Abstract
In an effort to understand the potential role of microRNAs (miRNAs) in mammary-gland stem or progenitor cells, miRNA microarrays were performed on subpopulations of the mouse mammary epithelial cell (MEC) line COMMA-DβGeo. This cell line contains a heterogeneous subpopulation of progenitors characterized by the expression of stem cell antigen 1 (Sca-1; encoded by Ly6a). Microarray analysis indicated that the Sca-1 subpopulations have distinct miRNA expression profiles. Functional studies were performed on miR-205, which was highly expressed in the Sca-1-positive (Sca-1+) cells. When miR-205 was overexpressed in vitro, the COMMA-DβGeo cells underwent several significant morphological and molecular changes. miR-205 overexpression led to an expansion of the progenitor-cell population, decreased cell size and increased cellular proliferation. In addition, the colony-forming potential of the two Sca-1 subpopulations was increased. Target prediction for miR-205 indicated that it might regulate the expression of the tumor-suppressor protein PTEN. Overexpression studies using reporter constructs confirmed that PTEN expression is regulated by miR-205. In addition to PTEN, several other putative and previously validated miR-205 targets were identified by microarray analysis, including the previously reported miR-205 targets ZEB1 and ZEB2. Additionally, in normal mouse MECs, high expression of miR-205 was observed in stem-cell-enriched cell populations isolated by FACS using established cell-surface markers.
Keywords: Mammary gland, miRNA, Stem cells
Introduction
Gene silencing by miRNAs is primarily achieved by targeting the 3′ untranslated region (UTR) of mRNAs and induces translational silencing, either by cleaving and degrading the target mRNA or blocking ribosomal translation of the targeted mRNA (Pillai et al., 2007), although examples of miRNAs targeting coding regions have recently been described (Forman et al., 2008; Grimson et al., 2007). miRNAs have been shown to be expressed in a lineage-specific manner both in normal development (Hino et al., 2008; Ramkissoon et al., 2006) and cancer (Blenkiron et al., 2007; Gaur et al., 2007; Mattie et al., 2006; Sempere et al., 2007).
The mammary gland consists of two distinct cell lineages, basal and lumenal, which together form the ductal mammary epithelium. Many studies have provided evidence for a common bipotent progenitor that gives rise to ductal lumenal, alveolar lumenal and myoepithelial (basal) cells. However, the molecular mechanisms that regulate lineage commitment at each stage of mammary-gland development remain ill-defined (reviewed by Wagner and Smith, 2005). In the present study, the COMMA-DβGeo cell line was used as an in vitro model system to study adult mammary stem or progenitor cells. This cell line was originally derived from mammary epithelium isolated from midpregnant Balb/c mice (Campbell et al., 1988; Danielson et al., 1984). COMMA-DβGeo cells consist of a heterogeneous population of cells, including a subpopulation enriched for putative progenitor-like cells as defined by the expression of stem cell antigen 1 (Sca-1; encoded by Ly6a). Sca-1, originally identified as a marker of hematopoietic stem cells (Spangrude et al., 1988), has been used to identify mammary-gland progenitor cells (Welm et al., 2002), as well as a variety of stem or progenitor cells in other tissues, including skin (Montanaro et al., 2003), muscle (Torrente et al., 2001) and testis (Falciatori et al., 2004; Xin et al., 2005). More recently, it has been suggested that, in the normal mammary gland, lumenal Sca-1-positive (Sca-1+) cells are estrogen-receptor positive, and, although most of these are terminally differentiated, there is a subset that might be lumenal progenitors (Sleeman et al., 2007; Stingl, 2009).
The Sca-1+ and Sca-1− cell populations of COMMA-DβGeo display different morphogenic phenotypes and growth potentials, in both in vitro clonogenic assays and in vivo mammary-gland repopulation assays. The Sca-1+ subpopulation displays an undifferentiated basal phenotype, on the basis of staining negative for lumenal cytokeratins (K) K8 and K18, positive for basal cytokeratins K5 and K14, and negative for myoepithelial α-smooth-muscle actin (Deugnier et al., 2006). When transplanted into the cleared fat pad of the mouse mammary gland, the Sca-1+ population gives rise to branched ductal epithelium containing lumenal and myoepithelial cells, whereas the Sca-1− population displays a reduced outgrowth potential (Deugnier et al., 2006; Welm et al., 2002). In addition, Sca-1+ cells form colonies when plated on Matrigel, whereas the same number of Sca-1− cells fail to do so (Chen et al., 2007; Woodward et al., 2007). This clonogenicity is reflective of the self-renewal potential of Sca-1+ cells. Sca-1+ cells will also give rise to both Sca-1+ and Sca-1− cells when grown in culture, which is indicative of their multipotency (Chen et al., 2007).
The COMMA-DβGeo cell line was used as representative of mammary-gland progenitor cells to identify miRNAs that might be important in lineage determination during mammary-gland development. We observed discrete miRNA expression profiles in these different Sca-1 subpopulations. We focused our studies on miR-205, which was observed to be the most highly upregulated miRNA in the Sca-1+ population as compared with the Sca-1− population. We used the well-characterized phenotypes attributed to the Sca-1+ and Sca-1− cells to assess the effect of overexpression of miR-205. We demonstrated that this miRNA targets the previously reported regulators of epithelial-mesenchymal transition (EMT) ZEB1 and ZEB2, as well as several targets not previously described, including the Na+-K+ transporter Atp1a1, the docking protein Dok4 and the tumor-suppressor gene phosphatase and tensin homolog (PTEN). In addition, when miR-205 was overexpressed, we observed changes in replicative competence, cell size, cell proliferation and the percentage of progenitor-cell populations. Finally, miR-205 was shown to be selectively upregulated in stem and/or progenitor cells isolated from normal MECs using FACS and established cell-surface markers, demonstrating the relevance of using the COMMA-DβGeo model.
Results
The Sca-1 subpopulations of the COMMA-DβGeo cell line express a distinct set of miRNAs
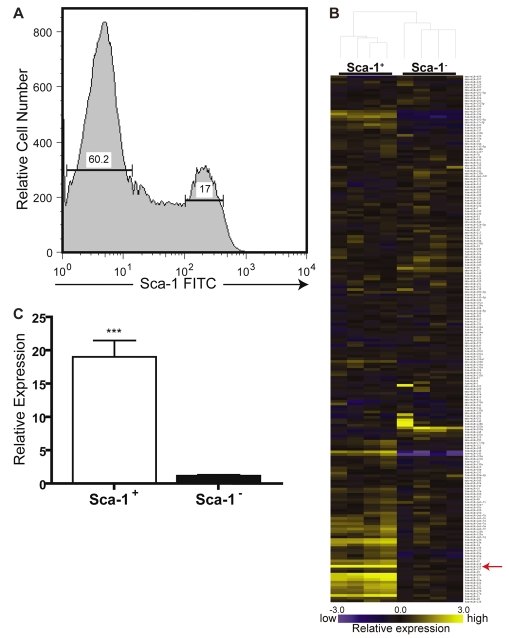
The COMMA-DβGeo cell line maintains a heterogeneous population of cells, on the basis of the presence or absence of the cell-surface marker Sca-1; presence of this marker can easily be distinguished by FACS (Fig. 1A). This cell line typically retains between 13-20% Sca-1+ cells in culture: Sca-1+ cells are characterized as multipotent progenitor cells by in vitro and in vivo assays (Chen et al., 2007; Deugnier et al., 2006). Because Sca-1+ and Sca-1− cells are functionally different, we sought to identify differentially expressed miRNAs between the two cell populations using miRNA microarrays (Fig. 1B). Two different miRNA microarray platforms were used (custom microarrays and μParaflo microarrays), revealing a consistent set of differentially expressed miRNAs. Results from both microarray platforms were analyzed for fold change and compared. A set of 15 upregulated miRNAs and six downregulated miRNAs between the two populations of cells were consistently observed (Table 1). Array data for several miRNAs were subsequently verified by quantitative real-time reverse-transcriptase (RT)-PCR (qPCR) and fold change was confirmed (Fig. 1C; supplementary material Fig. S1A-C).
Fig 1.
Identification of miRNAs differentially expressed in the Sca-1 subpopulations of the COMMA-DβGeo cell line. (A) Representative FACS histogram showing Sca-1 expression in COMMA-DβGeo cells. Cells stained with a FITC-conjugated antibody against Sca-1 analyzed by FACS display two distinct populations based on Sca-1—FITC fluorescence: 17% are Sca-1+ and 60.2% are Sca-1−. Shown are 75,000 events. (B) Representative heat map showing the differential expression of miRNAs in the Sca-1+ and Sca-1− subpopulations. Each row represents a different mature miRNA sequence. Shown are median values from normalized, log-ratio (base 2) data sets and plotted as a heat map from two biological and technical replicates. Yellow indicates high miRNA expression; blue indicates low miRNA expression. Differentially expressed miRNAs are detailed in Table 1 from three independent microarray experiments performed on three biological replicates each. Expression values produced a cladogram reflecting a clear distinction between the Sca-1+ and Sca-1− cells. The red arrow indicates the location of miR-205 on the array, where the average fold change was 6.56 higher in the Sca-1+ cells versus Sca-1−. (C) TaqMan qPCR of miR-205 verify the microarray data. miR-205 is approximately 19-fold higher in the Sca-1+ population compared with the Sca-1− population. Data represent the mean expression levels of two different biological replicates calculated by ΔΔCT normalized to the endogenous snoRNA55 control, of two independent experiments performed in triplicate. ***P<0.0001.
