Abstract
The folding of proinsulin, the single-chain precursor of insulin, ensures native disulfide pairing in pancreatic β-cells. Mutations that impair folding cause neonatal diabetes mellitus. Although the classical structure of insulin is well established, proinsulin is refractory to crystallization. Here, we employ heteronuclear NMR spectroscopy to characterize a monomeric analogue. Proinsulin contains a native-like insulin moiety (A- and B-domains); the tethered connecting (C) domain (as probed by {1H}-15N nuclear Overhauser enhancements) is progressively less ordered. Although the BC junction is flexible, residues near the CA junction exhibit α-helical-like features. Relative to canonical α-helices, however, segmental 13Cα/β chemical shifts are attenuated, suggesting that this junction and contiguous A-chain residues are molten. We propose that flexibility at each C-domain junction facilitates prohormone processing. Studies of protease SPC3 (PC1/3) suggest that C-domain sequences contribute to cleavage site selection. The structure of proinsulin provides a foundation for studies of insulin biosynthesis and its impairment in monogenic forms of diabetes mellitus.
Keywords: Diabetes, Hormones, Metabolism, NMR, Protein Structure, Beta-Cell, Preproprotein, Protein Biosynthesis
Introduction
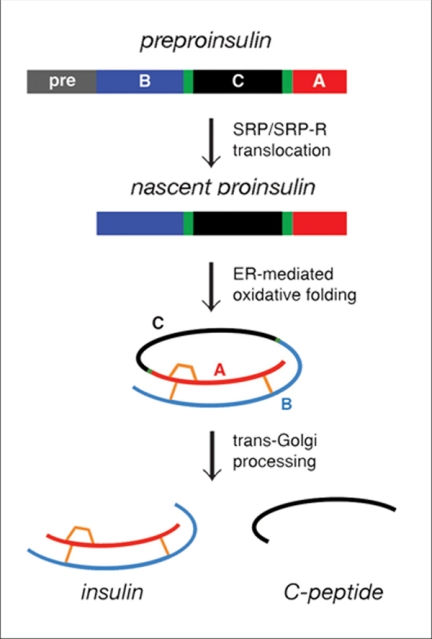
Insulin plays a central role in metabolic regulation. The hormone derives from single-chain precursors preproinsulin and proinsulin in β-cells (Fig. 1). The nascent polypeptide contains a signal peptide, which is cleaved on translocation into the endoplasmic reticulum. Folding is coupled to specific pairing of three disulfide bridges (Fig. 1, gold). On trafficking from the Golgi apparatus to glucose-regulated secretory granules (1), the connecting (C) domain is excised by specific proteases (2), liberating the C-peptide (Fig. 1, bottom). The mature hormone thus contains discrete A- and B-domains (3).
FIGURE 1.
Biosynthetic pathway of proinsulin. Insulin biosynthesis in β-cells begins with preproinsulin (top) as follows: signal peptide (pre, gray), B-domain (blue), dibasic BC junction (green), C-domain (black), dibasic CA junction (green), and A-domain (red). Disulfide bridges (residues B7–A7, B19–A20, A6–A11) are shown in gold (middle and bottom). In endoplasmic reticulum (ER), the unfolded prohormone undergoes specific disulfide pairing to yield native proinsulin (middle). Cleavage of BC and CA junctions by prohormone convertases SPC3 and SPC2 (PC1/3 and PC2) and carboxypeptidase E leads to mature insulin and the C-peptide in secretory granules (bottom). SRP and SRP-R designate the signal recognition particle and its receptor.
Here, we report the structure of proinsulin. Although insulin itself has been characterized at atomic resolution (4), proinsulin is refractory to crystallization, and its NMR study has been limited by self-association (5). To circumvent these obstacles, we undertook heteronuclear NMR analysis (6) of an engineered monomer (DKP-proinsulin)3 (7). A structural ensemble was obtained by distance geometry (DG) and simulated annealing (SA); associated subnanosecond disorder was probed by measurement of {1H}-15N nuclear Overhauser enhancements (hetNOEs) (8). Proinsulin contains a well organized insulin moiety whose folding contrasts with the flexibility of the C-domain. To assess the biological significance of C-domain sequences, we investigated effects of C-domain deletions and modifications on junctional cleavage by convertase SPC3 (PC1/PC3; Ref. 9). The structure of proinsulin provides a foundation for studies of insulin biosynthesis with potential application to diabetes-associated mutations in the insulin gene (10, 11).
