Abstract
Human cytomegalovirus (HCMV) depends upon a five-protein complex, gH/gL/UL128-131, to enter epithelial and endothelial cells. A separate HCMV gH/gL-containing complex, gH/gL/gO, has been described. Our prevailing model is that gH/gL/UL128-131 is required for entry into biologically important epithelial and endothelial cells and that gH/gL/gO is required for infection of fibroblasts. Genes encoding UL128-131 are rapidly mutated during laboratory propagation of HCMV on fibroblasts, apparently related to selective pressure for the fibroblast entry pathway. Arguing against this model in the accompanying paper by B. J. Ryckman et al. (J. Virol., 84:2597-2609, 2010), we describe evidence that clinical HCMV strain TR expresses a gO molecule that acts to promote endoplasmic reticulum (ER) export of gH/gL and that gO is not stably incorporated into the virus envelope. This was different from results involving fibroblast-adapted HCMV strain AD169, which incorporates gO into the virion envelope. Here, we constructed a TR gO-null mutant, TRΔgO, that replicated to low titers, spread poorly among fibroblasts, but produced normal quantities of extracellular virus particles. TRΔgO particles released from fibroblasts failed to infect fibroblasts and epithelial and endothelial cells, but the chemical fusogen polyethylene glycol (PEG) could partially overcome defects in infection. Therefore, TRΔgO is defective for entry into all three cell types. Defects in entry were explained by observations showing that TRΔgO incorporated about 5% of the quantities of gH/gL in extracellular virus particles compared with that in wild-type virions. Although TRΔgO particles could not enter cells, cell-to-cell spread involving epithelial and endothelial cells was increased relative to TR, apparently resulting from increased quantities of gH/gL/UL128-131 in virions. Together, our data suggest that TR gO acts as a chaperone to promote ER export and the incorporation of gH/gL complexes into the HCMV envelope. Moreover, these data suggest that it is gH/gL, and not gH/gL/gO, that is present in virions and is required for infection of fibroblasts and epithelial and endothelial cells. Our observations that both gH/gL and gH/gL/UL128-131 are required for entry into epithelial/endothelial cells differ from models for other beta- and gammaherpesviruses that use one of two different gH/gL complexes to enter different cells.
Human cytomegalovirus (HCMV) infects a broad spectrum of cell types in vivo, including epithelial and endothelial cells, fibroblasts, monocyte-macrophages, dendritic cells, hepatocytes, neurons, glial cells, and leukocytes (6, 28, 36). Infection of this diverse spectrum of cell types contributes to the multiplicity of CMV-associated disease. HCMV infection of hepatocytes and epithelial cells in the gut and lungs following transplant immunosuppression is directly associated with CMV disease (3, 44). HCMV can be transported in the blood by monocyte-macrophages, and virus produced in these cells can infect endothelial cells, leading to virus spread into solid tissues such as the brain, liver, and lungs, etc. (16). Despite the broad spectrum of cells infected in vivo, propagation of HCMV in the laboratory is largely limited to normal human fibroblasts because other cells produce little virus. HCMV rapidly adapts to laboratory propagation in fibroblasts, losing the capacity to infect other cell types, i.e., epithelial and endothelial cells and monocyte-macrophages (9, 16, 18, 43). This adaptation to fibroblasts involves mutations in the unique long b′ (ULb′) region of the HCMV genome, which includes 22 genes (9). Targeted mutation of three of the ULb′ genes, UL128, UL130, and UL131, abolished HCMV infection of endothelial cells, transmission to leukocytes, and infection of dendritic cells (17, 18). Restoration of UL128-131 genes in HCMV laboratory strain AD169 (which cannot infect epithelial and endothelial cells) produced viruses capable of infecting these cells (18, 48). There is also evidence that the UL128-131 proteins are deleterious to HCMV replication in fibroblasts, resulting in rapid loss or mutation of one or more of the UL128-131 genes during passage in fibroblasts (2).
A major step forward in understanding how the UL128-131 genes promote HCMV infection of epithelial and endothelial cells involved observations that the UL128-131 proteins assemble onto the extracellular domain of the membrane-anchored HCMV glycoprotein heterodimer gH/gL (1, 49). Antibodies to UL128, UL130, and UL131 each neutralized HCMV for infection of endothelial or epithelial cells (1, 49). All herpesviruses express gH/gL homologues and, where this has been tested, all depend upon gH/gL for replication and, more specifically, for entry into cells (14, 15, 31, 38). Indeed, we showed that the gH/gL/UL128-131 complex mediated entry into epithelial and endothelial cells (40). All five members of the gH/gL/UL128-131 complex were required for proper assembly and export from the endoplasmic reticulum (ER) and for function (39, 41). In addition, the expression of gH/gL/UL128-131, but not gH/gL or gB, in epithelial cells interfered with HCMV entry into these cells (39). This interference suggested that there are saturable gH/gL/UL128-131 receptors present on epithelial cells, molecules that HCMV uses for entry. There was no interference in fibroblasts expressing gH/gL/UL128-131, although some interference was observed with gH/gL (39). As noted above, gH/gL/UL128-131 plays no obvious role in entry into fibroblasts and, in fact, appears to be deleterious in this respect (2, 18, 40).
HCMV also expresses a second gH/gL complex, as follows: gH/gL/gO (20, 21, 22, 30, 48). Fibroblast-adapted HCMV strain AD169 expresses a gO protein that is a 110- to 125-kDa glycoprotein (21). Pulse-chase studies suggest that gH/gL assembles first in the ER before binding and forming disulfide links with gO (21, 22). The 220-kDa immature gH/gL/gO complex is transported from the ER to the Golgi apparatus and increases in size to ∼280 to 300 kDa before incorporation into the virion envelope (21). gH/gL/gO complexes are apparently distinct from gH/gL/UL128-131 complexes because gO-specific antibodies do not detect complexes containing either UL128 or UL130 and UL128-specific antibodies do not precipitate gO (49). Towne and AD169 gO-null mutant laboratory strains can produce small plaques on fibroblasts, leading to the conclusion that gO is not essential. However, the AD169 and Towne mutants produced ∼1,000-fold less infectious virus than wild-type HCMV (14, 19), which might also be interpreted to mean that gO is very important or even essential for replication. Thus, the prevailing model has been that wild-type HCMV particles contain the following two gH/gL complexes: gH/gL/gO, which promotes infection of fibroblasts, and gH/gL/UL128-131, which promotes entry into epithelial and endothelial cells. Supporting this model, there are two different entry mechanisms, as follows: HCMV enters fibroblasts by fusion at the plasma membrane at neutral pH (12), whereas entry into epithelial and endothelial cells involves endocytosis and a low pH-dependent fusion with endosomes (40). This model of HCMV entry parallels models for Epstein-Barr virus (EBV) entry that use gH/gL to enter epithelial cells and gH/gL/gp42 to enter B cells (24). Similarly, HHV-6 uses gH/gL/gO and gH/gL/gQ, which bind to different receptors (33).
Many of the studies of gH/gL/gO have involved the fibroblast-adapted HCMV strain AD169, which fails to express UL131 and assemble gH/gL/UL128-131 or AD169 recombinants in which UL131 expression was restored (20, 21, 22, 48, 49). It seemed possible that the adaptation of AD169 to long-term passage in fibroblasts might also involve alterations in gO. HCMV gO is unusually variable (15 to 25% amino acid differences) among different HCMV strains compared with other viral genes (13, 34, 35, 37, 46). In recent studies, Jiang et al. (26) described a gO-null mutant derived from the HCMV strain TB40/E, a strain that can infect endothelial cells following extensive passage on these cells. The TB40/E gO-null mutant spread poorly on fibroblasts compared with wild-type TB40/E, and there was little infectious virus detected in fibroblast culture supernatants. However, the few TB40/E gO-null mutant particles produced by fibroblasts that could initiate infection of endothelial cells were able to spread to form normal-sized plaques on endothelial cells. These results further supported the model for which gH/gL/gO is required for infection of fibroblasts but not for epithelial/endothelial cells. Those authors also concluded that gO is important for the assembly of enveloped particles in fibroblasts, based on observations of few infectious virus particles in supernatants and cytoplasmic accumulation of unenveloped capsids (26).