Table 1.
miRNAs differentially expressed in the Sca-1+ versus Sca-1− populations of the COMMA-D Geo cell line
Microarray data revealed that miR-205 was the most upregulated miRNA in the Sca-1+ cells and this expression was verified by qPCR (Fig. 1C). This miRNA has been shown to be upregulated in non-metastatic human breast cancers (Iorio et al., 2005); however, until very recently, very little was known about the function or targets of this miRNA. Interestingly, a recent study also observed high expression of miR-205 in a different progenitor-cell population of the COMMA-DβGeo cell line using a cloning method (Ibarra et al., 2007), although Aldefluor, the method they employed for isolating progenitor cells in the mouse, has yet to be functionally validated in vivo. In addition to miR-205, Ibarra et al. also detected several of the same miRNAs in the Aldefluorhi/Sca-hi population that we observed to be highly expressed in the Sca-1+ cells, miR-21, miR-22 and miR-31, indicating that these two populations, Sca-1+ and Aldefluorhi/Sca-1hi, might partially overlap.
Mouse miR-205 affects the progenitor-cell compartment, cell size and proliferation
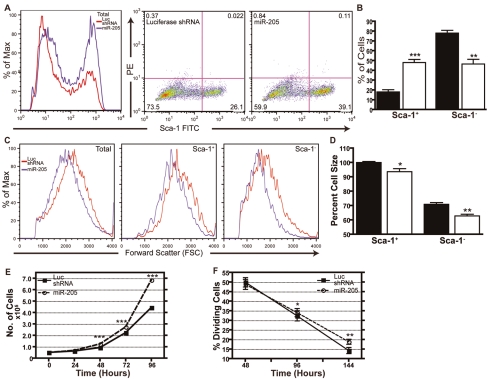
To determine whether miR-205 plays a functional role in the progenitor-cell population of COMMA-DβGeo cells, we first overexpressed it using the pSM2 retroviral vector. Under the pSM2 U6 promoter, miR-205 expression was increased between two- to fourfold over the levels observed in cells transduced with the luciferase shRNA negative control vector in the COMMA-DβGeo cells. When further separated into the Sca-1 subpopulations, we found that miR-205 levels in Sca-1+ cells were approximately twofold higher than the luciferase shRNA cells. Whereas the Sca-1− cells did not obtain the high levels of miR-205 expression observed in the Sca-1+ cells, expression of miR-205 was increased nearly fivefold in the Sca-1− population when transduced with the pSM2—miR-205 vector as compared to the luciferase shRNA control (supplementary material Fig. S1D). In cells overexpressing miR-205, we observed a significant expansion of the Sca-1+ cell population, on the basis of FACS analysis for Sca-1—FITC, compared with the luciferase shRNA control (Fig. 2A,B). Typically, an increase from the expected 13-20% of Sca-1+ cells to 39-53% was observed following overexpression of miR-205.
Fig 2.
Overexpression of miR-205 affects cell size, cell proliferation and expands the progenitor-cell compartment. (A) Representative FACS analysis of COMMA-DβGeo cells indicates that overexpression of miR-205 expands the Sca-1+ percentage compared with cells expressing the luciferase shRNA control. Cells were sorted for Sca-1—FITC and separated into Sca-1+ and Sca-1− cells. Shown are 25,000 events. (B) Quantitation of the percent of cells that are either Sca-1+ or Sca-1− based on FACS analysis. Populations that overexpress miR-205 (white bars) have more Sca-1+ cells compared with populations expressing the luciferase shRNA control (black bars). ***P<0.001; **P<0.01. (C) COMMA-DβGeo cells overexpressing miR-205 are smaller than cells expressing the control luciferase shRNA as determined by FACS analysis. Cells were sorted for Sca-1—FITC and separated into Sca-1+ and Sca-1− cells. In all three compartments — total (viable) cells, Sca-1+ and Sca-1− — miR-205-overexpressing cells are all smaller than the luciferase shRNA cells. Shown are 25,000 events. (D) Quantitation of mean FSC percent, which is proportional to cell size, based on FACS analysis. miR-205-overexpressing cells (white bars) are significantly smaller in the Sca-1+ and Sca-1− cell populations compared with cells expressing the luciferase shRNA control (black bars). Cell size is given in percentage relative to the control, set at 100%. *P<0.05; **P<0.005. (E) Growth curves of miR-205-overexpressing cells and luciferase shRNA cells. Cells were plated at the same density and counted every 24 hours for 6 days (shown are timepoints up to 96 hours). Cells overexpressing miR-205 are proliferating at significantly higher rates than the control. At 144 hours post-plating, there are still significantly more miR-205-overexpressing cells (P<0.0001) compared with the luciferase shRNA cells (not shown). ***P<0.0001 compared with luciferase shRNA. (F) Cell-proliferation assay, where percent dividing cells represents the fraction of cells that are or have synthesized DNA in preparation for cell division (equivalent to the sum of the percentages of cells in S+G2-M), as determined by the Click-iT EdU Flow Cytometry Assay. Cells overexpressing miR-205 have significantly higher proliferation rates (P<0.005) compared with the luciferase shRNA cells. All data represent the mean of two independent experiments performed in duplicate with two biological replicates.
In addition to the expansion of the progenitor-cell compartment, overexpression of miR-205 resulted in a significantly smaller cell size as compared with the luciferase shRNA control cells (Fig. 2C,D). Results are expressed as percentage change in forward scatter (FSC) compared with the luciferase shRNA cells. The total population of viable cells decreased in size by approximately 4-5% when overexpressing miR-205. Because Sca-1− cells are generally smaller than Sca-1+ cells, these differences were further highlighted when cells were separated into the two Sca-1 subpopulations. Sca-1+ cells were 6-7% smaller, and Sca-1− cells were 8-9% smaller, when overexpressing miR-205 compared with the Sca-1+ and Sca-1− luciferase shRNA control cells, respectively (Fig. 2D).
Overexpression of miR-205 also led to increased cell proliferation both in growth-curve (Fig. 2E) and cell-proliferation (Fig. 2F) assays. When plated at the same density and growth conditions, miR-205-overexpressing cells proliferated at a significantly higher rate for 6 days (P<0.0001) than the luciferase shRNA control cells in a growth-curve assay. Similarly, in a cell-proliferation assay, cells overexpressing miR-205 continued to proliferate at a significantly (P<0.005) higher rate once cell density reached confluency, in contrast to the luciferase shRNA control cells, that exhibited contact-mediated growth inhibition. These results suggest that miR-205 regulates the self-renewal and/or proliferation of Sca-1+ progenitor cells.
Overexpression of miR-205 changes the colony-forming potential of Sca-1+ and Sca-1− cells
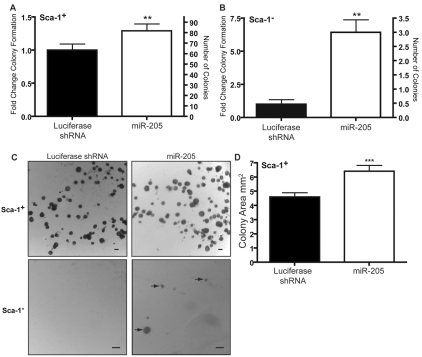
Sca-1+ COMMA-DβGeo cells are capable of forming colonies when plated on Matrigel, whereas Sca-1− cells are unable to do so (Chen et al., 2007; Deugnier et al., 2006; Woodward et al., 2007). As a measure of replicative competence, colony-forming assays were performed on miR-205-overexpressing cells. Sca-1+ cells overexpressing miR-205 formed significantly more colonies than control cells (Fig. 3A). Interestingly, whereas normal Sca-1− cells lack the ability to form colonies on Matrigel, they gained the ability to form colonies on Matrigel when overexpressing miR-205 (Fig. 3B). Representative images of both the Sca-1+ and Sca-1− colony-forming assays are shown in Fig. 3C.
Fig 3.
Overexpression of miR-205 increases the colony-forming capacity of Sca-1− COMMA-βGeo cells. COMMA-βGeo cells transduced with the pSM2 miRNA overexpression vectors were sorted into Sca-1+ and Sca-1− populations. (A) Left y-axis: fold change of Sca-1+ colonies compared with the luciferase shRNA control. Right y-axis: number of colonies formed. When overexpressing miR-205, the clonogenic potential of the Sca-1+ COMMA-βGeo significantly increases. **P<0.0001. (B) Left y-axis: fold change of Sca-1− colonies compared with the luciferase shRNA control. Right y-axis: average number of colonies formed. When Sca-1− cells overexpress miR-205 they gain significant potential to form colonies when normally they could not. **P<0.0001. (C) Representative images of colonies formed on Matrigel when sorted for Sca-1. Scale bars: 100 μm. Arrows highlight colonies formed. (D) Average area of colonies formed by Sca-1+ cells. Colonies were measured using Image-Pro Plus (Media Cybernetics). The average colony area formed from Sca-1+ cells overexpressing miR-205 were statistically larger than those formed from the luciferase shRNA control cells. An average of 130 colonies measured from each group were measured. ***P<0.0005. Data represent the mean of three independent experiments with two biological replicates.
In addition to changes in colony-forming ability, Sca-1+ colonies formed from miR-205-overexpressing cells were larger than colonies formed from the luciferase shRNA control cells (P<0.0005). Furthermore, Sca-1+ colonies formed from miR-205-overexpressing cells were composed of more cells than the luciferase shRNA cells. Fewer cells overall contribute to the Sca-1− miR-205 colonies, accounting for their smaller size (data not shown). These changes in Sca-1+ colony size are probably attributed to the increase in proliferation that was observed in the miR-205-overexpressing cells.
Mouse miR-205 targets the tumor-suppressor gene PTEN
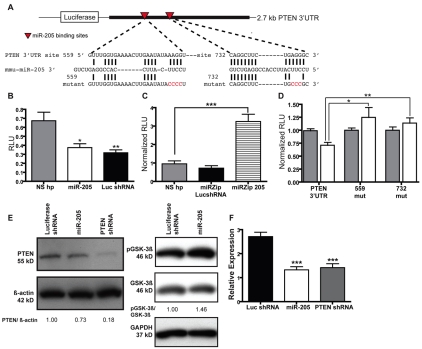
To elucidate potential targets of miR-205, we performed computational analysis using all available target prediction algorithms. Several computational algorithms exist for the prediction of miRNA targets (reviewed by Rajewsky and Socci, 2004), but often these algorithms will predict different targets for the same miRNA. PTEN was identified as a predicted target of miR-205 by the miRanda algorithm at the miRNA Viewer website (http://cbio.mskcc.org/mirnaviewer/). The miRanda algorithm predicts two putative miR-205-binding sites in the coding region of the PTEN mRNA. By other computational analyses, there were also two predicted miR-205-binding sites in the PTEN 3′UTR (Fig. 4A).
