Abstract
Ovarian cancer ranks the most lethal among gynecologic neoplasms in women. To develop potential bio-markers for diagnosis, we have identified five novel genes (CYP39A1, GTF2A1, FOXD4L4, EBP, and HAAO) that are hypermethylated in ovarian tumors, compared with the non-malignant normal ovarian surface epithelia, using the quantitative methylation-specific polymerase chain reactions. Interestingly enough, multivariate Cox regression analysis has identified hypermethylation of CYP39A1 correlated with an increase rate of relapsing (P=0.032, hazard ratio >1). Concordant hypermethylation in at least three loci was observed in 50 out of 55 (91%) of ovarian tumors examined. The test sensitivity and specificity were assessed to be 96 and 67% for CYP39A1; 95 and 88% for GTF2A1; 93 and 67% for FOXD4L4; 81 and 67% for EBP; 89 and 82% for HAAO, respectively. Our data have identified, for the first time, GTF2A1 alone, or GTF2A1 plus HAAO are excellent candidate biomarkers for detecting this disease. Moreover, the known functions of these gene products further implicate dys-regulated transcriptional control, cholesterol metabolism, or synthesis of quinolinic acids, may play important roles in attributing to ovarian neoplasm. Molecular therapies, by reversing the aberrant epigenomes using inhibitory agents or by abrogating the upstream signaling pathways that convey the epigenomic perturbations, may be developed into promising treatment regimens.
Keywords: ovarian cancer, epigenetics, DNA methylation, biomarkers, quantitative methylation-specific polymerase chain reaction
Introduction
Ovarian cancer remains the most lethal malignancy of the female reproductive tract, and is ranked as the fourth leading cause of cancer death in women (1). While there are over 20,000 new cases diagnosed annually, approximately 15,000 deaths are reported in the United States each year (1). Typically, ovarian cancer is asymptomatic in the early stages and hence three-fourths of women are diagnosed after the disease has metastasized beyond the primary site which frequently causes poor prognostic outcomes. Investigation of early molecular events resulting in ovarian cancer has been hampered by the lack of identifiable precursor lesions as well as poor biomarkers for laboratorial tests. Currently, the only clinically accepted biomarker is CA125, which is found elevated in the serum of the majority of ovarian cancers (reviewed in ref. 2). Although CA125 has high clinical sensitivity, its use for early detection and screening is limited as it is also frequently elevated in women with benign disease (reviewed in ref. 3). As of today, intensive efforts have been undertaken to identify biochemical or genetic biomarkers for improved diagnosis and for better stratification of disease subtypes.
One of the modern biomarkers is methylated DNA, which has been shown to be a common epigenetic alteration in solid tumors, including ovarian cancer (reviewed in refs. 4,5). This aberrant event frequently occurs in the promoter CpG islands of a gene and is associated with transcriptional silencing (6). In normal cells, the majority of CpG islands are generally refractory to chemical modification and remain unmethylated. In contrast, de novo DNA methylation (or DNA hypermethylation) has so far been observed in >200 loci with CpG islands in many different types of cancers (7). In addition, increased numbers of loci with elevated methylation in CpG islands were observed during tumorigenesis (8). Because this chemical modification is somatically stable, cancer-associated, and readily amplified by robust PCR-based technologies, hypermethylated DNA in exfoliate tumor cells has been detected in body fluids such as urine, cerebral spinal fluid, plasma and whole blood (9-11). Likewise, epigenetic alterations may potentially become a valuable biomarker resource for detection and diagnosis of ovarian cancer.
Toward this end, we have developed a microarray-based technique, known as differential methylation hybridization (DMH), for a global screening of hypermethylated loci in primary ovarian tumors (12,13). This initial effort has identified more than 180 candidate loci, which can be used for cancer detection as well as predicting patients' treatment responses (unpublished data). As an ongoing effort, we conducted a quantitative validation of individual locus, known as real-time methylation-specific polymerase chain reactions (qMSP), to quantify their methylation level in an independent set of primary ovary tumors. Here we report five loci, CYP39A1, GTF2A1, FOXD4L4, EBP, and HAAO, whose methylation status was found to be specific to ovarian tumor, in relation to the unrelated non-cancerous ovarian surface epithelia. Among them, GTF2A1 and HAAO are excellent candidate biomarkers, due to their high sensitivity and specificity in the qMSP tests. Moreover, our data further suggest reversing aberrant epigenomes may become a promising molecular therapy regimen for treating ovarian cancer.
Materials and methods
Tumor specimens from patients with ovarian cancer
Table I summarizes the clinicopathological information of 77 patients diagnosed with stage III or IV epithelial ovarian cancer. These tumor samples were generously provided by the Western Infirmary and Stobhill General Hospital (Glasgow, UK) and the University Medical Centre Groningen (Groningen, The Netherlands). As controls, normal ovarian surface epithelia (nOSE) were obtained from 9 patients undergoing procedures for benign conditions. All experimental protocols were approved by local institutional review boards and ethics committee.
Table I.