MATERIALS AND METHODS
Bacterial Expression
DKP-human proinsulin, containing substitutions HisB10 → Asp, ProB28 → Lys, and LysB29 → Pro, was expressed in Escherichia coli and purified as described (7). The protein was labeled by fermentation in M9 minimal medium containing [13C]glucose and [15N]ammonium chloride as sole carbon and nitrogen sources. 13C,15N-labeled C-peptide was obtained from DKP-proinsulin by tryptic digestion (12). Receptor binding activity and stability were characterized as described (13).
NMR Spectroscopy
Samples were prepared in H2O (7% D2O) in 50 mm sodium phosphate (pH 7.1) and 50 mm NaCl; the protein concentration was 0.3 mm. Spectra, acquired at 25 °C and 700 MHz, were processed with nmrPipe (14) and analyzed with pipp (15). Assignments were obtained from three-dimensional HNCACB, CBCA(CO)NH, C(CO)NH, and H(CCO)NH spectra4 and extended by analyses of three-dimensional 13C- and 15N-separated NOE spectroscopy-heteronuclear single-quantum coherence spectra (6). Interproton distance constraints were derived from 1H-1H nuclear Overhauser effects (NOEs) (16). To probe subnanosecond fluctuations, hetNOEs were measured as described (8, 17) with a relaxation delay of 6 s. Secondary-structure propensity (SSP) scores were calculated based on 13Cα and 13Cβ chemical shifts (18).
Structure Calculations
Structures were calculated using XPLOR-NIH (19, 20) (see supplemental Methods). Root mean square deviations (r.m.s.d. values) were calculated with respect to both mean coordinates and insulin structures in the Protein Data Bank.
Proinsulin Processing
To probe sequence requirements of prohormone processing, 14 variants of DKP-proinsulin were expressed and purified (7). Human SPC3 (PC1/3) was purified as described (supplemental Methods and supplemental Figs. S1 and S2). Following incubation of enzyme and substrate, junctional cleavages were analyzed by reverse-phase HPLC and mass spectrometry (supplemental Methods and supplemental Fig. S3).
RESULTS
DKP-proinsulin is 8–10-fold more active than wild-type proinsulin (supplemental Table S1). Its thermodynamic stability is also greater than that of wild-type proinsulin (ΔΔGu 0.9 ± 0.2 kcal/mol; supplemental Table S2). Enhanced receptor binding and stability are presumably the result of the AspB10 substitution (21).
Insulin Moiety
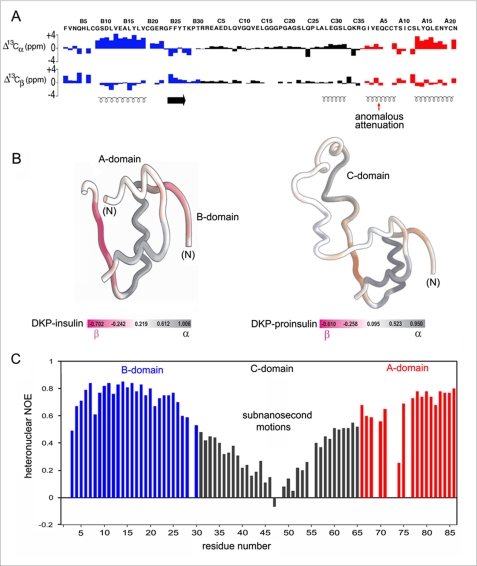
The 1H-15N heteronuclear single-quantum coherence spectrum of DKP-proinsulin resembles that of DKP-insulin with the addition of C-domain cross-peaks (supplemental Fig. S4). Further insight was provided by analysis of secondary 1Hα, 13Cα, and 13Cβ chemical shifts (differences between observed and random-coil values), which are sensitive to secondary structure (22). Trends in such shifts indicate that DKP-proinsulin and DKP-insulin exhibit similar structural elements within the A- and B-domains (Fig. 2A, blue and red; supplemental Table S3). These include N- and C-terminal A-domain α-helices (residues A1–A8 and A12–A20), central B-domain α-helix (B9–B18), and B-domain β-strand (B24–B27). In accord with canonical features of an α-helix, the C-terminal A-domain α-helix and central B-domain α-helix each exhibit large positive secondary 13Cα shifts and negative secondary 13Cβ shifts (Fig. 2A). Helical secondary structure was corroborated by characteristic NOEs (HN(i, i+1) and HN(i, i+3) contacts; Ref. 23). The N-terminal A-domain α-helix by contrast exhibits attenuated 13C secondary shifts (Fig. 2A) (24) despite retention of helix-related NOEs. Residue-specific SSP scores of DKP-insulin and DKP-proinsulin are illustrated by color-coded ribbon models (Fig. 2B).5
FIGURE 2.