Our studies of gH/gL/UL128-131 have involved the clinical HCMV strain TR (39, 40, 41, 47). HCMV TR was originally an ocular isolate from an AIDS patient (45) and was passaged only a few times on fibroblasts before being genetically frozen in the form of a bacterial artificial chromosome (BAC) (34, 40). HCMV TR infects epithelial and endothelial cells (40) and monocyte-macrophages (D. Streblow and J. Nelson, unpublished results) well. In the accompanying paper (42), we characterized the biochemistry and intracellular trafficking of TR gO. TR gO expressed either in TR-infected cells or by using adenovirus vectors (expressed without other HCMV proteins) was largely retained in the ER. Coexpression of gO with gH/gL promoted transport of gH/gL beyond the ER. Importantly, TR gO was not found in extracellular virions. In contrast, AD169 gO was present in extracellular virus particles, as described previously (20, 21). We concluded that TR gO is a chaperone that promotes ER export of the gH/gL complex, but gO dissociates prior to incorporation into the virus envelope. Moreover, these differences highlight major differences between gO molecules expressed by fibroblast-adapted strain AD169 and low-passage TR.
To extend these results and characterize how TR gO functions, whether in virus entry or virus assembly/egress, we constructed a TR gO-null mutant. TRΔgO exhibited major defects in entering fibroblasts, as evidenced by increased virus infection following treatment with the chemical fusogen polyethylene glycol (PEG). Unexpectedly, the mutant also failed to enter epithelial and endothelial cells, and again, PEG partially restored entry. Relatively normal numbers of TRΔgO particles were produced and released into cell culture supernatants, although even with PEG treatment, most of these virus particles remained defective in initiating immediate-early HCMV protein synthesis. Western blot analyses of TRΔgO extracellular particles demonstrated very low levels of gH/gL incorporated into virions, which likely explains the reduced entry of TRΔgO. However, the small amounts of gH/gL complexes that were present in TRΔgO virions were associated with increased quantities of UL130, and these TRΔgO particles spread better than wild-type HCMV on epithelial cell monolayers. Together with the results shown in the accompanying paper (42), we concluded that HCMV TR gO functions as a chaperone to promote ER export of gH/gL to HCMV assembly compartments and the incorporation of gH/gL into the virion envelope. The highly reduced quantities of gH/gL in virions are apparently responsible for the inability of HCMV to enter fibroblasts and epithelial and endothelial cells. These results suggest a modified version of our model, in which gH/gL, not gH/gL/gO, mediates entry into fibroblasts and both gH/gL and gH/gL/UL128-131 are required for entry into epithelial and endothelial cells.
MATERIALS AND METHODS
Cells and viruses.
MRC-5 fibroblasts were obtained from the American Type Culture Collection (ATCC CCL-171), neonatal normal human dermal fibroblasts (NHDF) were obtained from Cascade Biologics (Portland, OR), and they were grown in minimum essential medium (MEM; Invitrogen) and Dulbecco's minimum essential medium (DMEM, Invitrogen), respectively, that was supplemented with 5% fetal bovine serum (FBS; HyClone) and 5% bovine growth supplement (BGS; HyClone). The retinal pigment epithelial cell line ARPE-19 was obtained from the ATCC and was grown in a 1:1 dilution mix of DMEM and Ham's F-12 medium (Invitrogen) supplemented with 10% FBS. Human papillomavirus (HPV) E6/E7-transformed aortic endothelial cells (HPV-AEC) were provided by Ashlee Moses (Oregon Health & Science University) and were grown in Medium 200 (Cascade M-200-500) supplemented with a low-serum-growth supplement (Cascade). HCMV strain TR was derived from the ocular vitreous fluid sample from an AIDS patient (45) and was stabilized in the form of a bacterial artificial chromosome (BAC) (34). Wild-type TR was propagated on NHDF cells by infection at a multiplicity of less than 0.1 in DMEM plus 5% FBS for 10 to 16 days. Virus particles were concentrated from infected cell supernatants by centrifugation at 50,000 × g for 1 h through a 20% sorbitol cushion.
Construction of recombinant HCMV TRΔgO.
The UL74 gene of BAC-TR was mutated by using lambda phage linear recombination as previously described (5, 29, 51). Briefly, a 1.5-kb PCR product containing a kanamycin resistance (Kanr) gene cassette flanked by Flp recombination target (FRT) sites and with HCMV sequences near the 5′ end of the UL74 gene (60 nucleotides [nt], corresponding to nt 144593 to 144534 of the published TR sequence; GenBank accession no. AC146906) and sequences near the 3′ end of the UL74 gene (61 nt, corresponding to nt 143393 to 143333) was generated with the following primers using a PCR template from pCP015: GTACACGCAGGCAAGCCAAACCACAAGGCAGACGGACGGTGCGGGGTCTCCTCCTCTGTCGTAAAACGACGGCCAGT and TAGGTGTAGTTTCGGAAGCCGTTTTGTTTTCCACGAACATGGTTTCGTTGTAATATAAGGATTACAGGAAACAGCTATGAC (11). Following transformation of EL250 bacteria containing BAC-TR with the Kanr PCR product, bacteria were selected for growth on kanamycin. Kanr clones were analyzed by Southern blot analyses using probes specific for the UL74 gene and for the Kanr sequences following EcoRI restriction. Clones were also confirmed by direct DNA sequence analysis. The Kanr cassette was removed from BAC-TRΔgO Kanr clones by inducing Flp recombinase expression in EL250 host bacteria (29). Colonies were replica plated on chloramphenicol- and kanamycin-containing plates to screen for BAC lacking Kanr sequences, and clones were confirmed by Southern blotting. Infectious HCMV TRΔgO was derived by electroporation of BAC DNA (2 μg) into MRC-5 cells (5 × 106 cells). Following electroporation, the MRC-5 cells were washed, plated in 100-mm dishes in DMEM containing 10% serum, and allowed to spread on the plastic, and fresh medium was added after 24 h. When these cells became confluent, the cells were trypsinized and replated at a lower cell density, until TRΔgO produced a cytopathic effect (CPE) in ∼20% of the cells after 42 days.
Production of TRΔgO virus stocks and PEG treatment to enhance entry.
Initially, TRΔgO was propagated using BAC-TRΔgO-transfected MRC-5 cells by trypsinizing the cells and plating infected cells with other, uninfected MRC-5 cells. Cell culture supernatants were harvested, and viruses were concentrated either by centrifugation at 50,000 × g for 1 h or through 20% sorbitol at 50,000 × g for 1 h. Later, TRΔgO virus stocks were produced using NHDF infected with extracellular TRΔgO virus particles by enhancing virus entry using a combination of low-speed centrifugation and PEG treatment. Briefly, cells and virus were mixed and immediately centrifuged at 800 × g for 1 h at 15°C. Cells were washed once with warm phosphate-buffered saline (PBS), then treated with 44% (wt/wt) PEG (Fluka) in PBS at 37°C for 30 s, and then washed immediately four times with warm PBS.
Quantitative PCR (qPCR) of HCMV DNA.
Cell culture supernatants from HCMV-infected cells were treated with DNase I (Roche) to remove any DNA not protected within viral capsids. Capsids were disrupted by using the detergents and proteinase K in the QIAamp MinElute virus spin kit (Qiagen). DNA was resuspended in 50 μl nuclease-free water, and genomes were quantified by real-time qPCR using the following UL115 primers and TaqMan probes (Applied Biosystems): forward primer, GTAGCCATAACCTGTCAATCATCCTGTA; reverse primer, GTATTGTAGCGTGTAATTTAGGTTGCACT; and probe, 6-carboxyflu-orescein-TTGCGGTGGATGAAGAAGAGCCAGAACTG. PCR samples were made to a total volume of 25 μl with a 2× TaqMan Universal PCR master mix (Applied Biosystems), 10 μl of 1:100, 1:1,000, or 1:10,000 dilutions of cDNA, 600 nM forward and reverse primers, 150 nM probe, and nuclease-free water. Sequences were detected using an ABI Prism 7700 sequence detection system. Samples were compared to a plasmid pcDNA3.1(+)gLSP standard.
Antibodies.
Mouse monoclonal antibodies (MAb) specific for the HCMV major capsid protein (28-4), tegument protein pp65 (28-19), and glycoproteins gB (27-156) and gN (14-16A) were kindly provided by Bill Britt (University of Alabama, Birmingham, AL) (7, 8, 10). Rabbit polyclonal anti-IE86 antibody (R683) has been described previously (25). Rabbit polyclonal antisera directed against HCMV gH, gL, and UL130 were previously described (41). Polyclonal antisera directed against a peptide (KRKQAPVKEQSEKKSKKSC) derived from HCMV TR gO was produced by coupling the peptide to keyhole limpet hemocyanin using m-maleimidobenzoyl-N-hydroxysuccinimide ester (Sigma, St. Louis, MO) and by vaccinating rabbits as described previously (41).