Clinical and pathological characteristics of patients with ovarian cancer (n=77).
| Patient characteristics | No. | (%) |
|---|---|---|
| Age (years) | ||
| ≤60 | 38 | 49 |
| >60 | 39 | 51 |
| Histology | ||
| Serous | 58 | 75 |
| Mucinous | 3 | 4 |
| Endometrioid | 7 | 9 |
| Clear cell | 3 | 4 |
| Unknown | 6 | 8 |
| FIGO stage | ||
| III | 60 | 78 |
| IV | 17 | 22 |
| Response to chemotherapy | ||
| Complete response (CR) | 18 | 23 |
| Partial response (PR) | 19 | 25 |
| Stable disease (SD) | 8 | 10 |
| Progressive disease (PD) | 13 | 17 |
| Unable to evaluate | 19 | 25 |
| Progression-free survival | ||
| <6 months | 14 | 18 |
| 6-12 months | 23 | 30 |
| 1-2 years | 25 | 32 |
| 2-5 years | 9 | 12 |
| >5 years | 6 | 8 |
| Overall survival | ||
| <6 months | 7 | 9 |
| 6-12 months | 16 | 21 |
| 1-2 years | 15 | 19 |
| 2-5 years | 26 | 34 |
| >5 years | 13 | 17 |
| Status at last follow-up | ||
| No evidence of disease (NED) | 9 | 12 |
| Alive with disease (AWD) | 14 | 18 |
| Died of other causes (DWOD) | 0 | 0 |
| Died with/from disease (DWD/DOD) | 54 | 70 |
Bisulfite conversion, qMSP, and conventional real-time polymerase chain reactions (PCR) to assess the copy numbers of genes of interest. Genomic DNA was extracted from ovarian tumors or from cell lines, purified by using QIAamp DNA Mini Kit (Qiagen, Germantown, MD), and subjected to bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research, Orange, CA). The resultant DNA was subjected to real-time qMSP using Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) as described previously (37,38). The sequences of DNA oligonucleotide primers utilized in qMSP reactions are shown in Table II. PCR comprising SYBR-Green as the reporting molecule (Applied Biosystems), were carried out at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec, annealing temperature (60-64°C) for 30 sec, and 72°C for 35 sec. To equalize the amount of DNA in each reaction, the threshold cycle number (Ct) for each sample was first normalized in a PCR amplification utilizing primers devoid of CpG dinucleotides but interrogating COL2A1. Because COL2A1 is frequently in low- or single-copy in human genomes, it hence commonly serves as a methylation-independent control to normalize the quantity of DNA specimens (39). Next, relative methylation level in the locus of interest was assessed by comparing Ct of qMSP from each sample to that from a methylation standard curve generated by SssI-treated sperm DNA (Chemicon, Temecula, CA) in which nearly all CpG dinucleotides are methylated by CpG methylase in this study (data not shown). The level of methylation in each sample is expressed as a percentage, in relation to SssI standard which was set as 100%. Thus, a percentage of methylation >100 may be partly due to an elevated copy number in the tumor samples compared to the sperm DNA. To assess the DNA copy number of a given locus in relation to the one of SssI, DNA was amplified by a conventional real-time PCR method (40), in a methylation-independent manner using specific primers to interrogate CYP39A1 locus: forward primer (GGAGTTTAGGAGGT TATTGGG) and reverse primer (CTCCCTCCCACCTC CACT).
Table II.
DNA sequences of oligonucleotide primers for qMSP and for RT-PCR.
| qMSP forward | qMSP reverse | RT-PCR forward | RT-PCR reverse | |
|---|---|---|---|---|
| CYP39A1 | GTGGCGGTTTCGGGTGC | GCGAGAAGGAGTGCGTTATCG | TGCTCTGTTAGAGGTTCAGATGTGT | CCACCAAATGGAGATAACTCTGTTT |
| GTF2A1 | CGCGAAAGACCCACCCTC | CTGGCGCGTTCCTATTCG | GGAAGGACAGGAACTCTTTGACA | TGCCATCCTTGAGATGAAATTTC |
| FOXD4L4 | CGGCGGTTTTTGAAGATGTTC | GCAAATACCGCTAAACGTAAAACG | CTCGTACATCGCGCTCATCA | AGTATGGGAAGCGGCCACTA |
| EBP | TTTCGTTTTATTATTGGTTCGTTTC | ACGACAACGACCTCCCACG | CCCCTCCGCTTCATTCTACA | CAGCTCTCCGTGCTGGAATC |
| HAAO | AGGTGGGGAGGGTTCGC | CCAACGAAAAACCGCACG | CTCAGGTACTATGTGGGCGACA | GGGCTTTCCTGTTCTGTACTGC |
Ovarian cancer cell lines and epigenomic treatments
Immortalized ovarian surface epithelial cells (IOSE), derived from the primary culture transduced with the catalytic subunit of hTERT and the human papiloma virus E7 subunit (41), were grown in Medium MCDB105:199 (1:1) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS). The human ovarian cancer A2780 cell line was originally isolated from an ovarian adenocarcinoma of an untreated patient and is relatively sensitive to cisplatin in vitro (42). Independent, multiple selected lines (MCP3 and MCP8) derived from A2780, were obtained by seven cycles of selection, with increasing concentrations of cisplatin (43,44). Nevertheless, A2780, MCP3 and MCP8 were cultivated in RPMI-1640 (Invitrogen) containing 10% FBS as previously described (44). To reverse the aberrant epigenomes, sub-confluent cells were seeded on cultivation vessels and were treated with 2.5 μM 5-aza-2′-deoxycytidine (ADC) (Sigma Inc., St. Louis, MO) alone, 0.5 μM trichostatin A (TSA) (Sigma Inc.) alone, or with both drugs. Mock control cells were grown concurrently, but treated with vehicle DMSO. Forty-eight hours after drug treatments, cells were harvested and subjected to RNA extraction.