Fig 4.
miR-205 targets the mRNA of the tumor-suppressor protein PTEN. (A) Representation of the pGL3-PTEN 3′UTR reporter plasmid. Shown are the computationally predicted binding sites for mmu-miR-205 in the PTEN 3′UTR (red arrowheads). Mutations were made to the seed region of the miR-205-binding sites to disrupt miRNA-target binding (mutated nucleotides shown in red). (B) Luciferase reporter activity of the PTEN 3′UTR in NIH/3T3 cells expressing miR-205, the non-silencing hairpin negative control or the luciferase shRNA positive control. Reporter activity decreases by 30% when cells are overexpressing miR-205. Each sample was normalized to Renilla luciferase activity. *P<0.01 or **P<0.005 relative to the non-silencing hairpin. RLU, relative luciferase units (firefly luciferase/Renilla luciferase). (C) PTEN 3′UTR reporter activity of the in NIH/3T3 cells expressing the miR-205Zipper, the non-silencing hairpin negative control or the luiferaseZipper control. When expressing the miR-205Zipper, reporter activity increases 2.5-fold relative to the non-silencing control. Each sample was normalized to Renilla luciferase activity. ***P<0.0005 relative to the non-silencing hairpin. (D) The computationally predicted miR-205-binding sites in the PTEN 3′UTR were mutated in the seed region. Single seed-region mutations in the 559 and 732 sites significantly relieved silencing of the reporter. *P=0.013 or **P=0.0057. Results are representative of four individual experiments with two biological replicates for each overexpression cell line. (E, left) Western blot analysis of PTEN, phospho-GSK-3β (Ser9) and total GSK-3β levels in COMMA-DβGeo cells transduced with pSM2 miRNA overexpression vectors. When overexpressing either miR-205 or the PTEN shRNA, PTEN expression levels decrease compared to luciferase shRNA control. The blot was stripped and reprobed with β-actin as a loading control. (E, right) Levels of phospho-GSK-3β increase as a result of lowered PTEN levels when cells are overexpressing miR-205, compared with the luciferase shRNA control. The blot was stripped and reprobed with GAPDH as a loading control. Quantitation of relative expression levels of PTEN to β-actin and phospho-GSK-3β to total GSK-3β were determined with ImageJ. (F) TaqMan qPCR of PTEN expression. PTEN expression decreases approximately twofold when miR-205 is overexpressed, as well as when the PTEN shRNA is expressed, relative to the luciferase shRNA control. Data represent the mean expression levels of two biological replicates calculated by ΔΔCT normalized to endogenous 18S rRNA. ***P<0.0001.
To determine whether miR-205 regulates PTEN expression via the 3′UTR, we cloned the PTEN 3′UTR from a wild-type Balb/c mouse into the 3′UTR of a luciferase reporter (Fig. 4A). We used NIH/3T3 cells to overexpress miR-205 using the pSM2 vector, routinely obtaining miR-205 expression that was 20- to 30-fold higher, and in some cases up to 120-fold higher in some clones, as compared with cells transduced with the pSM2-non-silencing hairpin control or the pSM2-luciferase shRNA control (supplementary material Fig. S1E). Additionally, when the pmiRZip-205 anti-miRNA expression vector was transduced into NIH/3T3 cells, we were able to obtain an approximate five- to sixfold decrease in miR-205 expression compared with the pmiRZip-luciferase shRNA control (supplementary material Fig. S1F). We observed a statistically significant decrease (P<0.01) in luciferase reporter activity when cells overexpressed miR-205 compared with cells that expressed the non-silencing hairpin control (Fig. 4B). As expected, decreased reporter activity was observed in the cells expressing the luciferase shRNA (positive control). In the reciprocal experiment, when cells expressed the pmiRZip-205 miRNA inhibitor, we observed a significant increase (P<0.0005) in luciferase reporter activity compared with cells that expressed the non-silencing hairpin control (Fig. 4C).
Relative to the start of the PTEN 3′UTR, one miR-205-binding site was predicted at nucleotides 559-585 by a site prediction algorithm, STarMir (Long et al., 2008), as well as at nucleotides 732-764, which was predicted by MicroInspector (Rusinov et al., 2005). To further validate these miR-205 seed sites in the 3′UTR, we mutated these computationally predicted target sites in the miRNA-binding seed regions (nucleotides in red in Fig. 4A), to prevent miRNA binding. Accordingly, when either miR-205-binding site was mutated, we observed a relief of silencing of the reporter gene, suggesting that these are probably true miR-205 target sites (Fig. 4D).
For confirmation of PTEN expression at the protein level, western blot analysis was performed on protein extracts from COMMA-DβGeo cells transduced with the miR-205 overexpression vectors along with cells expressing an shRNA against PTEN as a positive control. These results showed a slight reduction in PTEN expression when miR-205 was overexpressed as compared with the luciferase shRNA control vector (Fig. 4E). This reduction in PTEN protein level was also correlated with increased phosphorylation of glycogen synthase kinase-3β (GSK-3β) at serine 9, one of the functional downstream PI3K-AKT substrates (Fig. 4E). This phosphorylation of the inhibitory N-terminal serine is mediated in part by the active AKT kinase. PTEN mRNA levels were also determined by qPCR. As in the western blot analysis, when miR-205 was overexpressed, PTEN mRNA levels decreased by approximately 50% as compared with the luciferase shRNA control (Fig. 4F). Collectively, these results indicate that miR-205 regulates PTEN expression.
Mouse miR-205 has many predicted and non-predicted targets
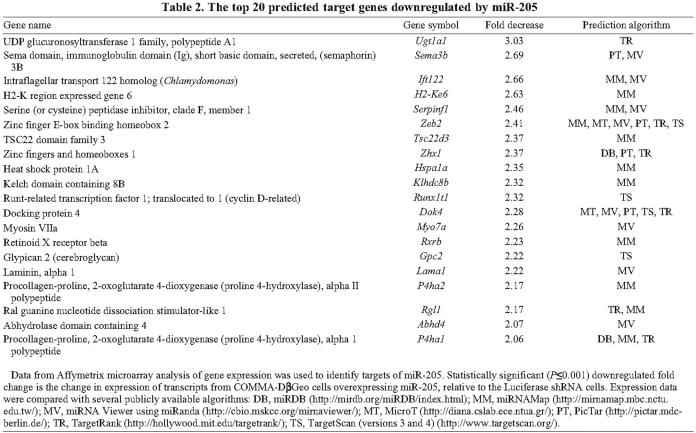
Because knockdown of PTEN did not completely recapitulate the miR-205-overexpression phenotype (data not shown), we sought to identify other targets of miR-205. If all available algorithms are taken into consideration, miR-205 can potentially target over 2000 genes. To determine the efficiency of miR-205 to target other mRNAs genome-wide, we performed a gene-chip analysis of miR-205-overexpressing COMMA-DβGeo cells. Of the ~45,000 probe sets represented on the Affymetrix Mouse Genome 430 2.0 array, 344 unique transcripts were decreased in cells overexpressing miR-205 compared with those expressing the luciferase shRNA control (using the criterion of >1.25-fold downregulation with P<0.01). Of these 344 transcripts, only 45 (13%) were identified by target prediction algorithms, most of which are directed at the 3′UTR. This is not totally unexpected because not all of the 344 mRNAs were anticipated to be direct primary miR-205 targets. Additionally, it was previously noted that target prediction algorithms accounted for only one-third of the actual targets revealed in proteomic analysis of a mouse knockout model (Baek et al., 2008), suggesting that there are probably primary targets not indicated by prediction algorithms as well as secondary effects. Table 2 lists the top 20 predicted targets of miR-205 with the fold change shown relative to the luciferase shRNA control, and target prediction algorithm. Supplementary material Table S1 lists all genes that were downregulated by miR-205 overexpression. Several predicted (Fig. 5A-D) and non-predicted (Fig. 5G-I) targets of miR-205 were confirmed by qPCR.
Table 2.
The top 20 predicted target genes downregulated by miR-205
Fig 5.
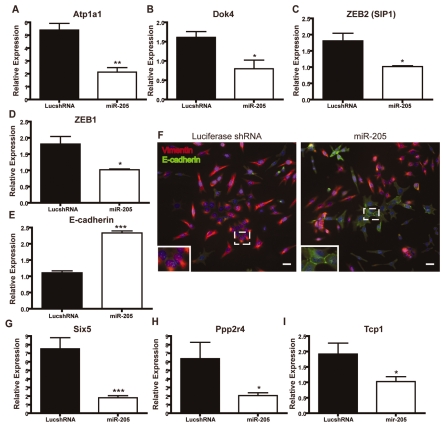
Targets of miR-205. (A,B,G-I) SYBR Green qPCR of putative targets of miR-205; (C-E) TaqMan qPCR of putative targets of miR-205. Data represent the mean expression levels of two biological replicates calculated by ΔΔCT normalized to either endogenous β-actin (A,B,G-I) or 18S rRNA (C-E), of two independent experiments performed in triplicate. (A-C) Atp1a1, Dok4 and ZEB2 are transcripts identified by microarray analysis that decreased when miR-205 was overexpressed. These transcripts were also predicted targets of miR-205 by several algorithms (Table 2). (A) Atp1a (ATPase, Na+-K+ transporting, α1 polypeptide) expression is 1.78-fold lower in miR-205-overexpressing cells (**P=0.005) and is a predicted miR-205 target by miRNAMap and miRNA Viewer. (B) Dok4 expression is 2.28-fold lower in miR-205-overexpressing cells (*P=0.007) and is a predicted miR-205 target by miRNA Viewer, TargetRank and TargetScan. (C) ZEB2 expression is 2.41-fold lower in miR-205-overexpressing cells (*P=0.005) and is a predicted miR-205 target by miRNAMap, TargetRank and TargetScan. (D) ZEB1 expression is 1.7-fold in miR-205-overexpressing cells (*P<0.05) and is a predicted miR-205 target. (E) Expression of E-cadherin increases 2.3-fold as a result of ZEB1 and ZEB2 decreasing following miR-205 overexpression (***P=0.0008). (F) Stably transduced COMMA-βGeo cells were stained for E-caherin (green) and vimentin (red). As a result of lower ZEB1 and ZEB2 expression following miR-205 overexpression, membrane-bound E-cadherin is upregulated. Some membrane-bound E-cadherin can be detected in cells expressing the luciferase shRNA, but only at areas of cell-cell junctions (inset). Scale bars: 25 μm. (G-I) Six5, Ppp2r4 and Tcp1 expression levels decrease when miR-205 is overexpressed; however they are not predicted miR-205 targets by any of the available algorithms. (G) Six5 (Six5 sine oculis-related homeobox 5 homolog) expression is 1.94-fold lower in miR-205-overexpressing cells (***P=0.009). (H) Ppp2r4 (protein phosphatase 2A, regulatory subunit B) expression is 1.46-fold lower in miR-205-overexpressing cells (*P=0.004). (I) Tcp1 (T-complex protein 1) expression is 1.64-fold lower in miR-205-overexpressing cells (*P=0.002).