{1H}-15N hetNOEs and 13C chemical shift index. A, 13C CSI values of B-domain (blue), C-domain (black), and A-domain (red). Upper and lower panels pertain to 13Cα and 13Cβ chemical shifts. Helical segments are indicated by spirals at the bottom; red arrow indicates CSI segmental attenuation. Horizontal arrow indicates B24–B27 β-strand. B, ribbon models are color-coded by 13C-derived SSP scores (18): left, DKP-insulin (12); right, DKP-proinsulin. Gray scale indicates α-helical scores; magenta scale indicates β-stand scores. C, {1H}-15N hetNOE enhancements in DKP-proinsulin at 25 °C and 700 MHz; the color code is as in panel A. Values were not obtained for prolines (residues B29, C18, and C25) and at HN sites not detected due to rapid solvent exchange (B1 and B2) or conformational broadening (A4, A7, A8, and A11).
C-domain
The BC junction and residues C1–C26 exhibit near random-coil chemical shifts (Fig. 2A; supplemental Table S4) and motional narrowing (supplemental Fig. S5). The absence of medium and long range contacts (despite retention of sequential and intraresidue NOEs) suggests disorder. Near the CA junction, however, residues C27–C31 (sequence ALEGS) exhibit helix-related NOEs. 13Cα/β secondary shifts and SSP scores are similar to those of the A1–A8 α-helix (Fig. 2, A and B; supplemental Tables S3 and S4). No segmental substructure was observed in control NMR studies of the isolated C-peptide.
Subnanosecond fluctuations were probed by analysis of {1H}-15N hetNOEs (Fig. 2C; supplemental Table S5). Residues B3, B8, B21, B27–B30, and A9 are less well ordered than the remainder of the insulin moiety.6 The V-shaped pattern of C-domain hetNOEs (Fig. 2C, black) is a signature of a flexible loop tethered at each end. Residues immediately adjoining the BC and CA junctions (including the dibasic cleavage sites) exhibit hetNOE values lower than those of adjoining residues in the insulin moiety in accord with their motional narrowing and absence of non-local interproton contacts.
Structure
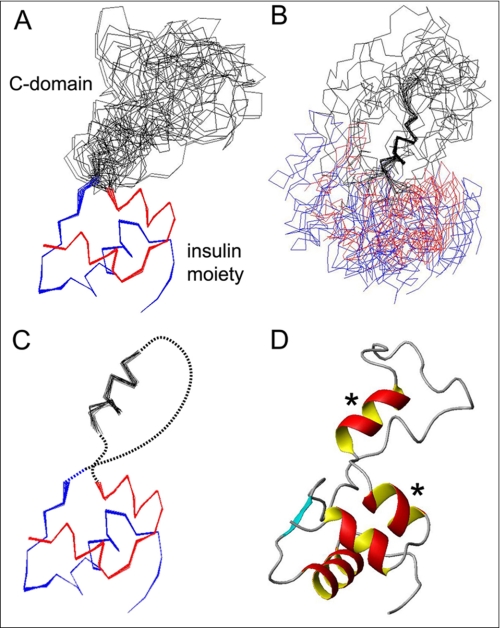
A DG/SA ensemble (Fig. 3A) was calculated based on 939 distance restraints and 97 dihedral angular restraints (supplemental Table S6). The number of restraints per residue in the insulin moiety (A1–A21 and B1–B28) and C-domain nascent α-helix (C27–C31) was 14. There were no distance violations >0.3 Å and no dihedral angle violations >5°. r.m.s.d. values within the insulin moiety are 0.24 Å (main chain) atoms and 0.89 Å (all heavy atoms); the C-domain α-helix exhibits segmental precisions 0.47 Å (main chain) and 0.91 Å (all heavy atoms) (supplemental Table S6). The A- and B-domains closely resemble the cognate chains of crystallographic T-state protomers (supplemental Fig. S6 and supplemental Table S7) (4). The structure of the C-domain is not well defined. The position of its nascent α-helix appears uncorrelated with that of the insulin moiety (Fig. 3B). Flexible tethering of the C-domain helix to the insulin moiety is shown in schematic form in Fig. 3C (dashed lines); a representative ribbon model is shown in Fig. 3D.