Quantify mRNA by quantitative reverse transcription PCR (qRT-PCR)
Total RNA from ovarian cancer cell lines was purified by using TRIzol reagents (Invitrogen). The resultant RNA was first reverse transcribed into cDNA, and later annealed to the corresponding primers (Table II) prior to PCR amplification using the following conditions: 95°C for 10 min, 40 cycles of 95°C for 15 sec, 58-60°C for 30 sec, and 72°C for 35 sec. In each sample, the level of mRNA was determined by subtracting the threshold cycle number (Ct) of the internal control GAPDH from that of test locus. The relative expression of mRNA in the locus of interest is expressed as fold changes in the treated cells, in relation to the untreated mock control.
Statistical analysis
The significance of DNA methylation in tumors, in relation to nOSE samples, was assessed by real-time qMSP followed by statistical analysis using the Wilcoxon rank-sum test. Pearson's correlation was used to calculate concurrent hypermethylation among multiple genes. Difference in mRNA level between the untreated and treated cell lines was determined by the paired Student's t-test and was considered to be significant, if the P-value is <0.05.
The clinical application of these five genes as biomarkers for detecting ovarian carcinomas was appraised by their specificity and sensitivity using a 100-fold cross-validation method (23). For a given locus, 44 specimens from the cancer group and 5 from nOSE, were randomly selected using the statistical analysis package R (24), and whose qMSP values were used for the following calculation. In principle, by collecting: i) the nOSE samples with methylation levels greater than the minimum in the cancer group; and ii) the cancer samples with methylation degree less than the maximum in nOSE specimens, the threshold value was set to be the median of all qMSP values of samples from this collection. By repeating the analysis for additional 99 times and each run encompasses a different random sampling of the specimens, the final threshold value was set as the average of median across the 100 iterations, and thereby served as a cut-off value for a given gene above which the test specimen is counted as hypermetylated. Further, sensitivity for a given locus is the percentage of the true positives (hypermethylated) in all cancer specimens studied, whereas the specificity is that of the true negatives (unmethylated) in nOSE (shown in Table III). On the other hand, Cox proportional hazard regression model was used to assess the affect of hypermethylation on clinical parameters including progression-free survival, using a software package R (17,24).
Table III.
qMSP detects hypermethylated loci with high sensitivity and specificity, in ovarian tumors.
| Gene(s) | Threshold | True positives in tumors | True negatives in nOSE | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| CYP39A1 | 32.73 | 53/55 | 6/9 | 96 | 67 |
| GTF2A1 | 41.89 | 52/55 | 7/8 | 95 | 88 |
| FOXD4L4 | 6.57 | 51/55 | 6/9 | 93 | 67 |
| EBP | 12.90 | 43/53 | 6/9 | 81 | 67 |
| HAAO | 0.06 | 51/57 | 7/9 | 89 | 82 |
Results
Identification of hypermethylated loci in ovarian cancer
Aberrant epigenomic regulation is a frequent event associated with human malignancy. While DNA hypermethylation and histone deacetylation contributes to tumor suppressor gene silencing, our laboratory has previously examined methylation patterns of ovarian cancer using DMH and has identified potential CpG island loci that can be developed as biomarkers for cancer diagnosis and prognosis (12,13). Briefly, amplicons derived from ovarian tumors or from normal controls were labeled with fluorescent Cy5 and Cy3 dyes, respectively, and then were co-hybridized to a CpG island microarray slide. Hybridized slides were scanned and candidate genes with signal ratios (Cy5/Cy3) ≥2 were scored as potential hypermethylation in tumor samples compared with nOSE. To continue validating the loci identified from our previous DMH microarray findings (12,13), we developed qMSP assays to quantify the methylation levels of candidate CpG island loci.
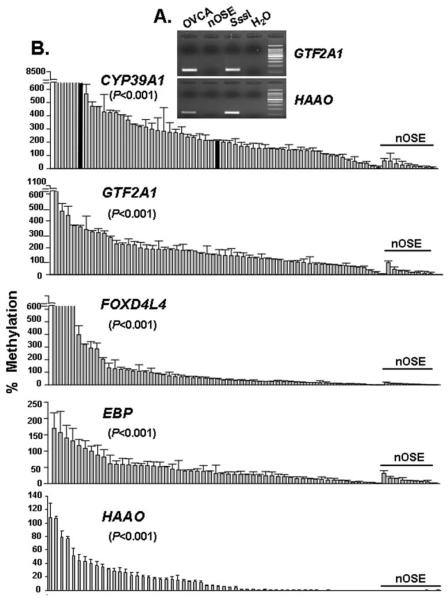
For each given locus, a study was conducted to determine amplification specificity of each methylation-specific primer pair. Pairs of oligonucleotides were utilized in the qMSP reactions (sequences are listed in Table II) to interrogate the CG-rich regions within the vicinity of the promoters and the first exon, of these genes. PCR products were viewed on agarose gels as an initial discriminatory testing to differentiate pooled ovarian tumors from the combined controls (nOSE; representative examples are shown in Fig. 1A). qMSP was then used to assess the methylation level of 5 CpG island loci (CYP39A1, GTF2A1, FOXD4L4, EBP, and HAAO) in 77 ovarian tumors and 9 control nOSEs.
Figure 1.