One gene verified by qPCR was Atp1a1 (Fig. 5A). Atp1a1, a Na+-K+-transporting ATPase, was predicted to be a miR-205 target by two algorithms, miRNAMap and miRNA Viewer. By array analysis, Atp1a1 expression was downregulated 1.8-fold in cells overexpressing miR-205, but this seems to be an underestimate because, by qPCR, an approximate fourfold change was detected, as compared with luciferase shRNA cells. As a Na+-K+ pump, Atp1a1 is responsible for detecting and maintaining ion gradients across the plasma membrane. Interestingly, colorectal tumors have been shown to have decreased levels of Atp1a1 (Cao et al., 1997) and also frequently overexpress miR-205 (Jiang et al., 2005).
We also detected a 1.5-fold decrease of docking protein 4 (Dok4) expression by qPCR (Fig. 5B), although array analysis indicated that Dok4 was downregulated to a slightly greater extent (2.2-fold) by miR-205. Dok4 is one of the most frequently predicted targets of miR-205, predicted by five different target prediction algorithms: MicroT, miRNA Viewer, PicTar, TargetScan and TargetRank. Dok4, and other Dok-family members, are downstream of several tyrosine kinases and are substrates of insulin and insulin-like growth factor (IGF)-1 receptors (Cai et al., 2003). Insulin and IGFs are required for the survival of normal MECs and for growth-factor-induced cellular differentiation. Activation of IGFs in breast cancer cell lines induces cellular proliferation and inhibits apoptosis, and it has been shown that insulin and IGFs signal through PI3K to regulate PTEN expression (Moorehead et al., 2003).
Not surprisingly, one of the previously confirmed targets of miR-205 to date, ZEB2, also known as SIP1 (Gregory et al., 2008), was identified in our array analysis as being downregulated 2.4-fold, and was a predicted miR-205 target by five algorithms: miRNAMap, miRNA Viewer, MicroT, PicTar, TargetRank and TargetScan. Gregory et al. found that miR-205 was downregulated in cells that have undergone an epithelial-to-mesenchymal transition (EMT), a key process in embryonic development as well as in tumor metastasis in response to growth factors. These authors also reported that miR-205 regulates EMT by silencing ZEB2, a repressor of E-cadherin and regulator of EMT. Accordingly, ZEB2 expression was subsequently verified by qPCR in our miR-205-overexpressing cells as being approximately twofold higher in the luciferase shRNA controls as compared with the miR-205-overexpressing cells (Fig. 5C). Gregory et al. also identified ZEB1 as a miR-205 target (Gregory et al., 2008). We therefore determined the expression levels of ZEB1 by qPCR on the miR-205 overexpression cells. Although the change in expression of ZEB1 was not determined to be significant by array analysis in our assay, expression did decrease by approximately 50% when miR-205 was overexpressed (Fig. 5D). Endogenous expression of ZEB1 was quite low, however, in our COMMA-DβGeo cells. As expected, expression of E-cadherin increased nearly 2.5-fold following miR-205 overexpression and the subsequent decrease of both ZEB1 and ZEB2 (Fig. 5E). We also detected by immunofluorescence a dramatic increase in cells expressing E-cadherin as a result of miR-205 overexpression (Fig. 5F). When initially plated in cell culture conditions, these epithelial cells typically express vimentin until the culture reaches confluency. E-cadherin expression was observed at cell-cell junctions in the luciferase shRNA cells, but at markedly lower levels than those detected in miR-205-overexpressing cells. Morphologically, more cells overexpressing miR-205 seem to have the round epithelial cell shape than the spindle-shaped luciferase shRNA cells.
We also determined, by array analysis, the expression levels of several non-predicted genes that were downregulated by miR-205 overexpression (Fig. 5G-I). Expressions of Six5, Ppp2r4 and Tcp1 were all downregulated by miR-205 in our array analysis. We detected a sevenfold decrease by qPCR of Six5, a sine oculis-related homeobox homolog, in miR-205-overexpressing cells (Fig. 5G).
A fivefold decrease in the expression of Ppp2r4, a serine/threonine phosphatase, was detected in miR-205-overexpressing cells by qPCR (Fig. 5H). Ppp2r4 has been shown to have competing effects on the MAPK and AKT pathways. Ppp2r4 dephosphorylates AKT, which should inhibit the AKT pathway (Ivaska et al., 2002). It also dephosphorylates Raf, which should result in activation of the MAPK pathway (Dhillon et al., 2002). Thus, it is conceivable that downregulation of Ppp2r4, in addition to PTEN, might in part account for the increased growth and cell survival of miR-205-overexpressing cells.
In miR-205-overexpressing cells, we observed a twofold decrease by qPCR of Tcp1 expression (Fig. 5I). Tcp1, a T-complex protein, has been shown to be upregulated in mammary-gland cells as a result of lactogenic hormone-stimulated differentiation (Desrivieres et al., 2003), so downregulation of Tcp1 might also account for the less differentiated phenotype that we observe in miR-205-overexpressing cells.
To understand the mechanisms involved in miR-205 overexpression, we performed gene-set enrichment assays (GSEA) on microarray expression profiles from the GSEA molecular signature database website (http://www.broad.mit.edu/gsea/msigdb/msigdb_index.html). Consistent with the hypothesis that forced miR-205 expression might be causing cells to assume more progenitor-cell-like properties (i.e. expansion in the progenitor-cell compartment, decreased cell size and increased cell proliferation), many downregulated genes overlapped with GSEA gene sets associated with hematopoietic stem cells (RUTELLA_HEMATOGFSNDCS_DIFF), stromal stem cells (BOQUEST_CD31PLUS_VS_CD31MINUS_DN) and genes downregulated in differentiation pathways (BRUNO_IL3_DN) (GSEA gene sets are indicated in supplementary material Table S1). Additionally, Ingenuity Pathway Analysis (IPA) (Ingenuity Systems, Redwood, CA) was used to identify biological pathways, networks and functions significantly altered by miR-205 overexpression. The genes identified by array analysis are classified by the IPA software into a variety of biological processes, including cellular growth and proliferation, cell death, cellular development, cancer, cell morphology, and DNA replication, recombination and repair, implying a diverse repertoire of miR-205 targets. The six most significant molecular and cellular processes affected by miR-205 identified by IPA are shown in supplementary material Fig. S2.