Immunofluorescence.
HCMV-infected cells were fixed with a 50:50 dilution of methanol-acetone for 10 min, washed three times with phosphate-buffered saline (PBS), and then permeabilized using IF buffer (0.5% Triton X-100, 0.5% deoxycholate, 1% bovine serum albumin [BSA], 0.05% sodium azide in PBS). The cells were stained with anti-IE86 rabbit sera diluted in IF buffer, washed with IF buffer, and then incubated with Alexa 594-conjugated goat anti-rabbit secondary antibody obtained from Molecular Probes (Eugene, OR). In some experiments, cells were also stained with Syto 13 green fluorescent nucleic acid stain (Invitrogen) in PBS for 10 min. The cells were analyzed using a Nikon Eclipse TS100 fluorescent microscope.
HCMV cell-to-cell spread assay.
NHDF, MRC-5, and ARPE-19 cells were grown to near confluence and infected with 100 to 200 PFU of wild-type TR or TRΔgO using centrifugation (800 × g for 1 h at 15°C) and PEG treatment. Cells were maintained in media containing 0.3% pooled human immunoglobulin (a source of HCMV neutralizing antibodies) for 10 and 20 days and then fixed and analyzed by immunofluorescence for IE86.
HCMV release assay.
NHDF cultured in 12-well dishes (Falcon) were infected with wild-type TR and TRΔgO using doses of virus that resulted in 20% infection by using PEG treatment. Cell culture medium was changed 1 day postinfection with 1 ml fresh DMEM containing 10% serum. Cell culture supernatants were collected after 2, 4, 6, and 8 days. Cell debris was removed from supernatants by centrifugation at 20,000 × g for 10 min. A total of 200 μl of each sample was DNase I treated, and then viral DNA was subsequently harvested with the QIAamp MinElute virus spin kit (Qiagen). Genomes were measured by qPCR (as described above). At these same times, the cells were fixed and analyzed by immunofluorescence for IE86 expression.
Immunoblotting.
Cell culture supernatants derived from HCMV-infected cells were clarified by centrifugation at 6,000 × g for 15 min, and then virus particles were pelleted at 50,000 × g for 1 h through 20% sorbitol cushions (40). Virus pellets or HCMV-infected cells were lysed in 50 mM Tris-HCl buffer, pH 6.8, containing 2% sodium dodecyl sulfate (SDS) and 2% β-mercaptoethanol, and then proteins were separated using SDS-polyacrylamide gels. Proteins were electrophoretically transferred to Immobilon membranes (Millipore) in a buffer containing 25 mM Tris, 192 mM glycine, and 20% methanol. HCMV proteins were detected with rabbit polyclonal antibodies or MAb, followed by horseradish peroxidase-conjugated secondary antibodies, and chemiluminescence imaging as described previously (23, 50).
Electron microscopy.
NHDF were infected with wild-type TR or TRΔgO by using centrifugation and PEG enhancement. After 7 days, the cells were washed with 0.1 M sodium cacodylate buffer, pH 7.2, and fixed in Ito and Karnovsky's fixative (1.6% paraformaldehyde, 2.5% glutaraldehyde, and 0.5% picric acid in 0.1 M sodium cacodylate; pH 7.2) for 30 min at 23°C. The samples were postfixed in 1.5% osmium tetroxide, rinsed, postfixed in 4% paraformaldehyde, dehydrated in a graded acetone series, and embedded in epoxy resin, and ultrathin sections were double stained in uranyl acetate and lead citrate and viewed with a Philips EM 300 electron microscope.
RESULTS
Construction of a HCMV TR mutant unable to express gO.
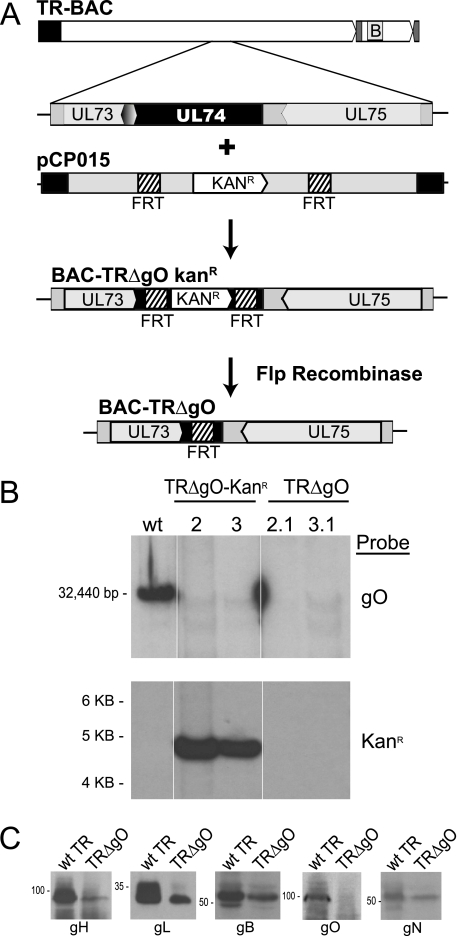
The previous evidence that HCMV gH/gL/UL128-131 promotes virus entry (40) suggested the possibility that gH/gL/gO might also participate in entry. This point has not been studied for any HCMV strain, neither AD169 nor the endotheliotropic TB40/E strain (26). We routinely use HCMV strain TR, which can infect fibroblasts and epithelial and endothelial cells well (34, 40, 45). To construct a gO-null mutant, a BAC containing TR sequences was used in conjunction with homologous recombination targeting the UL74 (gO) gene, as described previously (5, 40). First, the N-terminal 1,141 nucleotides of the UL74 (gO) gene beginning with the start codon were replaced with a kanamycin resistance (Kanr) cassette flanked by FRT sites (Fig. 1A). The gO sequences targeted were carefully designed to avoid disruption of the promoters and polyadenylation sites for flanking genes (gH and gN). Bacterial clones were selected for resistance to kanamycin, and then the Kanr cassette was removed by induction of Flp recombinase. Southern blot analysis revealed that the Kanr cassette replaced the UL74 gene in two BAC-TRΔgO Kanr clones, clones 2 and 3, and the Kanr cassette was removed in BAC-TRΔgO clones 2.1 and 3.1 (Fig. 1B).
FIG. 1.
Construction of a HCMV TR gO-null mutant. (A) BAC-TR containing the entire genome of HCMV clinical strain TR (except for where the BAC sequence replaces US2-US5) was previously described (34). The UL73 (gN), UL74 (gO), and UL75 (gH) genes are depicted. The N-terminal 1,141 nucleotides of UL74 (beginning at the gO start codon and extending to codon 380) were replaced by homologous recombination with a kanamycin resistance (Kanr) cassette flanked by FRT sites producing BAC-TRΔgO Kanr. The replacement did not affect the UL73 or UL75 promoters, coding sequences, or poly(A) sites. Following induction of Flp recombinase in bacteria, the Kanr cassette was excised, leaving a single FRT site in place of the N-terminal UL74 sequences (BAC-TRΔgO). (B) Southern blot analyses of BAC clones. BAC-TR (wild type [wt]), BAC-TRΔgO Kanr clones 2 and 3, and BAC-TRΔgO clones 2.1 and 3.1 were digested with EcoRI. This produces a 32.4-kb fragment for wild-type UL74 but a 4.5-kb fragment when the Kanr cassette is inserted. Flp recombination produced clones 2.1 and 3.1 that lacked both Kanr and UL74 sequences. The blots were probed with either gO sequences or Kanr sequences. (C) Expression of gH, gL, gB, gO, and gN proteins in wild-type TR and TRΔgO-infected NHDF after 8 days infection by Western blotting.