FIGURE 3.
NMR structure of DKP-proinsulin. A, superposition of 20 DG/SA structures of DKP-proinsulin aligned according the main-chain atoms of residues B2–B28 and A2–A20. The A, B, and C-domains are shown in red, blue, and black, respectively. B, ensemble aligned according to the main-chain atoms of the C-domain α-helix (C27–C31); color code is as in panel A. C, schematic representation of ensemble with dashed lines indicating unstructured segments within the C-domain; color code is as in panel A. D, representative ribbon structure of DKP-proinsulin. Asterisk indicates two α-helixes that exhibit conformational fluctuations as inferred from attenuation of 13C CSI and SSP scores (Fig. 2, A and B).
SPC3 Cleavage
To establish a model of prohormone processing, we investigated sequence determinants of cleavage by SPC3. Variant C-domains were introduced within DKP-proinsulin and tested as substrates (supplemental Table S8). Each variant retained native dibasic sites (Arg31-Arg32 and Lys64-Arg65; respective positions C1–C2 and C34–C35). These sites exhibit complementary structural contexts within a flexible extended strand (BC) or flanked by α-helices (CA). SPC3 cleaves the BC junction with higher efficiency than the CA junction (9). An HPLC/mass spectrometry assay enabled independent assessment of BC and CA cleavage.
Major determinants of substrate recognition by subtilisin/Kex-related proteases are mediated by residues N-terminal to the scissile bond (9). Surprisingly, however, deletion of residues C-terminal to the BC junction (residues C3–C6; sequence EAED) completely blocked BC cleavage; CA cleavage was impaired by less than 2-fold. The deletion shifted uncharged residues to positions C3–C6 (LQVG). Impaired BC cleavage is unlikely to be due to foreshortening of the C-domain as an extensive deletion (ΔC8–C32; lacking residues 38–62) impaired cleavage of either junction by <2-fold in accord with past studies (25). Deletions ΔC3 and ΔC3–C4 (leaving adjoining sequences AEDL and EDLQ) impaired BC cleavage by ∼4-fold. By contrast, activity was regained with successive deletion ΔC3–C5 (adjoining sequence DLQV). These results suggested that the P2′ residue (underlined above) contributes to SPC3 site selection. Indeed, a single acidic substitution at this position (AlaC4 → Glu; position 34) completely blocks BC cleavage. Ala substitutions at positions P1′, P3′, or P4′ (positions 33, 35, or 36) are by contrast well tolerated (supplemental Fig. S7A). The specific structural role of AlaC4 in the SPC3-proinsulin complex may enhance substrate binding as its substitution by Ser (in the context of HAED derived from glicentin; supplemental Table S8) likewise impairs BC cleavage (supplemental Fig. S7B). These findings suggest that the structural mechanism of prohormone processing has constrained C-domain sequences independently of its role in nascent protein folding.
DISCUSSION
The structure of an engineered proinsulin monomer, obtained by NMR methods, exhibits similarities to and differences from insulin. Alignment with a representative crystallographic insulin protomer (supplemental Fig. S6A) yielded average r.m.s.d. values of 1.04 Å (aligned atoms) and 1.78 Å (all heavy atoms). Although side-chain packing in the core is similar (supplemental Fig. S6B), salient differences occur in the B-domain β-turn (residues B20–B23), whose conformation may be influenced by lattice packing. Solution structures of DKP-proinsulin and DKP-insulin (26) are also similar. Following main-chain alignment (A1–A20 and B5–B28), r.m.s.d. values between mean structures are 1.06 Å (main chain) and 2.22 Å (all heavy atoms). Pronounced differences occur only in the A1–A8 segment (main-chain r.m.s.d. 1.41 Å) and its A1–A4 subsegment (main-chain r.m.s.d. 1.87 Å). This difference presumably reflects tethering of the C domain.