miR-205 is highly expressed in mouse mammary-gland stem-cell populations
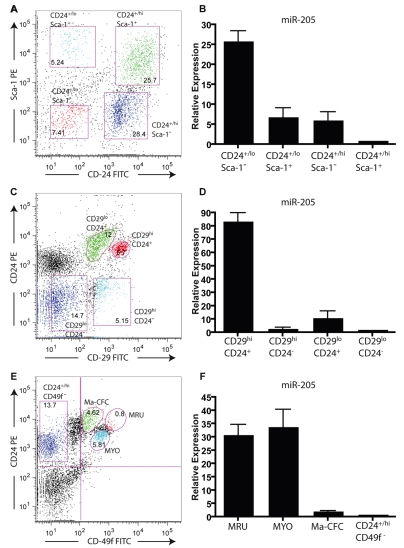
Although the Sca-1+ population of the COMMA-DβGeo cell line represents a subpopulation with basal cell characteristics retaining stem- and progenitor-cell capabilities, this is quite different from the Sca-1+ cells isolated from primary MECs, which are thought to represent primarily a lumenal estrogen-receptor (ER)+ population. For example, it was previously demonstrated that the virgin mouse mammary gland has a basal cell population represented by a Lin−CD24+/lo/Sca-1− profile that is enriched for mammary stem cells (Sleeman et al., 2006), in addition to populations that are lumenal ER− cells that are Lin−CD24+/hi/Sca-1− and lumenal ER+ cells that are Lin−CD24+/hi/Sca-1+ (Sleeman et al., 2007) as well as a Lin−CD24−/Sca-1+ non-epithelial population (Shackleton et al., 2006; Sleeman et al., 2006; Stingl et al., 2006). Surprisingly, FACS analysis demonstrated that the COMMA-DβGeo cell line is essentially homogeneous for CD29+ and CD24+ expression (supplementary material Fig. S3A). Thus, we sought to determine how the results obtained for miR-205 expression in the COMMA-DβGeo model compared with its expression in primary mammary stem or progenitor cells. Accordingly, we used adult virgin Balb/c female mammary glands to isolate MECs on the basis of the cell-surface markers Sca-1, CD29 (β-1 integrin), CD24 (heat-stable antigen) and CD49f (α6 integrin), which identifies populations that are enriched in mammary-gland stem cells (Fig. 6A-E). When sorted for these populations and assayed for miRNA expression, we observed high expression of miR-205 in the populations that were enriched for mammary-gland stem cells as well as myoepithelial cell populations. The CD24/Sca-1 mammary-gland stem-cell-enriched population (Lin−CD24+/lo/Sca-1−) (Fig. 6A) had approximately 25-fold higher expression of miR-205 than the lumenal ER− Lin−CD24+/hi/Sca-1− and lumenal ER+ Lin−CD24+/hi/Sca-1+ populations, whereas the myoepithelial and basal Lin−CD24+/lo/Sca-1+ population retained moderate expression of miR-205 (Fig. 6B). Furthermore, profiling and characterization of CD29/CD24 populations identified the Lin−CD29hi/CD24+ population as being enriched for slowly cycling mammary-gland stem cells such that a single Lin−CD29hi/CD24+ cell was capable of regenerating a mammary gland (Shackleton et al., 2006). This Lin−CD29hi/CD24+ population, although enriched for mammary-regenerating stem cells, also contains myoepithelial cells and is mostly Sca-1− (Shackleton et al., 2006). As in the Lin−CD24+/lo/Sca-1− stem-cell-enriched population, when mammary epithelial cells were sorted for CD29/CD24 (Fig. 6C), we also observed high expression, approximately 80-fold compared with other cell types, of miR-205 in the mammary stem-cell-enriched Lin−CD29hi/CD24+ population (Fig. 6D). The methods of enriching for stem and progenitor cells by isolating Lin−CD24+/lo/Sca-1− and Lin−CD29hi/CD24+ cells results in populations containing a mix of myoepithelial, basal and stem cells. To distinguish between myoepithelial cells and mammary epithelial stem cells, we also isolated cells on the basis of CD24/CD49f populations. Independent enrichment for mammary-gland stem cells using CD24/CD49f allows the isolation of a small population, referred to as the mammary repopulating units (MRUs), characterized as Lin−CD24med/CD49fhi, which is distinguished from the myoepithelial (MYO) Lin−CD24lo/CD49flo population. MRUs are enriched for stem cells capable of repopulating the mammary gland (Stingl et al., 2006). Again, when sorted for CD24/CD49f (Fig. 6E) and assayed for miR-205 expression, we observed a 30-fold increase in expression in the MRU population compared with non-epithelial cells, and additionally we observed high expression of miR-205 in the MYO population (Fig. 6F). Recent studies using in situ hybridization on normal and mammary tumor tissue samples found that miR-205 expression was restricted to the myoepithelial cell layer, which contains both myoepithelial cells and basal stem cells (Sempere et al., 2007), so the high expression of miR-205 that we observed in the MYO and Lin−CD24+/lo/Sca-1+ is consistent with these observations.
Fig 6.
miR-205 is highly expressed in mouse mammary-gland stem-cell populations. Primary mammary epithelial cells were isolated from wild-type Balb/c virgin females, age 8-9 weeks, and separated by FACS for cell-surface markers and analyzed by TaqMan qPCR for miR-205 expression. Data represent the mean expression levels of two different biological replicates calculated by ΔΔCT normalized to the endogenous snoRNA55 control, of two independent experiments performed in triplicate. (A) Representative FACS profile for CD24-FITC and Sca-1—PE. (B) Cells from different CD24/Sca-1 subpopulations were collected and assayed for expression of miR-205. In the basal and myoepithelial Lin−/CD24+/lo/Sca-1− stem-cell-enriched population, miR-205 expression is approximately 25-fold higher than in the lumenal Lin−/CD24+/hi/Sca-1+ and Lin−/CD24+/hi/Sca-1− populations. (C) Representative FACS profile for CD29-FITC and CD24-PE. (D) Cells from different CD29/CD24 subpopulations were collected and assayed for expression of miR-205. In the Lin−/CD29hi/CD24+ self-renewing stem-cell-enriched population, miR-205 expression is approximately 85-fold higher than in the non-stem-cell populations, including the lumenal progenitor Lin−/CD29lo/CD24+ population. (E) Representative FACS profile for CD24-PE and CD49f-FITC. (F) Cells from different CD24/CD49f subpopulations were collected and assayed for expression of miR-205. In the stem-cell-enriched MRU population (Lin−/CD24med/CD49fhi), miR-205 expression is approximately 45-fold higher, and the myoepithelial MYO population (Lin−/CD24lo/CD49flo) is approximately 32-fold higher, than in the non-stem-cell Lin−/CD24+/hi/CD49f− population.
Because it has been reported that miR-205 and the members of the miR-200 family might have overlapping functions in the regulation of EMT via ZEB1 and ZEB2 (Gregory et al., 2008), we assayed miRNA expression for members of the miR-200 family in these stem-cell-enriched populations. We found that members of the miR-200 family were expressed in the K18+ lumenal stem-cell-enriched Lin−CD29lo/CD24+ population (supplementary material Fig. S3B) but we did not observe significantly decreased expression in the Lin−CD29hi/CD24+ mammary stem-cell-enriched population. We also observed expression of miR-200-family members in the lumenal Lin−CD24+/hi/Sca-1− and the Lin−CD24+/hi/Sca-1+ populations, but not in the stem-cell-enriched Lin−CD24+/lo/Sca-1− (supplementary material Fig. S3C). This is consistent with recent findings that members of the miR-200 family are expressed at significantly lower levels in the MRU population relative to the colony-forming Ma-CFC population (Shimono et al., 2009). Additionally, as we previously observed by microarray analysis, miR-200b is expressed at lower levels in the Sca-1+ population compared with the Sca-1− population of the COMMA-DβGeo cells by miRNA analysis (Table 1) (supplementary material Fig. S3D). Importantly, these results suggest that miR-205 and members of the miR-200 family have unique functions and are not functionally redundant. Furthermore, miR-205 seems to display elevated expression in mammary MRUs in comparison to other subpopulations, which contrasts what has been reported for miR-200-family members. Thus, the identification of unique miR-205 targets in the current studies might help provide some insights into the selective effects of these miRNAs in stem and progenitor cells.
Discussion
When aberrantly expressed, either by deletions associated with frequent fragile sites or genomic amplification, miRNAs play an important role in carcinogenesis (Bartel, 2004; Calin et al., 2004; He et al., 2005; Kluiver et al., 2006) and accordingly are thought to represent crucial cancer biomarkers. In previous studies comparing miRNA expression with ER/progesterone-receptor (PR)/ErbB2 status in human breast cancers, miR-205 upregulation was shown to correlate with ER+/PR+ breast tumors (as compared to ER−/PR− tumors) but it did not show a significant correlation with ErbB2+ status (Mattie et al., 2006). In addition to breast cancers, miR-205 has been shown to be overexpressed in head and neck cancer cell lines (Jiang et al., 2005), and in cervical (Wang et al., 2008), bladder (Gottardo et al., 2007) and non-small cell lung (Markou et al., 2008) cancers, as compared with normal tissues. Additionally, miR-205 has been shown to regulate the process of EMT by means of silencing ZEB2 and ZEB1, a repressor of E-cadherin and regulator of EMT, as discussed previously. Interestingly, an association between low miR-205 expression in the myoepithelial and basal cells of ER−/PR−/Her2− basal breast cancer patient samples has been correlated with tumor progression (Sempere et al., 2007), possibly owing to its regulation of EMT via ZEB1 and ZEB2. High levels of miR-205 have also been observed in normal lobular and ductal mammary myoepithelial cells, implicating a potential role in normal development (Sempere et al., 2007).
In the COMMA-DβGeo cells overexpressing miR-205, we observed a significant 10-15% expansion of the Sca-1+ cell population. This is consistent with several previous observations that loss of PTEN leads to an expansion of stem cells. Loss of PTEN has been associated with stem-cell expansion of Flk-2−Sca-1+Lin−c-Kit+CD48− hematopoietic stem cells (Yilmaz et al., 2006), as well as expansion of the Sca-1+/BCL-2+ basal stem cells and progenitors in the prostate (Wang et al., 2006). Our results also indicated that miR-205 overexpression led to increased proliferation. This recapitulates other findings that loss of PTEN results in increased cell proliferation by regulating cell-cycle entry (Groszer et al., 2006). However, our observed decrease in cell size following overexpression of miR-205 has not been shown in other PTEN loss-of-function studies, although this effect is probably cell-type dependent as loss-of-function studies demonstrate a variety of phenotypes (reviewed in Knobbe et al., 2008). Thus, although PTEN might play a role in some of the phenotypes caused by miR-205 overexpression, there must be other targets involved. Cell size is controlled by the cell cycle and the process of S-phase initiation, which depends on the concentration of extracellular growth factors. It is of note that IPA determined that both cellular-growth and cell-morphology pathways are significantly affected by miR-205 overexpression. The genes involved in these pathways might give insight into the observed 4-9% decrease in cell size and increased proliferation following miR-205 overexpression. Additionally, it has been observed that cell size increased when normal mouse mammary epithelial cells were induced by TGFβ to undergo EMT, compared with untreated cells (Lamouille and Derynck, 2007). Thus, keeping cells in a more epithelial-like state by overexpressing miR-205 might be the cause of the decreased cell size we observed. Additionally, miR-205 overexpression by nearly fivefold in Sca-1− cells resulted in a gain in the ability of these cells to form colonies on Matrigel. Although this increase in expression is significant compared with the luciferase shRNA Sca-1− cells, this level of expression was not as high as that found in Sca-1+ cells. Although the number of colonies formed from the miR-205-overexpressing Sca-1− cells was significantly smaller than the number of colonies observed in Sca-1+ cells, this finding might indicate that, although expression of miR-205 might contribute to a gain of growth potential, expression of miR-205 alone does not seem to be sufficient to confer self-renewal to a non-progenitor cell population.