BAC-TRΔgO clones 2.1 and 3.1 were separately introduced into MRC-5 fibroblasts using electroporation. After 5 days, there was a visible cytopathic effect (CPE) in a small number of cells, and there was limited virus spread of both TRΔgO clones over the subsequent 5 to 7 days. This spread of the CPE was substantially (2 to 10%) less than that found in other dishes of MRC-5 cells that had been transfected with BAC-TR (wild type). Since the majority of MRC-5 cells transfected with both clones of BAC-TRΔgO did not display a CPE, we replated these cells with other, uninfected MRC-5 cells for several rounds to amplify virus. Over the course of 6 weeks, infections with both clones spread to ∼20% of the cells. Other attempts to transfect BAC-TRΔgO using NHDF (foreskin) produced virus, but the CPE was observed even later. Virus derived from one of these BAC clones, BAC-TRΔgO clone 3.1, was denoted HCMV TRΔgO and studied further by Western blot analysis and most other analyses. However, a second virus derived from BAC-TRΔgO clone 2.1 was also characterized in a more limited number of experiments described below. TRΔgO expressed gB, gH, gL, and gN, but not gO (Fig. 1C). There were reduced quantities of all HCMV proteins in TRΔgO-infected fibroblasts compared with those of wild-type TR, but these reductions in HCMV proteins were in line with reduced numbers of infected cells (not shown). Importantly, the reduced expression of gH and gN could be explained by these reduced numbers of cells and was similar to expression of gB, demonstrating that the adjacent (UL73 and UL75) genes were not negatively affected by mutation of the gO (UL74) gene.
TRΔgO particles are defective for entry into human fibroblasts.
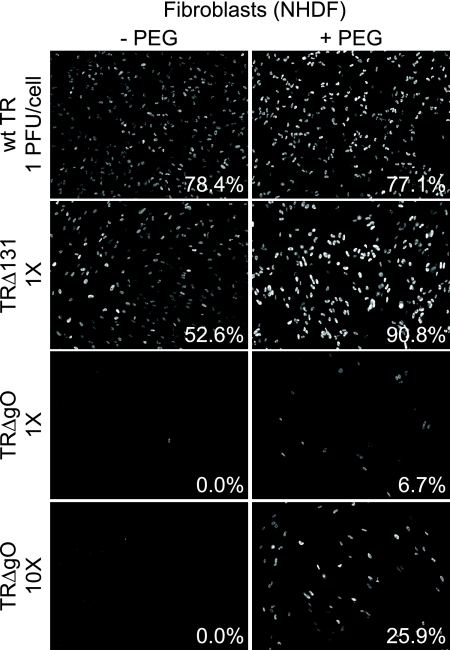
We attempted to characterize the entry of TRΔgO virus particles into fibroblasts by using particles derived from cell culture supernatants. Because infection and spread of TRΔgO in fibroblast monolayers were markedly reduced, compared with those of wild-type TR, many fewer cells were infected, and fewer virus particles were produced into cell culture supernatants. However, virus particles in supernatants could be concentrated by centrifugation and quantified by measuring HCMV DNA by quantitative PCR. Thus, by using the content of viral DNA, we could apply similar quantities of virus particles to cells. Wild-type TR and TRΔ131, a mutant unable to assemble gH/gL/UL128-131 complexes (41), were applied to NHDF, representing 1 PFU/cell of each virus. A similar quantity of TRΔgO particles (normalized using genome copies) was also incubated with fibroblasts. Infection of these fibroblasts was measured by staining cells using IE86-specific antibodies. Wild-type TR and TRΔ131 infected 78.4% and 52.6% of fibroblasts, respectively (Fig. 2). In contrast, we did not detect any IE86+ fibroblasts following inoculation with a comparable amount of TRΔgO particles (1× TRΔgO) (Fig. 2). When the TRΔgO inoculum was increased by 10-fold, we observed infection of 5 to 10 cells/well, representing 0.025 to 0.04% of the total cells in the dish (10× TRΔgO) (Fig. 2). Thus, extracellular TRΔgO virus particles cannot initiate infection of fibroblasts well. It is important to note that these infections involved extracellular TRΔgO particles and are different from our propagation of TRΔgO, which involved plating TRΔgO-infected fibroblasts with uninfected fibroblasts.
FIG. 2.
Entry of TRΔgO into fibroblasts with and without PEG treatment. Multiwell dishes of NHDF were incubated with extracellular HCMV particles concentrated by pelleting from fibroblast culture supernatants. Wild-type (wt) TR and TRΔUL131 were used at 1 PFU/cell. A similar quantity of TRΔgO extracellular virions (based on quantifying genomes using qPCR) or 10 times (10×) that amount of TRΔgO were also incubated with cells. These viruses were centrifuged with cells at 800 × g for 1 h. Some of the wells were subsequently treated with 44% PEG for 30 s and then immediately washed (+ PEG). After 48 h, cells were fixed, permeabilized, and stained for HCMV immediate-early (IE) protein 86 (IE86). Numbers indicate the average number of IE86+ cells in three replicate wells.
We previously showed that HCMV UL128-131 mutants failed to enter epithelial and endothelial cells, but this defect was reversed by treating cells and virus with the chemical fusogen PEG (40). PEG promotes entry of mutant viruses that are bound onto cell surfaces but unable to enter because the virus particles are blocked in the capacity to fuse with cellular membranes. Fibroblasts were inoculated with TRΔgO using quantities of virus particles corresponding to 1 PFU/cell of wild-type TR and then treated with PEG, and ∼7% of the cells were IE86+ (1× TRΔgO) (Fig. 2). Using 10-fold more TRΔgO and PEG, 26% of the fibroblasts expressed IE86 (10× TRΔgO) (Fig. 2). In separate experiments using concentrated stocks of TRΔgO, we were able to obtain 5 to 40% infection of fibroblasts using PEG treatment (not shown). Therefore, the markedly enhanced expression of IE86 following PEG treatment demonstrated that TRΔgO was severely defective for entry into human fibroblasts. However, the low levels of infection observed even with 10-fold more TRΔgO virions and PEG treatment suggested that many of the TRΔgO particles were defective in processes that preceded or followed fusion. This phenotype was different from that of the UL128-131 mutants (41).
A very similar phenotype was observed with HCMV derived from the second BAC clone, BAC-TRΔgO clone 2.1, described above, i.e., very few or no IE86+ fibroblasts were detected without PEG, and 5 to 40% IE86+ cells were detected with PEG (not shown). This provided evidence that this phenotype was not due to secondary mutations.
TRΔgO particles are defective for entry into epithelial and endothelial cells.
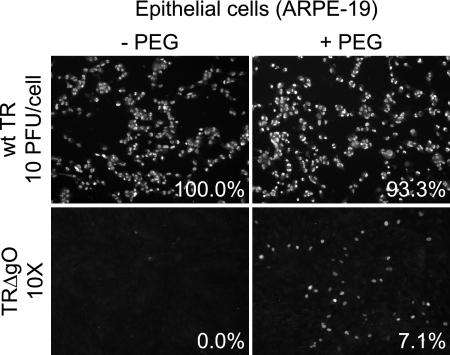
Our model suggested that gH/gL/gO might mediate infection of fibroblasts (12, 40). To test this model, we investigated whether TRΔgO could enter ARPE-19 retinal epithelial cells. Infection of ARPE-19 cells requires higher inputs of HCMV (12, 40), and consequently, cells were infected using 10 PFU/cell of wild-type TR and an equal quantity of TRΔgO particles (based on the quantities of viral DNA). All the epithelial cells were infected with wild-type TR (Fig. 3). In contrast, very few of the ARPE-19 cells were infected with TRΔgO. In this particular experiment, no IE86+ cells were detected in several wells infected with TRΔgO, although in other experiments, 1 to 10 IE86+ cells per well (0.005 to 0.04% of the cells) were detected. PEG treatment increased the number of IE86+ epithelial cells infected by TRΔgO to 7% (Fig. 3).
FIG. 3.
Entry of TRΔgO into epithelial cells with and without PEG treatment. Multiwell dishes of ARPE-19 epithelial cells were incubated with the following extracellular HCMV particles: wild-type (wt) TR corresponding to 10 PFU (defined using fibroblasts)/ARPE-19 cell or a similar quantity of TRΔgO virus particles (based on quantifying genomes using qPCR). The virus and cells were centrifuged at 800 × g for 1 h at 15°C. Some of the wells were subsequently incubated with 44% PEG for 30 s and then immediately washed. After 48 h, cells were fixed, permeabilized, and stained for IE86. Numbers indicate the average number of IE86+ cells in three wells.