Significant hypermethylation was observed in ovarian tumors, as opposed to nOSE. (A) Amplification of corresponding pairs of primers resulted in specific amplicons of the correct sizes. The target DNA employed in this assay was extracted from either pooled ovarian tumors (OVCA) or from nOSE. Noticeably, SssI DNA served as a positive control (SssI), while reactions devoid of target DNA (water) served as negative control. (B) After equalizing the quantity of DNA samples by normalization to the internal control loci COL2A1, the relative level of methylation was expressed as percentage (%), in relation to a standard curve generated from SssI in which methylation was set as 100%. All samples were assayed in triplicates and the tumor-specific methylation in ovarian tumors, in relation to nOSE samples (underlined), was analyzed by Wilcoxon rank-sum test and significance was granted if the P-value was <0.05. In the panel of CYP39A1, three samples denoted with black bars were further assessed for DNA copy numbers.
Due to limited amounts and the quality of DNA, not all patient specimens could be amplified by qMSP. However, the average methylation level of a given locus in 55 tumors successfully analyzed, in relation to the mean of 9 nOSE samples, was calculated as: 469.93 vs. 27.98 for CYP39A1; 182.02 vs. 29.43 for GTF2A1; 204.15 vs. 6.63 for FOXD4L4, 45.18 vs. 11.93 for EBP; and 17.35 vs. 0.05 for HAAO (Fig. 1B). Exceeding the 100% methylation level in four loci (CYP39A1, GTF2A1, FOXD4L4, and EBP) could likely be a result of increased genome copy number in ovarian tumors, as compared to the control DNA derived from the SssI-treated sperm sample and nOSE. For example, while we interrogated the meythylation levels of CYP39A1 by qMSP, we observed that hypermethylation was positively associated with increased DNA copy numbers. As the metyhylation levels in ovarian tumors samples named 5, Z112, and S05), were quantified to be 696, 210 and 10.61% of the SssI control (black bars in Fig. 1B), their corresponding DNA copy numbers were estimated as 4.00, 2.28, and 0.45, in relation to SssI derived from sperm DNA. In fact, growing body of evidence has demonstrated that elevated genome copy number is a common hallmark of advanced ovarian malignancy (14-16). Most importantly, the methylation levels of these five loci were significantly elevated in tumors (P<0.001). Interestingly enough, after adjusting for age, FIGO stage and response to chemotherapy, another advanced statistical method employing multivariate Cox regression analysis (17) has identified hypermethylation of CYP39A1 correlated with an increase rate of relapsing (P=0.032, hazard ratio >1). However, the same multivariate analysis cannot correlate hypermethylation of the remaining four genes with any of clinical parameters including progression-free survival.
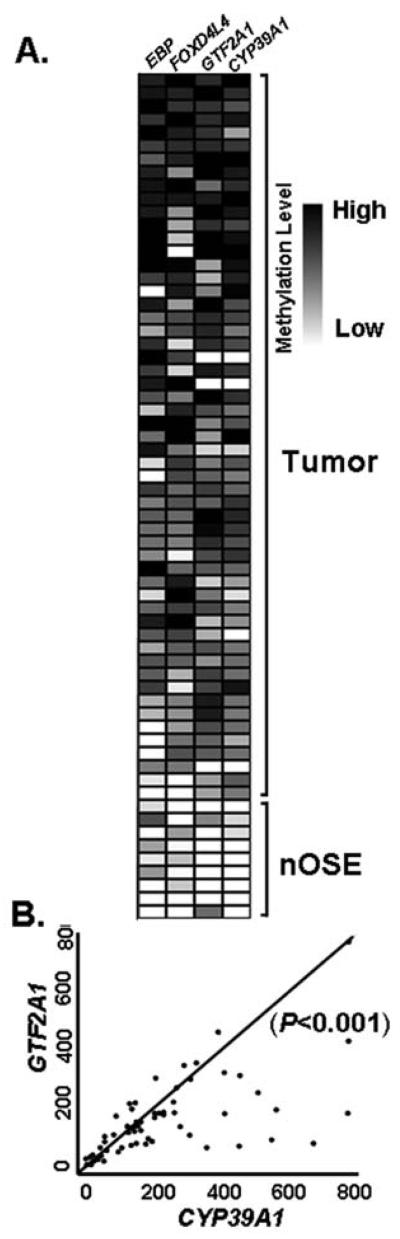
Next, we determined whether concordant hypermethylation of multiple genes could be simultaneously observed in these tumors. Based on the qMSP data shown in Fig. 1B, a methylation heat map of four loci was generated from 55 tumors and 9 nOSE specimens (Fig. 2A). Simultaneous hypermethylation of equal or greater than three loci was observed in 50 out of 55 (91%) of the tumors examined. Interestingly, two of these loci, CYP39A1 and GTF2A1, displayed statistically significant concurrent hypermethylation in 52 out of 55 (95%) ovarian tumors examined (P<0.001; Pearson's correlation coefficient, r=0.6408; Fig. 2B), suggesting the likelihood of the so-called CpG island methylator phenotype (CIMP) (18) in these ovarian tumors. CIMP refers to a notion that a subset of tumors has widespread methylation of CpG islands that leads to epigenetic inactivation of tumor suppressor genes. This phenomenon is most apparent in colon cancer and was first characterized by a concordant methylation of promoter regions of two separate genes, CDKN2A and TBSI (19). Subsequent studies have added several loci to the list of genes and the minimal number of genes is commonly ranged between 3 and 8 in colorectal cancer (20). Similar CIMP, with concurrent hypermethylation in between three to six loci, has been observed in ovarian cancer and is associated with adverse prognosis (21) or response to chemotherapy (22).