Two recent reports have identified new targets of miR-205. Protein kinase Cϵ was identified as a miR-205 target in prostate cells (Gandellini et al., 2009) and ErbB3 (also known as HER3) was shown to be a miR-205 target in breast cancer cells (Iorio et al., 2009). These studies indicated that, in both prostate cancer and breast cancer cells, miR-205 overexpression led to tumor-suppressor activity by reduction of cell migration and regulation of EMT, and inhibition of cell growth and increased sensitivity to anti-cancer agents. These studies demonstrated that overexpression of miR-205 also led to the decrease in phospho-ERK and phospho-AKT levels, both of which are also downstream of the PI3K pathway. We also observed a decrease in levels of phospho-AKT and phospho-ERK in our miR-205-overexpressing cells compared with the control, despite downregulation of PTEN (supplementary material Fig. S4). The different effect of miR-205 on cell growth might be due to the fact that these studies were performed in transformed cancer cell lines, whereas ours used normal epithelial cells. Thus, the high levels of expression of miR-205 found in both normal mammary myoepithelial and stem cells, as well as in cancer cells, might indicate its involvement in early cancer lesions.
To further investigate other targets of miR-205, we analyzed the potential for miR-205 to regulate other mRNAs genome-wide. These studies demonstrated that two recently identified targets of miR-205, ZEB1 and ZEB2, are also regulated in the COMMA-DβGeo epithelial cells. In addition to PTEN, ZEB1 and ZEB2, we have identified 45 putative targets of miR-205 that were predicted by several target prediction algorithms. Of the confirmed predicted targets, we demonstrated decreased mRNA levels of Atpa1a1 and Dok4 in association with overexpression of miR-205, in addition to several non-predicted targets. Although there were many potential targets identified by our array analysis, we found overlaps with gene sets associated with stem-cell differentiation pathways. Owing to the potential of miR-205 to regulate so many different mRNAs, the phenotypes associated with miR-205 overexpression are probably due to pleitropic effects on multiple gene targets and probably will depend on the cell context. This is also probable, because gene expression arrays usually underestimate the total number of miRNA targets, many of which might only be detected at the protein level (Baek et al., 2008).
Several groups have recently described enrichment strategies for mammary-gland stem and progenitor cells. Using cell-surface markers, Stingl et al. identified the MRU (CD24med/CD49f hi) epithelial-cell population as enriched for stem and progenitor cells with in vivo regenerative potential, whereas the MYO (CD24lo/CD49f lo) population was enriched for K14+ and α-SMA-expressing cells (Stingl et al., 2006). Interestingly, although the MRU population has low expression of miR-200-family members (Shimono et al., 2009), this population has high miR-205 expression, suggesting that other targets besides ZEB1 and ZEB2 are probably playing crucial roles in their unique functions. Shackleton et al. identified a cell population characterized by the Lin−CD29hi/CD24+ profile that was enriched for K14+ cells and had similar stem- and progenitor-cell capabilities (Shackleton et al., 2006). Furthermore, Sleeman et al. used the CD24lo/CD49f hi/Sca-1− population to enrich for basal stem cells (Sleeman et al., 2006). FACS isolation for Sca-1+/hi cells from these primary MECs did not retain stem and progenitor activity, and there was no enrichment for Sca-1 in the CD24med/CD49f hi or CD29hi/CD24+ stem-cell-enriched populations (Shackleton et al., 2006; Stingl et al., 2006), in contrast to the COMMA-DβGeo cell line and primary MECs grown in culture (Chen et al., 2007; Deugnier et al., 2006; Welm et al., 2002; Woodward et al., 2007). In fact, when freshly isolated MECs were grown in culture, Sca-1 expression was significantly upregulated, but there was no significant change in CD24 or CD49f expression (Matulka et al., 2007). The reasons for this discrepancy between a cell-line model and primary MECs are not apparent, but might reflect changes in Sca-1 expression reported to occur as a function of cell culture that has been observed by several groups (Matulka et al., 2007; Shackleton et al., 2006; Stingl et al., 2006). The precise genetic events that led to the immortalization of the COMMA-DβGeo cells are not known, but use of this model did facilitate the identification of miR-205 in the Sca-1+ population, which correlated with the high expression of miR-205 in MRUs and other populations enriched for mammary-gland stem and progenitor cells. Although miR-205 is frequently overexpressed in cancer compared with normal tissues, it has also been observed to be highly expressed in normal tissue (Volinia et al., 2006). This miRNA might not only be involved in normal cell function (proliferation, differentiation and growth), but its mis-regulation might contribute to cancer initiation, progression and metastasis. These current studies also suggest that miR-205 overexpression might affect progenitor-cell expansion, which might be important in the etiology of cancer. Generation of loss-of-function mouse models for miR-205 will be required to establish whether miR-205 is essential for normal development. Overexpression and loss-of-function mouse models would also allow for studies to determine the effect of miR-205 specifically on mammary stem cells. Additionally, generating miR-205-overexpression and loss-of-function mouse cancer models might demonstrate the mechanisms by which miR-205 mis-regulation contributes to cancer initiation and progression.
Materials and Methods
Cell lines
The COMMA-DβGeo cell line passages 21-27 was kindly provided by Daniel Medina at Baylor College of Medicine, Houston, TX. The cells were grown in DMEM/F12 (Invitrogen) at pH 7.6, with 2% adult bovine serum (ABS) (Gemini Bioproducts), 5 μg/ml gentamycin (Sigma), 10 μg/ml insulin (Invitrogen) and 5 ng/ml EGF (Invitrogen) in a 5% CO2 humidified chamber. NIH/3T3 cells were purchased from ATCC and cultured in DMEM (Invitrogen) with 10% bovine calf serum (BCS) (SAFC Biosciences) in a 5% CO2 humidified chamber. 293T cells were purchased from ATCC and maintained in DMEM with 10% fetal bovine serum (FBS) (SAFC Biosciences) in a 5% CO2 humidified chamber.
Custom miRNA microarrays
Total RNA (10 μg) purified from Sca-1+ and Sca-1− cells isolated by FACS was hybridized to custom miRNA microarrays as previously described (Thomson et al., 2004). Array analysis was performed with dChip (Li and Wong, 2001) and WebArray (Xia et al., 2005).
μParaflo miRNA microarray
This microarray assay was performed using a service provider (LC Sciences) through a custom array, as previously described (Gu et al., 2008). Custom microarrays were spotted with all of the mature miRNA sequences in miRBase at the time of assembly, approximately 200 probes, whereas μParaflo microarrays contained 328 human and approximately 300 mouse miRNAs, plus 2000 computationally predicted miRNA sequences from human and mouse.
Construction of pSHAG-MAGIC2c miRNA overexpression vectors
The endogenous mouse stem-loop pre-miR-205 sequence was obtained from miRBase. Oligonucleotides containing the full pre-miRNA sequence were synthesized, PCR amplified and TA cloned into pCR-2.1 TOPO (Invitrogen). Stem-loop sequences were subsequently verified by sequencing and subcloned into the XhoI-EcoRI site of the pSHAG-MAGIC2c (pSM2) vector (Open Biosystems) (Paddison et al., 2004). As non-miRNA controls, plasmids expressing an shRNA targeting the firefly luciferase cDNA (pSM2-luciferase shRNA RHS1705, Open Biosystems), as well as a non-silencing hairpin control (pSM2-NShp RHS1707, Open Biosystems), were used, in addition to a PTEN shRNA (RHS1764-9209859, Open Biosystems).
Retroviral transduction
COMMA-DβGeo cells
Passage 8 293T-packaging cells were transiently transfected with pSM2-miRNA vectors and pCL-Eco (Imgenex) using FuGENE 6 (Roche) according to the manufacturer's guidelines. Forty-eight hours after transfection (day 3), virus-containing medium was collected from transfected 293T cells, filtered through a 0.45-μm syringe filter, and applied to passage 20 COMMA-DβGeo with the addition of 10 mM HEPES (Invitrogen) and 5 mg/ml polybrene (Millipore). The cells were spun at 300 g in a swinging platform rotor for 30 minutes. After spinning, the retroviral supernatant was removed from COMMA-DβGeo cells and replaced with fresh medium. Forty-eight hours later, cells were trypsinized and split at a low density with the addition of 2 μg/ml puromycin (Sigma) to select for transduced cells.
NIH/3T3 cells
Transduction was performed on passage 9 NIH/3T3 cells as for the COMMA-DβGeo cells with the exception of the spin. Instead, cells were left with the virus-containing medium overnight and replaced with fresh medium 12 hours later.
Colony-forming assay
Retrovirally transduced COMMA-DβGeo were cells prepared for flow cytometry using an anti-Ly6A/E (Sca-1) antibody conjugated to FITC (BD Pharmingen). Cells were incubated with Sca-1 antibody for 15 minutes on ice, resuspended in HBSS+ [HBSS (Invitrogen), 2% FBS and 100 mM HEPES], and filtered through a 40-μm cell filter into polypropylene tubes containing 0.5 μg/ml propidium iodide (Sigma) to exclude dead cells. Analysis and sorting were performed on a triple laser MoFlo (Dako-Cytomation). Data analysis was performed on FlowJo version 8 (Tree Star). 500 cells per well were sorted as either Sca-1+ or Sca-1− into round-bottom 96-well plates containing 30 μl Matrigel Basement Membrane Matrix (BD Biosciences). After sorting, 100 μl of COMMA-DβGeo medium was added to each well and plates were incubated for 8 days. Plates were then fixed in glacial acetic acid and methanol (1:2; 10 minutes) and stained with crystal violet. Colonies were counted with ImageJ software.