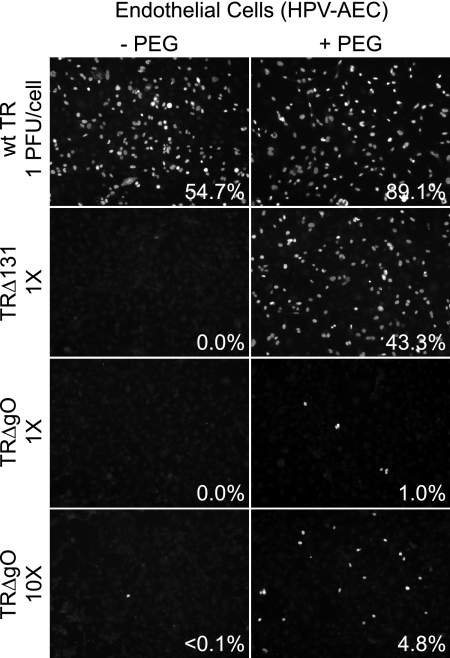
To determine whether TRΔgO could infect endothelial cells, HPV E6/E7-transformed aortic endothelial cells (HPV-AEC) were also infected with wild-type TR, TRΔ131, or TRΔgO. Again, similar quantities of TRΔgO particles were applied to cells using PCR to quantify viral genomes in preparations of supernatant virus particles. Wild-type TR infected 54.7% of the endothelial cells using 1 PFU/cell, whereas no cells were infected with either TRΔgO or TRΔ131 at this dose of virus (Fig. 4). Using 10-fold more TRΔgO (10× TRΔgO) (Fig. 4), less than 0.1% of the endothelial cells were infected in this experiment (Fig. 4), and no cells were infected in other experiments (not shown). PEG treatment increased TRΔ131 infection at the lower dose of virus (1 PFU/cell) to 43.3%. TRΔgO at the lower dose (1× TRΔgO) infected ∼1% of endothelial cells following PEG treatment, and 10-fold more TRΔgO particles infected 4.8% of the cells (10× TRΔgO) (Fig. 4). These observations suggested that TR gO is important for HCMV entry into both epithelial and endothelial cells. This conclusion was surprising given our model that showed gH/gL/gO functions primarily in fibroblasts and not in epithelial and endothelial cells. However, in contrast to TRΔ131, there were defects beyond entry fusion, because PEG enhancement did not restore infection to wild-type levels.
FIG. 4.
Entry of TRΔgO into endothelial cells with and without PEG treatment. Multiwell dishes of HPV-AEC endothelial cells were incubated with the following extracellular HCMV particles: wild-type (wt) TR or TRΔ131 using 1 PFU (defined using fibroblasts)/endothelial cell or a similar quantity (1×) of TRΔgO virus particles (based on quantifying genomes using qPCR) or 10 times that quantity of TRΔgO particles (10×). The cells and viruses were centrifuged at 800 × g for 1 h at 15°C. Some of the wells were subsequently incubated with 44% PEG for 30 s and then immediately washed. After 48 h, cells were stained for IE86. Numbers indicate the average number of IE86+ cells in three wells.
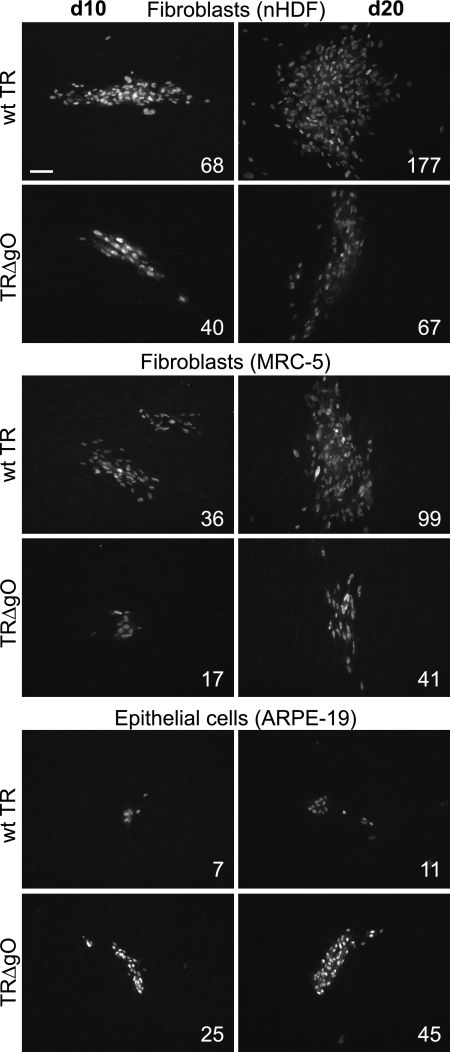
TRΔgO cell-to-cell spread.
Cell-to-cell spread of TRΔgO between fibroblasts and between epithelial cells was measured by using PEG to enhance virus entry into cells and then monitoring infected cells for IE86 expression over 20 days. TRΔgO produced plaques on NHDF and MRC-5 fibroblasts that included reduced numbers of infected cells per plaque, as follows: 40 to 65% infected compared with the numbers of infected cells per wild-type TR plaques (Fig. 5). In contrast, on ARPE-19 epithelial cells, TRΔgO formed larger plaques that included 350 to 440% of the number of infected cells per plaque compared with that per wild-type TR plaque (Fig. 5). We were unable to obtain plaques with several types of endothelial cells, as the cells did not remain attached to plastic dishes over the course of these long experiments. Given previous results (14, 19, 26), it was not unexpected that TRΔgO spread poorly on fibroblasts However, it was surprising that TRΔgO can spread between epithelial cells better than wild-type TR, especially given that the mutant largely failed to enter these epithelial cells as cell-free virus.
FIG. 5.
The cell-to-cell spread of TRΔgO in fibroblasts and epithelial cells. NHDF, MRC-5 fibroblasts, and ARPE-19 epithelial cells were infected with wild-type (wt) TR or TRΔgO using ∼100 to 200 PFU/well. Cells and virus were centrifuged at 800 × g for 1 h at 15°C, all wells were treated with 44% PEG for 30 s, and then the cells were washed and incubated in the presence of HCMV neutralizing antibodies for 10 or 20 days. Cells were then fixed and stained for IE86. The numbers of infected cells in 10 representative plaques were counted, and the average numbers are shown. Bar represents 100 μm.
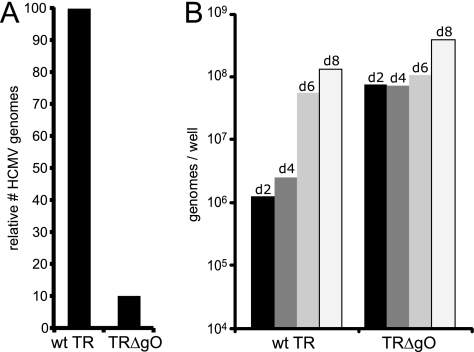
Quantification of TRΔgO virus particles in cell culture supernatants.
To determine whether the defects in TRΔgO virus particle entry and subsequent IE86 expression might be related to defects in assembly or release of virus particles, we quantified the numbers of viral genomes in cell culture supernatants. From the studies described above, it was clear that some TRΔgO particles were released from cells, and it was possible to concentrate these particles and enhance their entry by using PEG. However, these studies did not compare the numbers of particles released. In one set of experiments designed to quantify the numbers of particles present in cell culture supernatants, MRC-5 cells that had been transfected with BAC-TRΔgO or BAC-TR were trypsinized and plated with uninfected MRC-5. After two rounds of expansion of TRΔgO-infected fibroblasts, the numbers of infected cells were measured by IE86+ staining. In the experiment shown in Fig. 6A, ∼10% of the MRC-5 cells were infected with TRΔgO, and 100% of the cells were infected with wild-type TR. Cell culture supernatants were harvested from these cells, and following DNase treatment to remove DNA not protected by capsids, HCMV DNA was measured by quantitative PCR. In line with the numbers of infected cells, there was also ∼10% of the number of TRΔgO genomes compared with the number of wild-type TR genomes in these supernatants (Fig. 6A). To further determine whether there were defects in assembly or egress of HCMV with loss of gO, we attempted to increase the number of TRΔgO-infected cells by concentrating TRΔgO extracellular particles and used these in conjunction with PEG to enhance entry into cells. We also used fewer wild-type TR virions, so that after 2 days, ∼20% of the fibroblasts were infected by both TRΔgO and wild-type TR, as assessed by staining with IE86-specific antibodies. Cell culture supernatants were harvested after various times, and the quantities of HCMV genomes were measured by quantitative PCR. Given that there were higher input doses of TRΔgO particles, we observed larger quantities of extracellular virus particles (genomes) in TRΔgO supernatants after day 2 (Fig. 6B). This was related to the larger quantities of TRΔgO particles that stuck on cell surfaces (resisting washes) but failed to enter even with PEG enhancement. However, by day 8, differences between the amounts of viral DNA in cell culture supernatants comparing TRΔgO and wild-type TR narrowed so that there were comparable amounts of wild-type TR and TRΔgO DNA in supernatants. We concluded that TRΔgO assembles particles and that these particles are released into cell culture supernatants in relatively normal numbers.
