Abstract
Left unrepaired, DNA interstrand crosslinks represent impassable hurdles for DNA replication, and their removal is a complicated stepwise process involving participation of a variety of enzymes. In a recent paper published in Science, J. Walter and co-workers demonstrate that the Fanconi Anemia protein FANCD2 promotes multiple steps of the crosslink repair process.
Like good housekeepers, cells spend a lot of energy caring for their genetic material, mopping up DNA damage that may alter the reading of their genetic information. The most difficult to repair, and at the same time the most detrimental, are the DNA interstrand crosslinks (ICLs). Since ICLs block DNA replication, their removal is an essential requirement for cell survival. The severe clinical symptoms of patients with Fanconi Anemia (blood marrow failure, developmental abnormalities, and cancer predisposition, often leading to an early death) testify to the toxicity of ICLs (Moldovan and D'Andrea, 2009; Patel and Joenje, 2007). How the FA pathway protects against ICLs has long remained a mystery. Using a cell-free system, Knipscheer et al (Knipscheer et al., 2009) now show, for the first time, that FA proteins are directly involved, at several steps, in the ICL repair process.
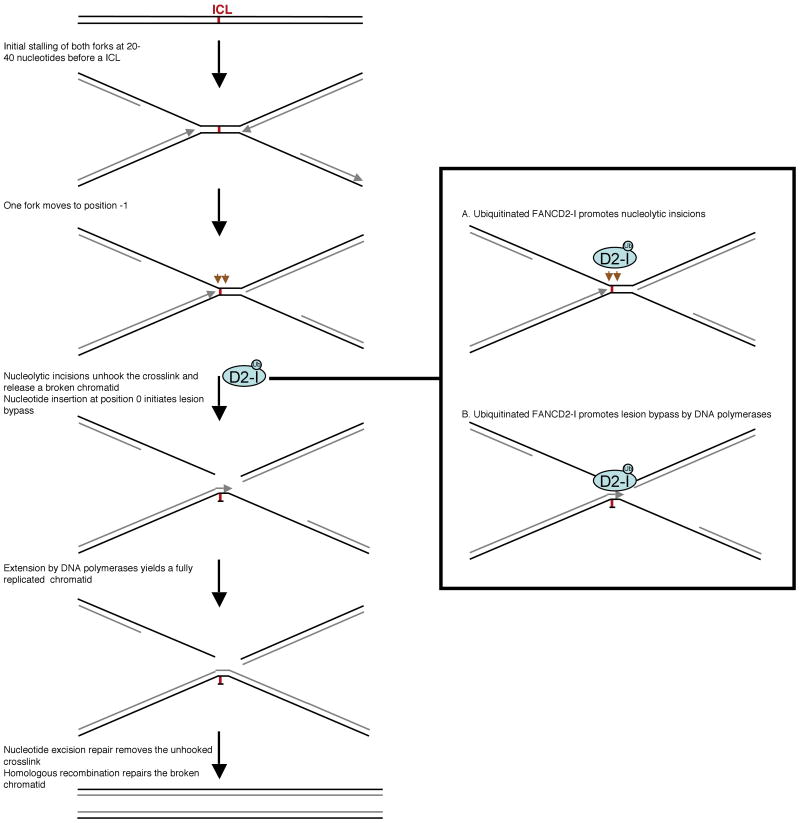
Removal of ICLs occurs mostly in S-phase and involves the stepwise involvement of nucleases, specialized lesion bypass DNA polymerases, and homologous recombination factors (Moldovan and D'Andrea, 2009). Using the tractable cell-free replication system of Xenopus egg extracts, the authors previously devised an experimental approach to investigate replication-dependent ICL repair (Raschle et al., 2008). As illustrated in Figure 1, their system reveals that replication forks, converging from both directions, initially stop 20-40 nucleotides from the crosslink. One of the forks subsequently moves further, and stops again just before the crosslink (at position -1). Nucleolytic incisions on both sides of the crosslink, and DNA polymerization across the lesion, restore one of the chromatids, while the other is most likely repaired through homologous recombination. In an elegant application of their crosslink repair system, Knipscheer et al now show that loss of the FA pathway can block both the incision and bypass steps.
Figure 1. FANCD2 monoubiquitination is required for the nucleolytic incision and translesion bypass steps of interstrand crosslink repair.
Crosslink repair is initiated by stalling of the two replication forks on either side of the crosslink. One of the forks subsequently moves to the crosslink site. By the time the fork reaches position -1, FANCD2 has been monoubiquitinated by the FA core complex. Ubiquitinated FANCI-FANCD2 then promotes nucleolytic incisions and crosslink unhooking, allowing bypass of the lesion by polymerases. Finally, homologous recombination concludes the repair reaction.
Monoubiquitinated FANCD2 may directly activate nucleolytic processes (A), recruit translesion synthesis polymerases (B), or both.
Thirteen FA proteins cooperate in the FA pathway. Eight of these proteins form an ubiquitin ligase complex that monoubiquitinates the substrates FANCD2 and FANCI. In the new study, immunodepletion of FANCD2 from the egg extracts (which also co-depletes its heterodimeric partner FANCI), dramatically inhibited crosslink repair, demonstrating that FA is a true DNA repair syndrome. Nucleotide insertion opposite the ICL was blocked in the absence of FANCD2, since the leading strand progressed only to position -1. Also, the two incisions required to unhook the crosslink could not be detected in extracts depleted of FANCD2. All defects could be rescued by adding back the recombinant FANCD2-FANCI complex, but not by adding back a complex containing a point mutant of FANCD2 that cannot by ubiquitinated. Accordingly, when the investigators examined the timing of FANCD2 ubiquitination, they found that this modification occurs precisely when the replication fork reaches the -1 position. FANCD2 ubiquitination, known to be essential for crosslink tolerance, is therefore required to advance the replication fork across the crosslink, by orchestrating crosslink unhooking and translesion synthesis.