Cell-proliferation assays
Cells were plated at 50,000 cells/plate in 60-mm plates and replenished with fresh media every 48 hours. Cells were trypsinized and counted every 24 hours for 6 days using a Vi-CELL XR Cell Viability Analyzer (Beckman Coulter) for a growth curve. For the proliferation assay, cells were trypsinized every 48 hours for 6 days and proliferation was determined using the Click-iT EdU Flow Cytometry Assay Kit (Invitrogen) with PI incorporation, and analyzed on an LSRII (BD Biosciences).
miRNA real-time RT-PCR
Total RNA was isolated using miRNeasy (Qiagen) and cDNA was synthesized from 10 ng of total RNA using the TaqMan MicroRNA Reverse Transcription Kit with miRNA-specific RT primer from the TaqMan MicroRNA Assay Mix. miRNA levels were measured using the miRNA-specific TaqMan probe provided in the MicroRNA Assays and the TaqMan Gene Expression Maser Mix (Applied Biosystems). Experiments were performed in triplicate. miRNA levels were normalized to snoRNA55 (Applied Biosystems) and fold change was determined by the comparative threshold method (ΔΔCT).
mRNA real-time RT-PCR
Total RNA was isolated from cells using miRNeasy (Qiagen). cDNA was made from 1 μg of total RNA with SuperScript II (Invitrogen) using random primers. cDNA was then treated with RNase H (Invitrogen) to remove RNA. For SYBR Green qPCR, amplification of cDNA was performed with SYBR Green PCR Maser Mix (Applied Biosystems), and normalized to β-actin. For TaqMan qPCR, amplification of cDNA was performed with TaqMan Gene Expression Maser Mix. Gene Expression Assays for PTEN and 18S rRNA were used (Applied Biosystems), in addition to custom PrimeTime TaqMan probe/primer sets for ZEB1, ZEB2 and E-cadherin (Integrated DNA Technologies). All qPCR was performed using the StepOnePlus Real-Time PCR System and software (Applied Biosystems). mRNA levels are shown as relative to 18S. Primer sets for qPCR are detailed in supplementary material Table S2.
Lentiviral miRNA inhibition
A lentiviral-based anti-miRNA expression vector, pmiRZip-205, was obtained from System Biosciences and used to stably transduce NIH/3T3 cells as per manufacturer recommendations. As a control, the luciferase shRNA was cloned from the pSM2-LucshRNA vector into the BamHI-EcoRI site of the pmiRZip vector (pmiRZip Luc shRNA).
Luciferase assay
The pGL3-PTEN 3′UTR plasmid was constructed with the 2.7 kb 3′UTR of PTEN amplified from genomic Balb/c tail DNA. PCR products were TA cloned into pCR-2.1 TOPO (Invitrogen), sequence verified and subcloned into the XbaI site of the pGL3-promoter vector (Promega). Stably transduced NIH/3T3 were co-transfected with pRL-null Renilla luciferase control (Promega) and pGL3-PTEN 3′UTR luciferase reporter plasmids (200 ng each) with FuGENE 6 transfection reagent (Roche) as per the manufacturer's protocol. Luciferase activity was measured 48 hours post-transfection with the Dual Luciferase Reporter Assay System (Promega) and reported as relative luciferase units (firefly luciferase/Renilla luciferase). miRNA-target-site mutants were generated with the Stratagene QuikChange II XL Site-Directed Mutagenesis Kit and sequence verified.
Western blotting
Cells were lysed in RIPA buffer with protease and phosphatase inhibitors (Roche). The protein concentration was assayed by BCA (Bio-Rad) and 25 μg of protein was separated on a 12% SDS-PAGE gel, transferred onto PVDF membrane and blotted with an anti-PTEN antibody (Santa Cruz Biotechnology), anti-phospho-GSK-3β (Ser9) and total GSK-3β (Cell Signaling) antibodies. The blot was stripped and reprobed with an anti-β-actin antibody (Sigma) or an anti-GAPDH antibody (AbD Serotech). Autoradiographic images were quantitated using ImageJ (Abramoff et al., 2004).
Immunofluorescence
Stably transduced cell lines were grown on poly-D-lysine-coated coverslips, fixed with 4% PFA for 15 minutes, followed by methanol permeabilization for 20 minutes at −20°C, then blocked using the M.O.M. Basic Kit (Vector Laboratories). Cells were stained with anti-E-cadherin (1:50) (BD Biosciences) and anti-vimentin (1:250) (Fitzgerald Industries) primary antibodies, and anti-mouse IgG—Alexa-Fluor-488 (1:100) and anti-guinea-pig IgG—Alexa-Fluor-594 (1:100) (Invitrogen) secondary antibodies. Coverslips were mounted with VECTASHIELD Mounting Medium with DAPI (Vector Laboratories).
Primary mammary epithelial cell preparations
Mouse mammary-gland cells were prepared as previously described (Zhang et al., 2008b) from 8- to 9-week-old Balb/c mice. Cells were labeled with antibodies for cell-surface markers: Sca-1-PE (1:300), CD29-FITC (1:200), CD24-PE (1:100), CD-24-FITC (1:200), CD49f-FITC (1:500) and separated by FACS using a BD FACSAria II cell sorter. Dead cells were excluded by Sytox Red (Invitrogen) and non-epithelial lineage-positive cells were excluded using a biotin-conjugated mouse lineage panel kit containing antibodies against CD3e, CD11b, CD45R/B220, Ly-6G/C and TER-119 (BD Bioscience), in addition to biotin-conjugated CD31 and CD140a antibodies.
miRNA SYBR Green qPCR
miRNA SYBR Green qPCR was performed as described previously (Shi and Chiang, 2005). Total RNA was isolated from cells with RNA-Bee (Tel-Test) and treated with DNaseI (Invitrogen). Total RNA (1 μg) was polyadenylated with Escherichia coli poly(A) polymerase (Ambion). Unincorporated nucleotides were removed by column purification using ChromaSpin-30 (BD Biosciences) columns. cDNA was transcribed with SuperScript II (Invitrogen) using a miRNA RT gene-specific primer. cDNA was then treated with RNase H (Invitrogen) to remove RNA. Amplification was performed with FastStart SYBR Green Master Mix (Rox) (Roche) using the ABI 7500 (Applied Biosystems). Primer sets for miRNA SYBR Green qPCR were obtained from Jiang et al. (Jiang et al., 2005).
Statistical analysis
Statistical significance was analyzed by unpaired Student's t-test, and P≤0.05 was considered to be statistically significant. All data are represented as the mean ± s.e.m.
Supplementary Material
Acknowledgments
The authors thank Amy Shore and Heather LaMarca for critical reading and discussion of this manuscript. We greatly appreciate Daniel Medina for providing the COMMA-DβGeo cell line. Microarray statistical analysis was performed with the assistance of Susan Hilsenbeck (The Breast Center, BCM), Chad Creighton and Dror Berel (The Cancer Center, BCM). Primary mammary epithelial preparations were done with the assistance of Jason Herschkowitz and Rachel Atkinson. We thank the Baylor College of Medicine Cytometry and Cell Sorting Facility and Chris Threeton with the Texas Children's Hospital Flow Laboratory for flow cytometry assistance. Affymetrix microarrays were performed by the BCM Microarray Core Facility. These studies were supported through the NIH R37-CA16303-33 (J.M.R.) Merit Award, and a Department of Defense Breast Cancer Program Predoctoral Fellowship DAMD W81XWH-06-1-0716 (S.B.G.). Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/4/606/DC1
References
- Abramoff M. D., Magelhaes P. J., Ram S. J. (2004). Image Processing with ImageJ. Biophotonics International 11, 36-42 [Google Scholar]
- Baek D., Villen J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008). The impact of microRNAs on protein output. Nature 455, 64-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281-297 [DOI] [PubMed] [Google Scholar]
- Benetti R., Gonzalo S., Jaco I., Munoz P., Gonzalez S., Schoeftner S., Murchison E., Andl T., Chen T., Klatt P., et al. (2008). A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 15, 268-279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blenkiron C., Goldstein L. D., Thorne N. P., Spiteri I., Chin S. F., Dunning M. J., Barbosa-Morais N. L., Teschendorff A. E., Green A. R., Ellis I. O., et al. (2007). MicroRNA expression profiling of human breast cancer identifies new markers of tumour subtype. Genome Biol. 8, R214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Dhe-Paganon S., Melendez P. A., Lee J., Shoelson S. E. (2003). Two new substrates in insulin signaling, IRS5/DOK4 and IRS6/DOK5. J. Biol. Chem. 278, 25323-25330 [DOI] [PubMed] [Google Scholar]
- Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., et al. (2004). Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 101, 2999-3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. M., Taha M. M., Medina D., Rosen J. M. (1988). A clonal derivative of mammary epithelial cell line COMMA-D retains stem cell characteristics of unique morphological and functional heterogeneity. Exp. Cell Res. 177, 109-121 [DOI] [PubMed] [Google Scholar]
- Cao J., Cai X., Zheng L., Geng L., Shi Z., Pao C. C., Zheng S. (1997). Characterization of colorectal-cancer-related cDNA clones obtained by subtractive hybridization screening. J. Cancer Res. Clin. Oncol. 123, 447-451 [DOI] [PubMed] [Google Scholar]
- Chen M. S., Woodward W. A., Behbod F., Peddibhotla S., Alfaro M. P., Buchholz T. A., Rosen J. M. (2007). Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J. Cell Sci. 120, 468-477 [DOI] [PubMed] [Google Scholar]
- Danielson K. G., Oborn C. J., Durban E. M., Butel J. S., Medina D. (1984). Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc. Natl. Acad. Sci. USA 81, 3756-3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrivieres S., Prinz T., Castro-Palomino Laria N., Meyer M., Boehm G., Bauer U., Schafer J., Neumann T., Shemanko C., Groner B. (2003). Comparative proteomic analysis of proliferating and functionally differentiated mammary epithelial cells. Mol. Cell Proteomics 2, 1039-1054 [DOI] [PubMed] [Google Scholar]
- Deugnier M. A., Faraldo M. M., Teuliere J., Thiery J. P., Medina D., Glukhova M. A. (2006). Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-Dbeta cell line. Dev. Biol. 293, 414-425 [DOI] [PubMed] [Google Scholar]
- Dhillon A. S., Meikle S., Yazici Z., Eulitz M., Kolch W. (2002). Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J. 21, 64-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatori I., Borsellino G., Haliassos N., Boitani C., Corallini S., Battistini L., Bernardi G., Stefanini M., Vicini E. (2004). Identification and enrichment of spermatogonial stem cells displaying side-population phenotype in immature mouse testis. FASEB J. 18, 376-378 [DOI] [PubMed] [Google Scholar]