FIG. 6.
Release of TRΔgO from fibroblasts into extracellular supernatants. (A) MRC-5 fibroblasts that had been transfected with BAC-TRΔgO or BAC-TR were trypsinized and plated with other MRC-5 cells. This allowed for the spread of TRΔgO, such that ∼10% of these cells showed a CPE and expressed IE86. After 10 days, culture supernatants from these TRΔgO-infected fibroblasts and supernatants from wild-type TR-infected fibroblasts (in which all the cells were infected) were subjected to quantitative PCR to enumerate HCMV genomes. (B) NHDF were infected with wild-type TR or TRΔgO using low-speed centrifugation, followed by PEG enhancement of entry, such that approximately 20% of the cells were infected by both viruses following 2 days of infection. Infected cell supernatants were collected 2, 4, 6, and 8 days postinfection and treated with DNase, and viral DNA was isolated and quantified by qPCR. d, day.
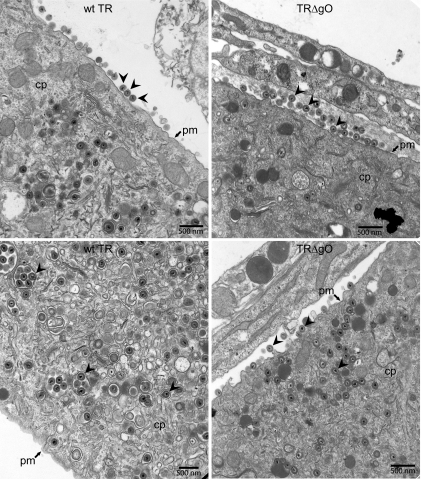
Electron microscopic analyses of TRΔgO-infected fibroblasts.
To further examine TRΔgO assembly and egress, we infected fibroblasts with wild-type TR or TRΔgO using PEG enhancement. Only 10 to 20% of the cells were infected with TRΔgO, whereas all cells were infected with wild-type TR. We harvested infected cells after 7 days and then fixed and stained the cells for electron microscopy. Enveloped virions were observed in the cytoplasm and on the surfaces of both wild-type TR- and TRΔgO-infected NHDF (Fig. 7). Due to the differences in the numbers of infected cells, it was difficult to compare (count) the absolute numbers of enveloped virions. However, among the TRΔgO-infected cells, cells that possessed capsids in the nucleus, there was no apparent reduction in the numbers of enveloped versus unenveloped capsids in the cytoplasm, and numerous TRΔgO virus particles were observed on cell surfaces. Together with the results described above involving quantification of viral DNA (Fig. 6), we concluded that TR gO is not required for assembly of enveloped virions or egress to cell surfaces.
FIG. 7.
Electron microscopy of wild-type TR- and TRΔgO-infected fibroblasts. NHDF were infected with wild-type TR or TRΔgO by using low-speed centrifugation and PEG treatment. Under these conditions, ∼10 to 20% of TRΔgO-infected cells displayed IE86 expression by day 2, whereas all the cells were infected with wild-type TR. After 7 days, cells were fixed, stained, and analyzed by transmission electron microscopy. cp, cytoplasm; pm, plasma membrane; arrowheads, enveloped virus particles.
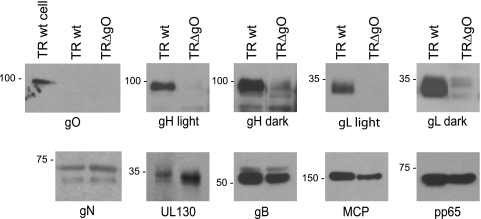
Loss of gO reduces incorporation of gH/gL complexes into the virion envelope but increases quantities of UL130.
In the accompanying paper (42), we showed that gO promotes ER export of gH/gL. Thus, it was of special interest to determine whether loss of gO altered the quantities of the gH/gL complexes present in extracellular virus particles. Fibroblasts were infected with wild-type TR or TRΔgO using PEG enhancement, and cell culture supernatants were harvested and concentrated by pelleting the particles through 20% sorbitol cushions. Viral proteins in these particles were analyzed by Western blotting. Again, we attempted to equalize the quantities of wild-type TR versus those of TRΔgO particles by using quantitative PCR to determine virus genomes. Several of the major structural proteins of HCMV particles, including the major capsid protein (MCP), tegument protein pp65, and glycoproteins gB and gN, were present in wild-type TR particles, as expected (Fig. 8). TRΔgO exhibited reduced quantities of gB, MCP, and pp65, although these proteins were reduced by 2-fold or less, likely reflecting differences in measuring viral DNA and proteins (Fig. 8). gO was detected in TR-infected cells, but not in wild-type TR particles, consistent with observations in the accompanying paper (42). Surprisingly, TRΔgO virions contained little gH and gL, despite being present in infected cell lysates at a level comparable to that of gB (Fig. 1). In lighter exposures, no gH or gL was observed, but darker exposures revealed a small quantity of gH and gL in TRΔgO particles. Even taking into account the reduced quantities of gB and MCP in TRΔgO particles, we estimate that gH and gL were reduced by ∼90 to 95% in TRΔgO particles compared with those levels in TR particles. Despite this decreased quantity of gH/gL, UL130 was increased in TRΔgO virions by 2- to 3-fold compared with that in wild-type TR. These studies were further confirmation that virus particles were released from TRΔgO-infected cells. We concluded that loss of TR gO leads to the production of virions with much less gH/gL, although the small quantities of gH/gL that are present contain more UL130 and, likely, gH/gL/UL128-131.
FIG. 8.
Analyses of HCMV proteins in TRΔgO extracellular virus particles. NHDF were infected with wild-type TR or TRΔgO using low-speed centrifugation and PEG to enhance entry. After 8 days, virus particles were prepared from cell culture supernatants by centrifugation through 20% sorbitol cushions. The quantities of the virus particles per sample were made similar by measuring viral genomes using qPCR. A cell lysate from wild-type TR-infected cells was loaded as a positive control for gO immunoblotting (TR wt cell). Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to Immobilon membranes, and Western blotted for gO, gH, gL, gN, UL130, gB, the major capsid protein (MCP), or tegument protein pp65. Lighter and darker exposures are shown for gH and gL.
DISCUSSION
Our studies of HCMV TR gO were prompted by observations that clinical and lab strains of HCMV differ with respect to expression of gH/gL/UL128-131, which mediates entry into epithelial and endothelial cells (41). Given these results and work with other herpesviruses, it seemed likely that gH/gL/gO might be involved in entry into fibroblasts. Consistent with a role for gH/gL/gO in HCMV entry into fibroblasts, we recently found that expression of gH/gL/gO in fibroblasts effectively interfered with HCMV entry into fibroblasts, while expression of gB, gH/gL, and gH/gL/UL128-131 produced much less or no interference (M. C. Chase, B. J. Ryckman, and D. C. Johnson, unpublished data). Thus, our working model suggested that HCMV uses gH/gL/UL128-131 to enter epithelial/endothelial cells and gH/gL/gO to enter fibroblasts, and adaptation to fibroblasts favors the gH/gL/gO entry pathway.
To test the model, a HCMV TR gO-null mutant was constructed using BAC mutagenesis. The use of BACs substantially reduces the chances of second-site mutations involving distant genes (4). However, the possibility of second-site mutations (affecting neighboring or more distant genes) was made extremely unlikely by three sets of observations. First, the deletion of the gO coding sequences did not affect either the promoters or polyadenylation sites for the adjacent gH and gN genes, and gH and gN proteins were expressed at levels comparable to that of gB. Second, two separate BAC clones were used to produce two different viruses, and both were found to be unable to enter fibroblasts and epithelial cells unless PEG enhancement was used. Third, our TR gO-null mutant was phenotypically similar to the previously characterized HCMV AD169, Towne, and TB40/E gO-null mutants with respect to production of small plaques on fibroblasts and relatively normal (or better) spread on epithelial and endothelial cells (14, 19, 26). It is very hard to conceive of the possibility that secondary mutations, i.e., mutations in genes other than the gO gene, would produce this exact phenotype on all cell types.