Figure 2.
Concordant DNA hypermethylation can be observed in ovarian cancer. (A) By aligning the level of DNA methylation to ovarian tumors and nOSE samples, a heat map was generated. Ovarian tumors harboring the highest magnitude of methylation (in black) observed in the greatest number of loci were shown at the top, as opposed to tumor samples with the lowest level of methylation (in white and grey), observed in the least number of genes, are shown in bottom panel. Because nOSE samples retain a negligible level of methylation in all four loci, they were placed on the farthest bottom section. (B) Scatterplot manifested concurrent hypermethylation (P<0.001; Pearson's correlation coefficient, r=0.6408) observed in two loci (CYP39A1 and GTF2A1) in ovarian tumors. Scales on X- and Y-axis represent % of methylation for CYP39A1 and GTF2A1, respectively (shown in Fig. 1B).
qMSP assays for hypermethylated genes bestow diagnostic tests with high sensitivity and specificity
The application of these hypermethylated loci as biomarkers was accessed by their sensitivity and specificity. By using a 100-fold cross-validation method (23) and the statistical analysis package R (24), we have set off the threshold cut-off value for hypermethylation of a given locus (first column, Table III) and demonstrated that qMSP tests on all five genes can offer high sensitivity in detecting ovarian carcinoma (with sensitivity values ranged between 81 and 96%) (Table III). Among them, the highest specificity was identified in GTF2A1 (88%), while HAAO ranked the second (82%). Given the advantage of high sensitivity plus specificity, testing for the methylation in GTF2A1 alone or in combination with HAAO, may comprise novel targets for molecular diagnostics.
DNA demethylation and/or histone acetylation reactivated the expression of hypermethylated genes
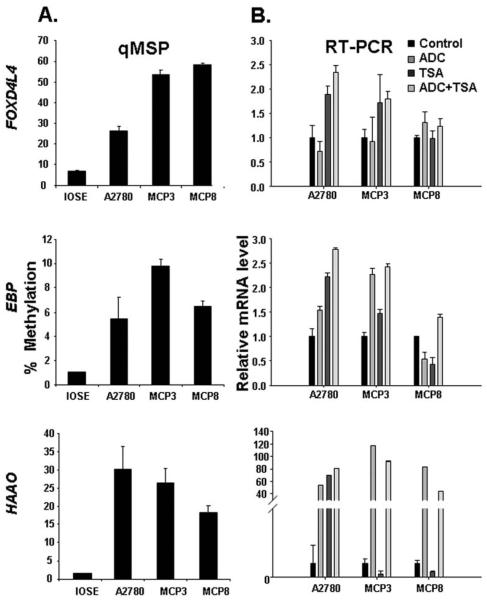
Our data demonstrated elevated methylation of five loci in ovarian tumors. To validate this finding in the laboratory cultivation system, ovarian cancer cell lines (A2780, MCP3 and MCP8) and the immortalized ovarian surface epithelia (IOSE) were subjected to epigenomic studies. We demonstrated that all three ovarian cancer lines retained remarkable methylation in three loci (FOXD4L4, EBP, HAAO) (Fig. 3A). However, methylation of two other loci (CYP39A1 and GTF2A1) was unnoticeable (data not shown) and hence both genes were excluded from further studies.
Figure 3.
Aberrant methylation in ovarian cancer cell lines represses target gene expression. (A) qMSP method has assessed elevated methylation in 3 loci observed in ovarian cancer cell lines, in relation to the IOSE. As described in the legend of the Fig. 1B, by comparing to SssI which was set as 100%, the methylation level of three loci was appraised in ovarian cancer cell lines. (B) To evaluate if an aberrant epigenome is one of the causes attributing to down-regulated expression, cells were treated with 5-aza-2′-deoxycytidine (ADC) alone, trichostatin A (TSA) alone, or both ADC plus TSA. Forty-eight hours later, mRNA expression was assessed by RT-PCR. Equal quantity of mRNA was assured by normalizing to the internal control GAPDH. Relative expression of a given locus in treated cells was obtained by comparing mRNA level in treated cells to that in vehicle DMSO treated cells (control) which was set as 1.*Indicates statistic significance between treated and control samples, which is denoted by the P-value (<0.05) derived from Student's t-test.
Our ultimate goal is to decipher the epigenetic gene silencing in ovarian cancer and to advance our current understanding about this malignancy. Because both DNA methylation and histone modifications are apparent epigenetic phenomena, we next performed a series of treatments to reverse the epigenomic suppression and to reveal the epigenetic regulatory mechanisms underlying gene silencing. Interestingly, DNA demethylating agent (ADC) caused prominent reactivation of HAAO (>60 fold increase) in all 3 ovarian cancer cell lines. However, inhibition of histone de-acetylation (by TSA treatment) resulted in variable outcomes. TSA induced greater than 70-fold activation in parental A2780 line, but conducted suppression, rather than activation, in the drug-resistant lines (MCP3 and MCP8). Yet, dual treatments failed to convey synergistic effect, suggesting DNA hypermethylation is far more critical than histone deacetylation in commanding HAAO repression in the drug resistant lines (MCP3 and MCP8), as opposed to the parental A2780 line that requires both mechanisms for regulation.