Dynamics of Proinsulin
{1H}-15N hetNOEs provide a sensitive probe for subnanosecond fluctuations. Such studies revealed a V-shaped pattern in the C-domain, indicating progressive flexibility from its tethered ends. Although the isolated C-peptide is a random coil (supplemental Table S4), the intact C-domain contains a short α-helix within its C-terminal subsegment. Its position is likely to be poorly correlated with that of the insulin moiety. {1H}-15N hetNOE-detected sites of flexibility within the insulin moiety are consistent with patterns of crystallographic B-factors (4).
13C NMR resonance assignment enabled calculation of residue-specific SSP scores (18). This score predicts at each residue the percentage of folded conformers (α-helix or β-strand). Limiting SSP scores of 1 or −1 imply formation of a fully formed α-helix or β-strand, respectively. In DKP-proinsulin, the mean segment-specific SSP scores are 18% (N-terminal A-domain segment), 77% (C-terminal A-domain α-helix), 100% (central B-domain α-helix), and 27% (nascent C-domain α-helix). The similar pattern in DKP-insulin (Fig. 2B) (12) implies that the C-domain does not “tighten” the secondary structure of the insulin moiety. Segmental SSP scores of the A1–A8 A-domain α-helix and C-domain α-helix imply conformational fluctuations (leading to averaging of chemical shifts; Ref. 24) despite maintenance of helix-related NOEs. Because A1–A8 hetNOE values are unremarkable, the time scale of such fluctuations must be longer than the rotational correlation time of the protein in accord with previous observations of conformational broadening on the millisecond time scale (26). Further characterization of such motions may be obtained by analysis of rotating-frame spin relaxation spectroscopy (27).
Biological Implications
Conversion of proinsulin to insulin occurs in the course of trafficking through the trans-Golgi network (1). C-peptide excision requires cleavage at Arg31-Arg32 (positions C1 and C2; the BC junction) and Lys64-Arg64 (positions C34 and C35; the CA junction). These cleavages are effected by prohormone convertases SPC3 (PC1/PC3) and SPC2 (PC2), members of a family of calcium-dependent endoproteases that process a wide variety of precursor proteins (9). The active sites of such enzymes accommodate extended and flexible peptide substrates as observed at the BC junction. Because excision of the C-domain occurs after folding of proinsulin, however, formation of enzyme-substrate complexes may require a conformational change at the CA junction to expose an extended peptide conformation (28). The attenuated SSP profile of the A1–A8 segment, evidence of a molten conformation, implies that kinetic and thermodynamic barriers to such distortion may be low. The dibasic residues at the CA junction are themselves flexible. The nascent C-domain α-helix would not in itself be expected to interfere with formation of an enzyme-substrate complex.
The molten character of the A1–A8 α-helix may reflect the anomalous presence of three β-branched residues (sequence GIVEQCCT; β-branched residues, underlined). IleA2, ordinarily buried in the core, must be exposed for both prohormone processing and binding of the mature hormone to the insulin receptor (29, 30). Flexibility of the N-terminal A-domain segment may also facilitate its disulfide-coupled folding in the β-cell by reducing kinetic barriers to pairing of cysteines A7–B7 and A6–A11. Oxidative folding of proinsulin proceeds through a defined series of intermediates (31). Pairwise substitution of cysteines A6–A11 by Ala or Ser leads to segmental unfolding of the A1–A11 segment (32), and more marked flexibility occurs on pairwise substitution of cysteine A7–B7 (33). Flexibility of the A1–A8 segment in protein-folding intermediates may facilitate transient deprotonation of thiolate moieties, their alignment for disulfide pairing, and interactions with protein disulfide isomerase (34). In the future, probes of protein dynamics (such as {1H}-15N hetNOEs and 13C CSI and SSP scores) may provide insight into the biophysical properties of populated disulfide intermediates (32) and perturbations caused by diabetes-associated mutations (10, 11).
A general mechanism of disease is mediated by proteotoxicity, ranging from aberrant intracellular aggregation of folding intermediates to formation of extracellular fibrils. Such pathological processes are exemplified by the toxic misfolding of insulin and proinsulin (31). Due to the presence of the C domain, proinsulin is markedly less susceptible to fibrillation than is insulin (35). We suggest that despite its flexibility, the C-domain sequence has evolved to minimize its propensity to fold or misfold. The extreme resistance of the isolated C-peptide to fibrillation (35), which is otherwise a universal property of polypeptides, highlights the special nature of such a non-folding sequence.