TRΔgO produced very little infectious virus into cell culture supernatants, i.e., virus that can initiate infection of fibroblasts. When TRΔgO particles from supernatants were concentrated and applied to fibroblast monolayers, only a few cells per dish (0 to 0.04%) were infected. However, when PEG was used to chemically promote membrane fusion, as much as 40% of the fibroblasts were able to be infected with TRΔgO. The striking increase in rates of infection with PEG demonstrated that a fraction of TRΔgO particles were markedly defective for entry into fibroblasts. However, PEG enhancement of TRΔgO failed to produce rates of infection similar to those produced by wild-type HCMV. When 10 times as many TRΔgO particles were used (compared to TR at 1 PFU/cell, a dose that infected ∼75% of cells), there was infection of only 5 to 40% of fibroblasts. These results also suggested that another fraction of TRΔgO particles possessed defects either preceding or following entry fusion.
Quantification of HCMV genomes in extracellular virus particle preparations demonstrated relatively normal numbers of TRΔgO particles compared with those demonstrated by wild-type TR, when numbers of infected cells were taken into account. These studies were complicated to perform in relation to defects in TRΔgO infection and spread between fibroblasts. With fewer fibroblasts infected by TRΔgO rather than wild-type TR, it was difficult to compare extracellular particles. PEG was used to boost the entry of TRΔgO, and using lower inputs of TR, we could roughly match infections so that after 2 days, there was ∼20% infection with both mutant and wild-type HCMV. After 8 days, TRΔgO produced comparable quantities of HCMV particles (measured by viral DNA analysis). Immunoblotting of extracellular TRΔgO particles also showed that there were relatively normal quantities of gB, the tegument protein pp65, and MCP. Furthermore, electron microscopy of TRΔgO-infected fibroblasts revealed morphologically normal enveloped virus particles in the cytoplasm and on cell surfaces.
Our analyses of extracellular TRΔgO particles derived from fibroblasts by immunoblotting demonstrated marked reductions in the amounts of gH and gL, compared with those of gB, pp65, and MCP. Related to these observations, in the accompanying paper (42), we showed that TR gO is largely ER retained but acts as a molecular chaperone to facilitate ER export of gH and gL. Previous studies had shown that coexpression of UL128-131 with gH/gL (without other HCMV proteins) also increases ER export of gH/gL, but importantly, the UL128-131 proteins remain bound onto gH/gL and become incorporated into virions as gH/gL/UL128-131 (41). The markedly reduced quantities of gH/gL present in extracellular TRΔgO virions probably explain the observed defects in entry into fibroblasts. It appears that gH/gL, and not gH/gL/gO, mediates entry into fibroblasts. This conclusion is partially based on the observations that HCMV UL128-131-null mutants can efficiently enter fibroblasts (18, 40), indicating that a second gH/gL complex must function in fibroblast entry. Furthermore, the highly reduced amounts of gH/gL present in TRΔgO particles might explain our observations that these particles are morphologically normal yet cannot enter cells efficiently, even with PEG treatment. Without normal amounts of gH/gL, these virions might be less able to bind onto cell surfaces. Reduced binding to cells would not be overcome by PEG treatment. This point is under investigation in ongoing studies.
Entry into epithelial and endothelial cells by TRΔgO (produced by fibroblasts) was also extremely poor. Concentrated stocks of extracellular TRΔgO particles infected only a few cells in the entire monolayer. PEG treatment increased TRΔgO infection to 5 to 10% of these cells. This inhibition of entry into epithelial and endothelial cells was not predicted from our working model in which gH/gL/UL128-131, rather than some other gH/gL complex, is required for entry into epithelial and endothelial cells. Instead, the highly reduced quantities of gH/gL in the envelope of TRΔgO apparently reduced entry into these cells. This supports a modified hypothesis in which HCMV entry into epithelial and endothelial cells requires both gH/gL and gH/gL/UL128-131.
Spread of TRΔgO in fibroblast monolayers was reduced, as plaques included 35 to 60% of the number of cells, compared with that of wild-type TR. In contrast, the defect in entering fibroblasts was much more pronounced, involving 104 to 105 fewer cells infected. Cell-to-cell spread of TRΔgO between epithelial cells was increased by 2.5 to 4-fold, compared with that of wild-type TR. Again, this spread was compared with 104 to 105 fewer cells infected by TRΔgO. These results were similar to those involving the TB40/E gO-null mutant, which could spread normally on endothelial cells (26). We found that there were increased amounts of UL130 present in TRΔgO extracellular virions. It seems probable that these increased amounts of UL130 were accompanied by increased amounts of UL128 and UL131 as well, although this was not directly tested. If this is the case, the increased quantities of gH/gL/UL128-131 found in TRΔgO virions were insufficient for entry into epithelial/endothelial cells but apparently promoted increased movement of TRΔgO particles between epithelial and endothelial cells.
Observations of enhanced HCMV cell-to-cell spread with virus particles that were highly defective for entry into cells were quite astonishing. These results underscore major differences between these two processes. One possibility is that cell-to-cell spread is much less sensitive to deficiencies in the quantities of gH/gL, compared with entry of extracellular particles. This may relate to the relative concentration of herpesvirus receptors at cell junctions where cell-to-cell spread likely occurs (reviewed in reference 27). Extracellular virus particles might require higher quantities of gH/gL complexes to find and bind receptors on cell surfaces. However, it is important to note that we did not purify extracellular TRΔgO virus particles from epithelial cells. Thus, we do not know whether these extracellular particles, or particles spreading between epithelial cells, have deficiencies in gH/gL, but this is likely.
Previous characterization of an HCMV strain TB40/E gO-null mutant by Jiang et al. (26) produced some results that are similar to ours but also some different conclusions about how gO functions. They observed very few infectious virus particles (100- to 1,000-fold decreased) produced into fibroblast culture supernatants, similar to our results with TRΔgO. The very small amounts of infectious TB40/E gO-null mutants in cell culture supernatants were concentrated and then applied to fibroblasts using 0.001 PFU/cell, which compares to the same amount of wild-type TB40/E. Both the mutant and the wild type produced rare cells expressing early and late HCMV genes. From this, they concluded that there were no defects in virus entry associated with loss of gO. However, because TB40/E gO-null particles in supernatants were not enumerated, it was not clear whether there were large quantities of noninfectious TB40/E gO-null mutants (based on the 100- to 1,000-fold decrease in infectious virus) that failed to enter these cells. Certainly, our studies involving PEG treatment directly demonstrated defects in TRΔgO entry into all cells tested. We also observed no defects in virus egress, normal numbers of virus particles were found in cell culture supernatants, and enveloped particles were observed in infected cells in normal numbers by electron microscopy. Jiang et al. observed substantial accumulation of unenveloped capsids in the cytoplasm and few enveloped particles produced in TB40/E gO-null-infected fibroblasts by electron microscopy and concluded that gO is required for virus assembly (26). The different conclusions about how TB40/E and TR gO function might also reflect strain differences, as gO is highly variable (13, 34, 35). In this regard, TB40/E was passaged extensively on endothelial cells, whereas TR is a low-passage strain. Whether our results involving TR gO apply to other HCMV strains is not clear. TR is a low-passage isolate that we consider the “wild type.” Alternatively, the concept of “wild type” may be misplaced here, so that different strains of HCMV make use of gO for different functions. Resolving these issues will require the characterization of other clinical strains of HCMV (e.g., strain Merlin).
In summary, the studies herein and in the accompanying paper (42) demonstrated that TR gO is not present in virions and, instead, acts to increase gH/gL export from the ER (42) and incorporation into extracellular virions (this study). Thus, it appears that gH/gL, rather than gH/gL/gO, mediates HCMV entry into fibroblasts. However, entry into epithelial and endothelial cells appears to require both gH/gL and gH/gL/UL128-131. These observations provide a molecular picture for why loss of UL128-131 proteins promotes adaptation of HCMV to propagation in fibroblasts. The increased levels of UL130 in TRΔgO particles suggest that gO and UL128-131 compete for binding to gH/gL. Consistent with this notion, coexpression of gO with gH/gL decreases binding of UL128-131 to gH/gL in experiments involving Ad vectors (B. J. Ryckman, unpublished results). Using this model, loss of UL128-131 might favor the production of gH/gL and the fibroblast entry pathway. It seems possible that adaptation to passage in fibroblasts produces changes in gO. Observations that both HCMV gH/gL and gH/gL/UL128-131 are required for entry into cells differ from models involving EBV and HHV-6, which use one of two gH/gL complexes to enter different cell types (24, 32).
Acknowledgments
This work was supported by grants from the National Institutes of Health, R01AI081517 (to D.C.J.) and RO1AI21640 (to J.A.N.).