Similar variability was observed in silencing of two loci, FOXD4L4 and EBP (Fig. 3B). DNA hypermethylation appeared to be more critical in two cases (FOXD4L4 in MCP8, and EBP in MCP3) while histone deacetylation seemed to be more prominant in three occurrence (FOXD4L4 in A2780 and MCP3; and EBP in A2780). As an exception, reactivation of EBP in MCP8 required combinatorial actions from both agents, suggesting both mechanisms simultaneously suppressed the expression of EBP. However, we cannot exclude the possibility that failed outcomes are due to ineffective uptake, increased efflux and metabolism, or unknown cytotoxicity that disabled their action.
Discussion
DNA methylation and covalent modifications of histone proteins are the two epigenetic modifications important in transcriptional control and conveying neoplasms. Given the silencing effect on tumor-suppressor genes by DNA methylation, the importance of examining hypermethylated genes in ovarian tumors cannot be overemphasized. In this study, we have identified five novel loci, CYP39A1, GTF2A1, FOXD4L4, EBP, and HAAO, with extraordinary methylation in ovarian carcinomas, as opposed to normal surface epithelial counterparts. Discovery of these hypermethylated genes has provided a greater understanding of cellular signaling pathways that promote the neoplastic phenotypes, or are the consequence of ovarian malignancy.
The known biological functions associated with these gene products include acting as transcription factors [GTF2A1 (25); and FOXD4L4 (26)], playing roles in drug binding, transport and anti-proliferative activities [EBP (27,28)], governing vertebrate neural crest development [FOXD4L4 (29)], modulating lipid and steroid biosynthesis and drug metabolism [CYP39A1 (30,31) and EBP (32)]; and encoding an enzyme 3-hydroxyanthranilate 3, 4-dioxygenase that harbors electron carrier activity and catalyzes the synthesis of the neurotoxin quinolinic acid from 3-hydroxyanthranilic acid [HAAO (33,34)]. Perhaps, their normal functions are involved in tumor suppression, and perturbation of these loci by DNA methylation is associated with ovarian malignancies. However, whether hypermethylated genes are the etiological causes or the consequence of ovarian malignancy remains unresolved. While abnormal expression of CYP39A1, GTF2A1, FOXD4L4, and EBP is linked to various neoplasms (25-29,32), HAAO is a novel gene whose methylation status has not been reported in any human disease, nor its relation to malignant features.
In addition to ovarian tumors, hypermethylation of three loci (FOXD4L4, EBP, and HAAO) was similarly validated in ovarian cancer cell lines in vitro, suggesting this chemical modification is stable and can be inherited during laboratory cultivation phase. However, methylation of two other genes (CYP39A1 and GTF2A1) was undetectable in ovarian cancer lines, indicating lack of tumor microenvironment niche might have paused the CpG island methylator phenotype, as both loci were concordantly methylated in 95% of tumors (52 out of 55 tumors) (Fig. 2B).
To substantiate in vitro epigenomic studies, we further investigated how DNA hypermethylation imparts transcriptional expression by performing a series of laboratory treatments using ADC or TSA Transcriptional repression of three loci (FOXD4L4, EBP, and HAAO) can be relieved by either a single agent alone (3 out of 9 occurrence: FOXD4L4 in MCP8; HAAO in MCP3 and MCP8) or synergized by combinatorial treatments using ADC plus TSA (6 out of 9 cases: FOXD4L4 in A2780 and MCP3; EBP in A2780, MCP3, and MCP8; HAAO in A2780) (Fig. 3B), suggesting the control of these three loci is commanded mostly by cooperative actions from dual pathways, while minimally by a single epigenomic mechanism. Yet, the degree of methylation in these ovarian cancer lines has not been proportionally correlated to their susceptibility to ADC (compare Fig. 3A and B), which strongly supports the notion that multiple epigenomic events beside DNA methylation can orchestrate gene expression in a cooperative manner, in consistent with we reported previously (35).
The ovarian cancer biomarkers identified in this study, are likely to be potentially developed into an innovative strategy for disease detection, prognosis, and monitoring the treatment outcomes. Although the current study is limited to the unavailable early stage ovarian cancer and can only manifest promoter hypermethylation of five loci in the later stages of tumors (stages III and IV), remarkable silencing of HAAO has been independently observed in the majority of early stage ovarian tumors. It is noted that 80 out of 83 ovarian tumors at FIGO stage I showed down-expression, compared with the controls derived from the pooled normal tissues (GEO database, accession number GSE8842). Because promoter hypermethylation is a progressive as well as cumulative event (36) and commonly leads to gene silencing, down-regulation of HAAO in the early stage of ovarian cancer supports a promising notion that this locus (and perhaps the remaining genes studied in this report) might have been hypermethylated in the lower stages of ovarian tumors. Nevertheless, the advantage of stability in methylated DNA and robust PCR amplification technology has shed light on the feasibility of detecting these epigenetic biomarkers in patients' body fluids. With its high sensitivity and specificity, qMSP method can be developed into a novel molecular diagnosis test. Combating these pathways to reverse aberrant epigenomes may provide a feasible option of target-specific molecular therapy, if the molecular pathways that orchestrate the abnormal epigenomic changes can be further identified and characterized in the future.