Concluding Remarks
The present study has demonstrated the utility of heteronuclear NMR spectroscopy in combination with protein engineering to investigate a protein refractory to crystallization. DKP-proinsulin provides a tractable model for both structural analysis and studies of prohormone processing. Although the organized insulin moiety foreshadows the structure of the mature hormone, its flexible tethering by the C-domain facilitates nascent folding. The further characterization of variant proinsulins may enable molecular rules governing dibasic target site selection to be deciphered. The solution structure of DKP-proinsulin thus provides a foundation for analysis of prohormone processing and comparative studies of non-foldable variants associated with neonatal diabetes mellitus.
Supplementary Material
Acknowledgments
We thank J. Whittaker and L. Whittaker for receptor-binding assays and K. Huang for participation in early stages of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant DK052085 (to R. B. M.) and Grants DK040949 and DK0697604 from the National Institutes of Health and American Diabetes Association (to M. A. W.). This work was also supported by American Heart Association Grant 0530067N (to Y. Y.).
The atomic coordinates and structure factors (code 2KQP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, supplemental Figs. S1–S7, supplemental Tables S1–S8.
NMR spectra are designated by nuclear coherence transfer.
Segmental SSP scores within the insulin moiety correlate with structural variability among crystallographic protomers of insulin (12).
Conformational fluctuations at these sites are in accord with their structural variability and higher thermal B-factors in crystal structures of insulin (4).
- DKP-insulin and DKP-proinsulin
- analogues containing B-chain substitutions AspB10, LysB28 and ProB29
- CSI
- chemical-shift index
- DG
- distance geometry
- SA
- simulated annealing
- hetNOE
- {1H}-15N nuclear Overhauser enhancement
- NOE
- nuclear Overhauser enhancement
- r.m.s.d.
- root mean square deviation
- SSP
- secondary structural propensity
- HPLC
- high performance liquid chromatography.
REFERENCES
- 1.Dodson G., Steiner D. (1998) Curr. Opin. Struct. Biol. 8, 189–194 [DOI] [PubMed] [Google Scholar]
- 2.Steiner D. F. (1998) Curr. Opin. Chem. Biol. 2, 31–39 [DOI] [PubMed] [Google Scholar]
- 3.Steiner D. F. (1967) Trans. N. Y. Acad. Sci. 30, 60–68 [DOI] [PubMed] [Google Scholar]
- 4.Baker E. N., Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. M., Hubbard R. E., Isaacs N. W., Reynolds C. D. (1988) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 319, 369–456 [DOI] [PubMed] [Google Scholar]
- 5.Weiss M. A., Frank B. H., Khait I., Pekar A., Heiney R., Shoelson S. E., Neuringer L. J. (1990) Biochemistry 29, 8389–8401 [DOI] [PubMed] [Google Scholar]
- 6.Bax A. (1994) Curr. Opin. Struct. Biol. 4, 738–744 [Google Scholar]
- 7.Mackin R. B., Choquette M. H. (2003) Protein Expr. Purif. 27, 210–219 [DOI] [PubMed] [Google Scholar]
- 8.Kay L. E., Torchia D. A., Bax A. (1989) Biochemistry 28, 8972–8979 [DOI] [PubMed] [Google Scholar]
- 9.Henrich S., Lindberg I., Bode W., Than M. E. (2005) J. Mol. Biol. 345, 211–227 [DOI] [PubMed] [Google Scholar]