We thank Michael Webb of the OHSU EM Core for excellent technical assistance with the electron microscopy experiments and Craig Kreklywich for assistance with qPCR. We are grateful to William J. Britt (University of Alabama—Birmingham) for supplying important monoclonal antibodies. Tiffani Howard prepared the graphics. Finally, we appreciate the advice and support from all the members of the Johnson and Nelson laboratories, especially Brent Ryckman, Adam Vanarsdall, and Marie Chase.
Footnotes
Published ahead of print on 23 December 2009.
REFERENCES
- 1.Adler, B., L. Scrivano, Z. Ruzcics, B. Rupp, C. Sinzger, and U. Koszinowski. 2006. Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol. 87:2451-2460. [DOI] [PubMed] [Google Scholar]
- 2.Akter, P., C. Cunningham, B. P. McSharry, A. Dolan, C. Addison, D. J. Dargan, A. F. Hassan-Walker, V. C. Emery, P. D. Griffiths, G. W. Wilkinson, and A. J. Davison. 2003. Two novel spliced genes in human cytomegalovirus. J. Gen. Virol. 84:1117-1122. [DOI] [PubMed] [Google Scholar]
- 3.Bissinger, A. L., C. Sinzger, E. Kaiserling, and G. Jahn. 2002. Human cytomegalovirus as a direct pathogen: correlation of multiorgan involvement and cell distribution with clinical and pathological findings in a case of congenital inclusion disease. J. Med. Virol. 67:200-206. [DOI] [PubMed] [Google Scholar]
- 4.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britt, W. J., M. Jarvis, J. Y. Seo, D. Drummond, and J. Nelson. 2004. Rapid genetic engineering of human cytomegalovirus by using a lambda phage linear recombination system: demonstration that pp28 (UL99) is essential for production of infectious virus. J. Virol. 78:539-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Britt, W. J., and C. A. Alford. 1996. Cytomegaloviruses, p. 2493-2523. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Press, Philadelphia, PA.
- 7.Britt, W. J. 1984. Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology 135:369-378. [DOI] [PubMed] [Google Scholar]
- 8.Britt, W. J., and D. Auger. 1985. Identification of a 65 000 dalton virion envelope protein of human cytomegalovirus. Virus Res. 4:31-36. [DOI] [PubMed] [Google Scholar]
- 9.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chee, M., S. A. Rudolph, B. Plachter, B. Barrell, and G. Jahn. 1989. Identification of the major capsid protein gene of human cytomegalovirus. J. Virol. 63:1345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 12.Compton, T., R. R. Nepomuceno, and D. M. Nowlin. 1992. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 191:387-395. [DOI] [PubMed] [Google Scholar]
- 13.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerna, G., F. Baldanti, and M. G. Revello. 2004. Pathogenesis of human cytomegalovirus infection and cellular targets. Hum. Immunol. 65:381-386. [DOI] [PubMed] [Google Scholar]
- 17.Gerna, G., E. Percivalle, D. Lilleri, L. Lozza, C. Fornara, G. Hahn, F. Baldanti, and M. G. Revello. 2005. Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131-128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J. Gen. Virol. 86:275-284. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huber, M. T., and T. Compton. 1997. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J. Virol. 71:5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, M. T., and T. Compton. 1999. Intracellular formation and processing of the heterotrimeric gH-gL-gO (gCIII) glycoprotein envelope complex of human cytomegalovirus. J. Virol. 73:3886-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber, M. T., T. W. Wisner, N. R. Hegde, K. A. Goldsmith, D. A. Rauch, R. J. Roller, C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and D. C. Johnson. 2001. Herpes simplex virus with highly reduced gD levels can efficiently enter and spread between human keratinocytes. J. Virol. 75:10309-10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutt-Fletcher, L. M. 2007. Epstein-Barr virus entry. J. Virol. 81:7825-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarvis, M. A., C. E. Wang, H. L. Meyers, P. P. Smith, C. L. Corless, G. J. Henderson, J. Vieira, W. J. Britt, and J. A. Nelson. 1999. Human cytomegalovirus infection of caco-2 cells occurs at the basolateral membrane and is differentiation state dependent. J. Virol. 73:4552-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang, X. J., B. Adler, K. L. Sampaio, M. Digel, G. Jahn, N. Ettischer, Y. D. Stierhof, L. Scrivano, U. Koszinowski, M. Mach, and C. Sinzger. 2008. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J. Virol. 82:2802-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landolfo, S., M. Gariglio, G. Gribaudo, and D. Lembo. 2003. The human cytomegalovirus. Pharmacol. Ther. 98:269-297. [DOI] [PubMed] [Google Scholar]
- 29.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 30.Li, L., J. A. Nelson, and W. J. Britt. 1997. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J. Virol. 71:3090-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori, Y. 2009. Recent topics related to human herpesvirus 6 cell tropism. Cell. Microbiol. 11:1001-1006. [DOI] [PubMed] [Google Scholar]
- 33.Mori, Y., P. Akkapaiboon, S. Yonemoto, M. Koike, M. Takemoto, T. Sadaoka, Y. Sasamoto, S. Konishi, Y. Uchiyama, and K. Yamanishi. 2004. Discovery of a second form of tripartite complex containing gH-gL of human herpesvirus 6 and observations on CD46. J. Virol. 78:4609-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paterson, D. A., A. P. Dyer, R. S. Milne, E. Sevilla-Reyes, and U. A. Gompels. 2002. A role for human cytomegalovirus glycoprotein O (gO) in cell fusion and a new hypervariable locus. Virology 293:281-294. [DOI] [PubMed] [Google Scholar]
- 36.Plachter, B., C. Sinzger, and G. Jahn. 1996. Cell types involved in replication and distribution of human cytomegalovirus. Adv. Virus Res. 46:195-261. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen, L., A. Geissler, C. Cowan, A. Chase, and M. Winters. 2002. The genes encoding the gCIII complex of human cytomegalovirus exist in highly diverse combinations in clinical isolates. J. Virol. 76:10841-10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roop, C., L. Hutchinson, and D. C. Johnson. 1993. A mutant herpes simplex virus type 1 unable to express glycoprotein L cannot enter cells, and its particles lack glycoprotein H. J. Virol. 67:2285-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryckman, B. J., M. C. Chase, and D. C. Johnson. 2008. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proc. Natl. Acad. Sci. U. S. A. 105:14118-14123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryckman, B. J., M. A. Jarvis, D. D. Drummond, J. A. Nelson, and D. C. Johnson. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80:710-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryckman, B. J., B. L. Rainish, M. C. Chase, J. A. Borton, J. A. Nelson, M. A. Jarvis, and D. C. Johnson. 2008. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 82:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryckman, B. J., M. C. Chase, and D. C. Johnson. 2010. Human cytomegalovirus TR strain glycoprotein O acts as a chaperone promoting gH/gL incorporation into virions but is not present in virions. J. Virol. 84:2597-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sinzger, C., M. Digel, and G. Jahn. 2008. Cytomegalovirus cell tropism. Curr. Top. Microbiol. Immunol. 325:63-83. [DOI] [PubMed] [Google Scholar]
- 44.Sinzger, C., and G. Jahn. 1996. Human cytomegalovirus cell tropism and pathogenesis. Intervirology 39:302-319. [DOI] [PubMed] [Google Scholar]
- 45.Smith, I. L., I. Taskintuna, F. M. Rahhal, H. C. Powell, E. Ai, A. J. Mueller, S. A. Spector, and W. R. Freeman. 1998. Clinical failure of CMV retinitis with intravitreal cidofovir is associated with antiviral resistance. Arch. Ophthalmol. 116:178-185. [DOI] [PubMed] [Google Scholar]
- 46.Stanton, R., D. Westmoreland, J. D. Fox, A. J. Davison, and G. W. Wilkinson. 2005. Stability of human cytomegalovirus genotypes in persistently infected renal transplant recipients. J. Med. Virol. 75:42-46. [DOI] [PubMed] [Google Scholar]
- 47.Vanarsdall, A. L., B. J. Ryckman, M. C. Chase, and D. C. Johnson. 2008. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J. Virol. 82:11837-11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, D., and T. Shenk. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 79:10330-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, D., and T. Shenk. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. U. S. A. 102:18153-18158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wisner, T., C. Brunetti, K. Dingwell, and D. C. Johnson. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J. Virol. 74:2278-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978- 5983. [DOI] [PMC free article] [PubMed] [Google Scholar]