Acknowledgements
Authors thank Daniel Deatherage and Chris Cleland for critical reading of this manuscript. This research is supported by the following funds: the American Cancer Society, and the Department of Defense BC073892 (H.J.L.L.); National Cancer Institute grants R21 CA110475 (T.H.M.H. and R.B.); R01 CA-85289 (K.P.N. and T.H.M.H.); U54 CA113001 (K.P.N., P.S.Y. and T.H.M.H.); program grant A2662 from Cancer Research, UK (R.B.); and the Ohio State University Comprehensive Cancer Center-Arthur G. James Cancer Hospital and Richard J. Solove Research Institute (P.S.Y. and T.H.M.H.).
Abbreviations
- DAC
5-aza-2′-deoxycytidine
- nOSE
normal ovarian surface epithelia
- qMSP
quantitative methylation-specific polymerase chain reactions
- qRT-PCR
quantitative reverse transcription followed by polymerase chain reactions
- TSA
trichostatin A
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Bast RC, Jr, Badgwell D, Lu Z, Marquez R, Rosen D, Liu J, Baggerly KA, Atkinson EN, Skates S, Zhang Z, Lokshin A, Menon U, Jacobs I, Lu K. New tumor markers: CA125 and beyond. Int J Gynecol Cancer. 2005;15(Suppl 3):274–281. doi: 10.1111/j.1525-1438.2005.00441.x. [DOI] [PubMed] [Google Scholar]
- 3.Urban N, McIntosh MW, Andersen M, Karlan BY. Ovarian cancer screening. Hematol Oncol Clin North Am. 2003;17:989–1005. doi: 10.1016/s0889-8588(03)00063-7. ix. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 5.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 6.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 8.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome - components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 9.Wong IH, Lo YM, Johnson PJ. Epigenetic tumor markers in plasma and serum: biology and applications to molecular diagnosis and disease monitoring. Ann NY Acad Sci. 2001;945:36–50. [PubMed] [Google Scholar]
- 10.Cairns P. Gene methylation and early detection of genitourinary cancer: the road ahead. Nat Rev Cancer. 2007;7:531–543. doi: 10.1038/nrc2170. [DOI] [PubMed] [Google Scholar]
- 11.Melnikov A, Scholtens D, Godwin A, Levenson V. Differential methylation profile of ovarian cancer in tissues and plasma. J Mol Diagn. 2009;11:60–65. doi: 10.2353/jmoldx.2009.080072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei SH, Chen CM, Strathdee G, Harnsomburana J, Shyu CR, Rahmatpanah F, Shi H, Ng SW, Yan PS, Nephew KP, Brown R, Huang TH. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin Cancer Res. 2002;8:2246–2252. [PubMed] [Google Scholar]
- 13.Wei SH, Balch C, Paik HH, Kim YS, Baldwin RL, Liyanarachchi S, Li L, Wang Z, Wan JC, Davuluri RV, Karlan BY, Gifford G, Brown R, Kim S, Huang TH, Nephew KP. Prognostic DNA methylation biomarkers in ovarian cancer. Clin Cancer Res. 2006;12:2788–2794. doi: 10.1158/1078-0432.CCR-05-1551. [DOI] [PubMed] [Google Scholar]
- 14.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, Kalloger SE, Carlson JW, Ginzinger DG, Celniker SE, Mills GB, Huntsman DG, Gray JW. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res. 2007;13:5745–5755. doi: 10.1158/1078-0432.CCR-06-2882. [DOI] [PubMed] [Google Scholar]
- 16.Nanjundan M, Nakayama Y, Cheng KW, Lahad J, Liu J, Lu K, Kuo WL, Smith-McCune K, Fishman D, Gray JW, Mills GB. Amplification of MDS1/EVI1 and EVI1, located in the 3q26.2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res. 2007;67:3074–3084. doi: 10.1158/0008-5472.CAN-06-2366. [DOI] [PubMed] [Google Scholar]
- 17.Klein J, Moeschberger M. Survival Analysis Techniques for Censored and Truncated Data. Springer; New York: 1997. [Google Scholar]
- 18.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 19.Ahuja N, Mohan AL, Li Q, Stolker JM, Herman JG, Hamilton SR, Baylin SB, Issa JP. Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- 20.Ogino S, Kawasaki T, Kirkner GJ, Ohnishi M, Fuchs CS. 18q loss of heterozygosity in microsatellite stable colorectal cancer is correlated with CpG island methylator phenotype-negative (CIMP-0) and inversely with CIMP-low and CIMP-high. BMC Cancer. 2007;7:72. doi: 10.1186/1471-2407-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang JW, Karlan BY, Cass L, Baldwin RL. BRCA1 promoter methylation predicts adverse ovarian cancer prognosis. Gynecol Oncol. 2006;101:403–410. doi: 10.1016/j.ygyno.2005.10.034. [DOI] [PubMed] [Google Scholar]
- 22.Teodoridis JM, Hall J, Marsh S, Kannall HD, Smyth C, Curto J, Siddiqui N, Gabra H, McLeod HL, Strathdee G, Brown R. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 2005;65:8961–8967. doi: 10.1158/0008-5472.CAN-05-1187. [DOI] [PubMed] [Google Scholar]
- 23.Kohavi R. In: Kaufmann M, editor. A study of cross-validation and bootstrap for accuracy estimation and model selection; Proceedings of the Fourteenth International Joint Conference on Artificial Intelligence; CA: San Mateo; 1995. pp. 1137–1143. [Google Scholar]