- 10.Støy J., Edghill E. L., Flanagan S. E., Ye H., Paz V. P., Pluzhnikov A., Below J. E., Hayes M. G., Cox N. J., Lipkind G. M., Lipton R. B., Greeley S. A., Patch A. M., Ellard S., Steiner D. F., Hattersley A. T., Philipson L. H., Bell G. I. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15040–15044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colombo C., Porzio O., Liu M., Massa O., Vasta M., Salardi S., Beccaria L., Monciotti C., Toni S., Pedersen O., Hansen T., Federici L., Pesavento R., Cadario F., Federici G., Ghirri P., Arvan P., Iafusco D., Barbetti F.Early Onset Diabetes Study Group of the Italian Society of Pediatric Endocrinology and Diabetes (2008) J. Clin. Invest. 118, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y., Petkova A., Huang K., Xu B., Hua Q. X., Ye I. J., Chu Y. C., Hu S. Q., Phillips N. B., Whittaker J., Ismail-Beigi F., Mackin R. B., Katsoyannis P. G., Tycko R., Weiss M. A. (January27, 2010) J. Biol. Chem. DOI 10.1074/jbc.M109.067850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu M., Wan Z. L., Chu Y. C., Aladdin H., Klaproth B., Choquette M., Hua Q. X., Mackin R. B., Rao J. S., De Meyts P., Katsoyannis P. G., Arvan P., Weiss M. A. (2009) J. Biol. Chem. 284, 35259–35272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 15.Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. (1991) J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 16.Omichinski J. G., Pedone P. V., Felsenfeld G., Gronenborn A. M., Clore G. M. (1997) Nat. Struct. Biol. 4, 122–132 [DOI] [PubMed] [Google Scholar]
- 17.Stone M. J., Fairbrother W. J., Palmer A. G., 3rd, Reizer J., Saier M. H., Jr., Wright P. E. (1992) Biochemistry 31, 4394–4406 [DOI] [PubMed] [Google Scholar]
- 18.Marsh J. A., Singh V. K., Jia Z., Forman-Kay J. D. (2006) Protein Sci. 15, 2795–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 20.Schwieters C. D., Kuszewski J. J., Clore G. M. (2006) Progr. NMR Spectroscopy 48, 47–62 [Google Scholar]
- 21.Weiss M. A., Hua Q. X., Lynch C. S., Frank B. H., Shoelson S. E. (1991) Biochemistry 30, 7373–7389 [DOI] [PubMed] [Google Scholar]
- 22.Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 23.Wuthrich K. (1986) NMR of Proteins and Nucleic Acids, pp. 117–199, John Wiley & Sons, New York, NY [Google Scholar]
- 24.Berjanskii M. V., Wishart D. S. (2005) J. Am. Chem. Soc. 127, 14970–14971 [DOI] [PubMed] [Google Scholar]
- 25.Taylor N. A., Docherty K. (1992) Biochem. J. 286, 619–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hua Q. X., Hu S. Q., Frank B. H., Jia W., Chu Y. C., Wang S. H., Burke G. T., Katsoyannis P. G., Weiss M. A. (1996) J. Mol. Biol. 264, 390–403 [DOI] [PubMed] [Google Scholar]
- 27.Palmer A. G., 3rd, Massi F. (2006) Chem. Rev. 106, 1700–1719 [DOI] [PubMed] [Google Scholar]
- 28.Lipkind G., Steiner D. F. (1999) Biochemistry 38, 890–896 [DOI] [PubMed] [Google Scholar]
- 29.Xu B., Hua Q. X., Nakagawa S. H., Jia W., Chu Y. C., Katsoyannis P. G., Weiss M. A. (2002) J. Mol. Biol. 316, 435–441 [DOI] [PubMed] [Google Scholar]
- 30.Xu B., Huang K., Chu Y. C., Hu S. Q., Nakagawa S., Wang S., Wang R. Y., Whittaker J., Katsoyannis P. G., Weiss M. A. (2009) J. Biol. Chem. 284, 14597–14608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss M. A. (2009) J. Biol. Chem. 284, 19159–19163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss M. A., Hua Q. X., Jia W., Chu Y. C., Wang R. Y., Katsoyannis P. G. (2000) Biochemistry 39, 15429–15440 [DOI] [PubMed] [Google Scholar]
- 33.Hua Q. X., Nakagawa S. H., Jia W., Hu S. Q., Chu Y. C., Katsoyannis P. G., Weiss M. A. (2001) Biochemistry 40, 12299–12311 [DOI] [PubMed] [Google Scholar]
- 34.Winter J., Klappa P., Freedman R. B., Lilie H., Rudolph R. (2002) J. Biol. Chem. 277, 310–317 [DOI] [PubMed] [Google Scholar]
- 35.Huang K., Dong J., Phillips N. B., Carey P. R., Weiss M. A. (2005) J. Biol. Chem. 280, 42345–42355 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.