] [Google Scholar]
- 37.Esteller, M., Fraga, M. F., Paz, M. F., Campo, E., Colomer, D., Novo, F. J., Calasanz, M. J., Galm, O., Guo, M., Benitez, J., et al. (2002) Science 297, 1807–1808. [DOI] [PubMed] [Google Scholar]
- 38.Herman, J. G., Latif, F., Weng, Y., Lerman, M. I., Zbar, B., Liu, S., Samid, D., Duan, D. S., Gnarra, J. R., Linehan, W. M., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 9700–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esteller, M., Hamilton, S. R., Burger, P. C., Baylin, S. B. & Herman, J. G. (1999) Cancer Res. 59, 793–797. [PubMed] [Google Scholar]
- 40.Herman, J. G., Umar, A., Polyak, K., Graff, J. R., Ahuja, N., Issa, J. P., Markowitz, S., Willson, J. K., Hamilton, S. R., Kinzler, K. W., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 6870–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katzenellenbogen, R. A., Baylin, S. B. & Herman, J. G. (1999) Blood 93, 4347–4353. [PubMed] [Google Scholar]
- 42.Suzuki, H., Gabrielson, E., Chen, W., Anbazhagan, R., van Engeland, M., Weijenberg, M. P., Herman, J. G. & Baylin, S. B. (2002) Nat. Genet. 31, 141–149. [DOI] [PubMed] [Google Scholar]
- 43.Krebs, D. L. & Hilton, D. J. (2003) Sci. STKE 169, PE6. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki, A., Hanada, T., Mitsuyama, K., Yoshida, T., Kamizono, S., Hoshino, T., Kubo, M., Yamashita, A., Okabe, M., Takeda, K., et al. (2001) J. Exp. Med. 193, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, E. J., Park, S. Y., Joe, E. H. & Jou, I. (2003) J. Biol. Chem. 278, 14747–14752. [DOI] [PubMed] [Google Scholar]