- 24.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 25.Kobayashi N, Boyer TG, Berk AJ. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freyaldenhoven BS, Fried C, Wielckens K. FOXD4a and FOXD4b, two new winged helix transcription factors, are expressed in human leukemia cell lines. Gene. 2002;294:131–140. doi: 10.1016/s0378-1119(02)00702-3. [DOI] [PubMed] [Google Scholar]
- 27.Moebius FF, Fitzky BU, Wietzorrek G, Haidekker A, Eder A, Glossmann H. Cloning of an emopamil-binding protein (EBP)-like protein that lacks sterol delta8-delta7 isomerase activity. Biochem J. 2003;374:229–237. doi: 10.1042/BJ20030465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berthois Y, Bourrie B, Galiegue S, Vidal H, Carayon P, Martin PM, Casellas P. SR31747A is a sigma receptor ligand exhibiting antitumoural activity both in vitro and in vivo. Br J Cancer. 2003;88:438–446. doi: 10.1038/sj.bjc.6600709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu JK, Holland ND, Holland LZ. An amphioxus winged helix/forkhead gene, AmphiFoxD: insights into vertebrate neural crest evolution. Dev Dyn. 2002;225:289–297. doi: 10.1002/dvdy.10173. [DOI] [PubMed] [Google Scholar]
- 30.Lathe R. Steroid and sterol 7-hydroxylation: ancient pathways. Steroids. 2002;67:967–977. doi: 10.1016/s0039-128x(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 31.Cheung C, Akiyama TE, Kudo G, Gonzalez FJ. Hepatic expression of cytochrome P450s in hepatocyte nuclear factor 1-alpha (HNF1alpha)-deficient mice. Biochem Pharmacol. 2003;66:2011–2020. doi: 10.1016/s0006-2952(03)00586-0. [DOI] [PubMed] [Google Scholar]
- 32.Braverman N, Lin P, Moebius FF, Obie C, Moser A, Glossmann H, Wilcox WR, Rimoin DL, Smith M, Kratz L, Kelley RI, Valle D. Mutations in the gene encoding 3 beta-hydroxysteroid-delta 8, delta 7-isomerase cause X-linked dominant Conradi-Hunermann syndrome. Nat Genet. 1999;22:291–294. doi: 10.1038/10357. [DOI] [PubMed] [Google Scholar]
- 33.Yates JR, Heyes MP, Blight AR. 4-chloro-3-hydroxyanthranilate reduces local quinolinic acid synthesis, improves functional recovery, and preserves white matter after spinal cord injury. J Neurotrauma. 2006;23:866–881. doi: 10.1089/neu.2006.23.866. [DOI] [PubMed] [Google Scholar]
- 34.Colabroy KL, Zhai H, Li T, Ge Y, Zhang Y, Liu A, Ealick SE, McLafferty FW, Begley TP. The mechanism of inactivation of 3-hydroxyanthranilate-3,4-dioxygenase by 4-chloro-3-hydroxyanthranilate. Biochemistry. 2005;44:7623–7631. doi: 10.1021/bi0473455. [DOI] [PubMed] [Google Scholar]
- 35.Chan MW, Huang YW, Hartman-Frey C, Kuo CT, Deatherage D, Qin H, Cheng AS, Yan PS, Davuluri RV, Huang TH, Nephew KP, Lin HJ. Aberrant transforming growth factor beta1 signaling and SMAD4 nuclear translocation confer epigenetic repression of ADAM19 in ovarian cancer. Neoplasia. 2008;10:908–919. doi: 10.1593/neo.08540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watts GS, Futscher BW, Holtan N, Degeest K, Domann FE, Rose SL. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med Genomics. 2008;1:47. doi: 10.1186/1755-8794-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan MW, Wei SH, Wen P, Wang Z, Matei DE, Liu JC, Liyanarachchi S, Brown R, Nephew KP, Yan PS, Huang TH. Hypermethylation of 18S and 28S ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin Cancer Res. 2005;11:7376–7383. doi: 10.1158/1078-0432.CCR-05-1100. [DOI] [PubMed] [Google Scholar]
- 38.Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, Centeno B, Weber F, Leu YW, Shapiro CL, Eng C, Yeatman TJ, Huang TH. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res. 2006;12:6626–6636. doi: 10.1158/1078-0432.CCR-06-0467. [DOI] [PubMed] [Google Scholar]
- 39.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ginzinger DG, Godfrey TE, Nigro J, Moore DH, II, Suzuki S, Pallavicini MG, Gray JW, Jensen RH. Measurement of DNA copy number at microsatellite loci using quantitative PCR analysis. Cancer Res. 2000;60:5405–5409. [PubMed] [Google Scholar]
- 41.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 42.Hamaguchi K, Godwin AK, Yakushiji M, O'Dwyer PJ, Ozols RF, Hamilton TC. Cross-resistance to diverse drugs is associated with primary cisplatin resistance in ovarian cancer cell lines. Cancer Res. 1993;53:5225–5232. [PubMed] [Google Scholar]
- 43.Anthoney DA, McIlwrath AJ, Gallagher WM, Edlin AR, Brown R. Microsatellite instability, apoptosis, and loss of p53 function in drug-resistant tumor cells. Cancer Res. 1996;56:1374–1381. [PubMed] [Google Scholar]
- 44.Vasey PA, Jones NA, Jenkins S, Dive C, Brown R. Cisplatin, camptothecin, and taxol sensitivities of cells with p53-associated multidrug resistance. Mol Pharmacol. 1996;50:1536–1540. [PubMed] [Google Scholar]