Bypass of DNA lesions is a potentially mutagenic process performed by specialized polymerases. The FA pathway is known to be required for mutagenesis, and the polymerase Rev1 is epistatic to FA, and may function in FA-dependent lesion bypass (Mirchandani et al., 2008; Niedzwiedz et al., 2004). The finding that FANCD2 ubiquitination promotes crosslink bypass supports a model in which Rev1 is recruited, via its ubiquitin binding domain, to the FANCD2-Ub protein. This model is analogous to the recruitment of Y-family polymerases, such as DNA polymerase η, by monoubiquitinated PCNA in order to bypass UV-induced lesions (Bienko et al., 2005). Rev1 depletion from egg extracts, together with Rev1 interaction studies, should now be able to directly test this hypothesis. Since human cells have at least fifteen polymerases, a different mutagenic polymerase might also be involved. Interestingly, only about 25% of crosslinked plasmids are recovered as repaired, error-free DNA molecules (Knipscheer et al., 2009), raising the possibility that some sites are repaired with errors. Investigating the mutation spectrum of the repaired templates may help identify the relevant polymerases. Alternatively, FANCD2 may not directly recruit a polymerase, but rather stabilize some structure at the crosslink site, thus indirectly allowing a nucleotide insertion event.
How the FANCD2 protein regulates the DNA incisions flanking the crosslink is even more intriguing. FANCD2 has no known enzymatic activity, possesses no obvious nuclease domains, and has no known associated nucleases. Perhaps a cryptic nuclease activity of FANCD2 is activated by monoubiquitination when a replication fork encounters a crosslink. Alternatively, the FANCD2-Ub may transiently recruit some known nuclease complex, such as the Mus81-Eme1, Xpf-Ercc1, or the newly-described Slx4 complex.
At present, the Xenopus experimental system cannot temporally dissociate the incision and bypass steps. Although it is logical to assume that incision occurs first, allowing bypass by a polymerase such as Rev1, the alternative possibility should not be excluded: nucleotide insertion opposite the ICL may create a DNA structure that promotes incision. Also, it is possible that the proximal incision occurs first, followed by the lesion bypass step and the distal incision. In either case, FANCD2 may only be required for the first activity in the stepwise process. Depletion of FANCD2 may thus only indirectly affect the subsequent steps. Alternatively, FANCD2 may directly activate both incision and bypass, irrespective of their order of occurrence.
Recent studies indicate that the regulated deubiquitination of FANCD2 by the ubiquitin protease complex, USP1/UAF1, is also required for crosslinker tolerance (Moldovan and D'Andrea, 2009). Whether deubiquitination is required for incision and/or lesion bypass is also unknown. The Xenopus system may be useful in resolving these questions as well.
An apparent shortcoming of this model system is its inability (to date) to decipher the roles of the FA pathway in the last part of the ICL repair process -namely, the homologous recombination (HR) step. Only the repair of the chromatid with the unhooked crosslink can be investigated, probably due to degradation of the other sister chromatid that is broken by incisions. Therefore, the repair process in this system terminates before the final recombination step. It is possible that FANCD2 plays only an indirect role in the HR step, simply “handing off” the product of the incision and insertion steps to the HR machinery. This most likely involves the recombination mediator BRCA2, identified as the FA factor FANCD1 (Howlett et al., 2002), and its binding partner, FANCN. Alternatively, the FA pathway may be actively involved in HR. This possibility is supported by evidence that FANCD2 and FANCI are required for double strand break-induced HR repair (Smogorzewska et al., 2007), suggesting a general role of the FA pathway in promoting recombination. Whether this function represents a direct participation (for example, recruitment of HR factors), or a more indirect one, such as inhibition of alternative competing repair pathways (such as the error prone non-homologous end joining pathway), is currently unclear.
Despite these shortcomings, Xenopus egg extracts should allow the identification and characterization of the nucleases and polymerases involved in ICL repair, and elucidate how these activities are controlled by the FA pathway. This knowledge may, over time, lead to better therapeutic strategies for FA patients.
Together with the recently described role of the FA pathway in crosslink-induced checkpoint signaling (Ben-Yehoyada et al., 2009), these new findings paint the picture of a dynamic, multileveled cellular pathway that is involved at multiple steps in the DNA crosslink response.
Acknowledgments
G.L.M. is supported by a Human Frontiers Science Program Organization postdoctoral fellowship. Work in D'Andrea lab is supported by NIH grants RO1DK43889, R01HL52725, PO1CA092584, and U19A1067751.
References
- Ben-Yehoyada M, Wang LC, Kozekov ID, Rizzo CJ, Gottesman ME, Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Molecular cell. 2009;35:704–715. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, Kannouche P, Wider G, Peter M, Lehmann AR, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science (New York, NY. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science (New York, NY. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand crosslink repair. Science. 2009 doi: 10.1126/science.1182372. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirchandani KD, McCaffrey RM, D'Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA repair. 2008;7:902–911. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, D'Andrea AD. How the Fanconi Anemia Pathway Guards the Genome. Annual review of genetics. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Molecular cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA repair. 2007;6:885–890. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]