- Forman J. J., Legesse-Miller A., Coller H. A. (2008). A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. USA 105, 14879-14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandellini P., Folini M., Longoni N., Pennati M., Binda M., Colecchia M., Salvioni R., Supino R., Moretti R., Limonta P., et al. (2009). miR-205 Exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 69, 2287-2295 [DOI] [PubMed] [Google Scholar]
- Gaur A., Jewell D. A., Liang Y., Ridzon D., Moore J. H., Chen C., Ambros V. R., Israel M. A. (2007). Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 67, 2456-2468 [DOI] [PubMed] [Google Scholar]
- Gottardo F., Liu C. G., Ferracin M., Calin G. A., Fassan M., Bassi P., Sevignani C., Byrne D., Negrini M., Pagano F., et al. (2007). Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 25, 387-392 [DOI] [PubMed] [Google Scholar]
- Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell. Biol. 10, 593-601 [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszer M., Erickson R., Scripture-Adams D. D., Dougherty J. D., Le Belle J., Zack J. A., Geschwind D. H., Liu X., Kornblum H. I., Wu H. (2006). PTEN negatively regulates neural stem cell self-renewal by modulating G0-G1 cell cycle entry. Proc. Natl. Acad. Sci. USA 103, 111-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., Reid J. G., Gao X., Shaw C. A., Creighton C., Tran P. L., Zhou X., Drabek R. B., Steffen D. L., Hoang D. M., et al. (2008). Novel microRNA candidates and miRNA-mRNA pairs in embryonic stem (ES) cells. PLoS ONE 3, e2548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P. B., Mani S., Yang J., Hartwell K., Weinberg R. A. (2005). The evolving portrait of cancer metastasis. Cold Spring Harb. Symp. Quant. Biol. 70, 291-297 [DOI] [PubMed] [Google Scholar]
- He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., et al. (2005). A microRNA polycistron as a potential human oncogene. Nature 435, 828-833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino K., Tsuchiya K., Fukao T., Kiga K., Okamoto R., Kanai T., Watanabe M. (2008). Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA 14, 1433-1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra I., Erlich Y., Muthuswamy S. K., Sachidanandam R., Hannon G. J. (2007). A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev. 21, 3238-3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V., Ferracin M., Liu C. G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., et al. (2005). MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065-7070 [DOI] [PubMed] [Google Scholar]
- Iorio M. V., Casalini P., Piovan C., Di Leva G., Merlo A., Triulzi T., Menard S., Croce C. M., Tagliabue E. (2009). microRNA-205 regulates HER3 in human breast cancer. Cancer Res. 69, 2195-2200 [DOI] [PubMed] [Google Scholar]
- Ivaska J., Nissinen L., Immonen N., Eriksson J. E., Kahari V. M., Heino J. (2002). Integrin alpha 2 beta 1 promotes activation of protein phosphatase 2A and dephosphorylation of Akt and glycogen synthase kinase 3 beta. Mol. Cell. Biol. 22, 1352-1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Lee E. J., Gusev Y., Schmittgen T. D. (2005). Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 33, 5394-5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluiver J., Haralambieva E., de Jong D., Blokzijl T., Jacobs S., Kroesen B. J., Poppema S., van den Berg A. (2006). Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer 45, 147-153 [DOI] [PubMed] [Google Scholar]
- Knobbe C. B., Lapin V., Suzuki A., Mak T. W. (2008). The roles of PTEN in development, physiology and tumorigenesis in mouse models: a tissue-by-tissue survey. Oncogene 27, 5398-5415 [DOI] [PubMed] [Google Scholar]
- Lamouille S., Derynck R. (2007). Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 178, 437-451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wong W. H. (2001). Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98, 31-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D., Chan C. Y., Ding Y. (2008). Analysis of microRNA-target interactions by a target structure based hybridization model. Pac. Symp. Biocomput. 64-74 [PubMed] [Google Scholar]
- Markou A., Tsaroucha E. G., Kaklamanis L., Fotinou M., Georgoulias V., Lianidou E. S. (2008). Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin. Chem. 54, 1696-1704 [DOI] [PubMed] [Google Scholar]
- Mattie M. D., Benz C. C., Bowers J., Sensinger K., Wong L., Scott G. K., Fedele V., Ginzinger D., Getts R., Haqq C. (2006). Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 5, 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulka L. A., Triplett A. A., Wagner K. U. (2007). Parity-induced mammary epithelial cells are multipotent and express cell surface markers associated with stem cells. Dev. Biol. 303, 29-44 [DOI] [PubMed] [Google Scholar]
- Montanaro F., Liadaki K., Volinski J., Flint A., Kunkel L. M. (2003). Skeletal muscle engraftment potential of adult mouse skin side population cells. Proc. Natl. Acad. Sci. USA 100, 9336-9341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorehead R. A., Hojilla C. V., De Belle I., Wood G. A., Fata J. E., Adamson E. D., Watson K. L., Edwards D. R., Khokha R. (2003). Insulin-like growth factor-II regulates PTEN expression in the mammary gland. J. Biol. Chem. 278, 50422-50427 [DOI] [PubMed] [Google Scholar]
- Paddison P. J., Cleary M., Silva J. M., Chang K., Sheth N., Sachidanandam R., Hannon G. J. (2004). Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Meth. 1, 163-167 [DOI] [PubMed] [Google Scholar]
- Pillai R. S., Bhattacharyya S. N., Filipowicz W. (2007). Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell. Biol. 17, 118-126 [DOI] [PubMed] [Google Scholar]
- Rajewsky N., Socci N. D. (2004). Computational identification of microRNA targets. Dev. Biol. 267, 529-535 [DOI] [PubMed] [Google Scholar]
- Ramkissoon S. H., Mainwaring L. A., Ogasawara Y., Keyvanfar K., McCoy J. P., Jr, Sloand E. M., Kajigaya S., Young N. S. (2006). Hematopoietic-specific microRNA expression in human cells. Leuk. Res. 30, 643-647 [DOI] [PubMed] [Google Scholar]
- Rusinov V., Baev V., Minkov I. N., Tabler M. (2005). MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res. 33, W696-W700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere L. F., Christensen M., Silahtaroglu A., Bak M., Heath C. V., Schwartz G., Wells W., Kauppinen S., Cole C. N. (2007). Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 67, 11612-11620 [DOI] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K. J., Stingl J., Smyth G. K., Asselin-Labat M. L., Wu L., Lindeman G. J., Visvader J. E. (2006). Generation of a functional mammary gland from a single stem cell. Nature 439, 84-88 [DOI] [PubMed] [Google Scholar]
- Shi R., Chiang V. L. (2005). Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39, 519-525 [DOI] [PubMed] [Google Scholar]
- Shimono Y., Zabala M., Cho R. W., Lobo N., Dalerba P., Qian D., Diehn M., Liu H., Panula S. P., Chiao E., et al. (2009). Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman K. E., Kendrick H., Ashworth A., Isacke C. M., Smalley M. J. (2006). CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 8, R7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman K. E., Kendrick H., Robertson D., Isacke C. M., Ashworth A., Smalley M. J. (2007). Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J. Cell Biol. 176, 19-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude G. J., Heimfeld S., Weissman I. L. (1988). Purification and characterization of mouse hematopoietic stem cells. Science 241, 58-62 [DOI] [PubMed] [Google Scholar]
- Stingl J. (2009). Detection and analysis of mammary gland stem cells. J. Pathol. 217, 229-241 [DOI] [PubMed] [Google Scholar]
- Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H. I., Eaves C. J. (2006). Purification and unique properties of mammary epithelial stem cells. Nature 439, 993-997 [DOI] [PubMed] [Google Scholar]
- Tavazoie S. F., Alarcon C., Oskarsson T., Padua D., Wang Q., Bos P. D., Gerald W. L., Massague J. (2008). Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. M., Parker J., Perou C. M., Hammond S. M. (2004). A custom microarray platform for analysis of microRNA gene expression. Nat. Meth. 1, 47-53 [DOI] [PubMed] [Google Scholar]
- Torrente Y., Tremblay J. P., Pisati F., Belicchi M., Rossi B., Sironi M., Fortunato F., El Fahime M., D'Angelo M. G., Caron N. J., et al. (2001). Intraarterial injection of muscle-derived CD34(+)Sca-1(+) stem cells restores dystrophin in mdx mice. J. Cell Biol. 152, 335-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 103, 2257-2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner K. U., Smith G. H. (2005). Pregnancy and stem cell behavior. J. Mammary Gland Biol. Neoplasia 10, 25-36 [DOI] [PubMed] [Google Scholar]
- Wang S., Garcia A. J., Wu M., Lawson D. A., Witte O. N., Wu H. (2006). Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc. Natl. Acad. Sci. USA 103, 1480-1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tang S., Le S. Y., Lu R., Rader J. S., Meyers C., Zheng Z. M. (2008). Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE 3, e2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welm B. E., Tepera S. B., Venezia T., Graubert T. A., Rosen J. M., Goodell M. A. (2002). Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev. Biol. 245, 42-56 [DOI] [PubMed] [Google Scholar]
- Woodward W. A., Chen M. S., Behbod F., Alfaro M. P., Buchholz T. A., Rosen J. M. (2007). WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc. Natl. Acad. Sci. USA 104, 618-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X., McClelland M., Wang Y. (2005). WebArray: an online platform for microarray data analysis. BMC Bioinformatics 6, 306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin L., Lawson D. A., Witte O. N. (2005). The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc. Natl. Acad. Sci. USA 102, 6942-6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O. H., Valdez R., Theisen B. K., Guo W., Ferguson D. O., Wu H., Morrison S. J. (2006). Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 441, 475-482 [DOI] [PubMed] [Google Scholar]
- Zhang L., Volinia S., Bonome T., Calin G. A., Greshock J., Yang N., Liu C. G., Giannakakis A., Alexiou P., Hasegawa K., et al. (2008a). Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl. Acad. Sci. USA 105, 7004-7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Behbod F., Atkinson R. L., Landis M. D., Kittrell F., Edwards D., Medina D., Tsimelzon A., Hilsenbeck S., Green J. E., et al. (2008b). Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 68, 4674-4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.