Abstract
Eukaryotic genomes are packaged into a nucleoprotein complex known as chromatin, which affects most processes that occur on DNA. Along with genetic and biochemical studies of resident chromatin proteins and their modifying enzymes, mapping of chromatin structure in vivo is one of the main pillars in our understanding of how chromatin relates to cellular processes. In this review, we discuss the use of genomic technologies to characterize chromatin structure in vivo, with a focus on data from budding yeast and humans. The picture emerging from these studies is the detailed chromatin structure of a typical gene, where the typical behavior gives insight into the mechanisms and deep rules that establish chromatin structure. Important deviation from the archetype is also observed, usually as a consequence of unique regulatory mechanisms at special genomic loci. Chromatin structure shows substantial conservation from yeast to humans, but mammalian chromatin has additional layers of complexity that likely relate to the requirements of multicellularity such as the need to establish faithful gene regulatory mechanisms for cell differentiation.
Keywords: epigenetics, gene regulation, histone modifications, microarray, nucleosome
INTRODUCTION: CHROMATIN STRUCTURE
In vivo eukaryotic genomes are organized in chromatin, a DNA-protein complex whose basic repeating unit is the nucleosome (1). The nucleosome consists of 147 base pairs of DNA wrapped 1.7 times around an octamer of histone proteins (two each of histones H2A, H2B, H3, and H4). Polynucleosomal tracts appear by electron microscopy as “beads on a string,” where nucleosomes are seen as beads, and the intervening linker DNA is the string (2). A number of features distinguish individual nucleosomes from one another. First, the location of a nucleosome relative to underlying genomic sequence affects accessibility of regulatory sequences, so precise translational positioning of nucleosomes can be of great regulatory consequence. Additionally, there are multiple isoforms of the histones, which combine to form a number of distinct octamers. Finally, histones are subject to a bewildering array of covalent modifications. Since the description of the nucleosome’s basic composition in 1974, chromatin structure has been of increasing interest as it has been implicated in processes ranging from recombination to transcription to cell cycle control and cancer. Furthermore, it is almost universally believed that at least some aspects of chromatin architecture are epigenetically inherited, although this is not as firmly established as many think (3).
For three decades, most of our knowledge of chromatin structure came from intensive single-gene approaches on loci such as the chicken β-globin locus or the yeast PHO5, GAL1-10, and HIS3 promoters. However, since the advent of the genomics era, brought about by the availability of whole-genome sequences and technologies such as microarrays and high-throughput sequencing, we can now measure many aspects of chromatin structure over entire genomes in a single experiment. This review describes the use of genome-scale technologies to study chromatin structure. We start with a brief overview of genomics technologies used to study chromatin structure. We then review chromatin structure in budding yeast, the best-characterized model organism, with an eye toward (a) describing a “typical” yeast gene, (b) enumerating hypotheses for the establishment of the typical gene’s chromatin structure, and (c) noting where departures from typical behavior indicate potentially interesting regulatory mechanisms at work. Next, genomic studies on mammalian chromatin structure are reviewed, with an emphasis on aspects of chromatin structure that are unique to metazoans.
TECHNICAL APPROACHES
Genomic measurements of chromatin structure consist of two phases—isolation/separation of DNA associated with a particular type of chromatin, and characterization of the isolated nucleic acid pool. Fractionation techniques used in genomics experiments are often the same as those used for single-gene studies, but the measurement technology used is an “omics” technology rather than PCR or blotting. The two major types of fractionation used to study chromatin structure are nuclease digestion to enrich for protected genomic regions, and affinity techniques such as chromatin immunoprecipitation.
As an example of the first, DNase I has long been known to preferentially cleave regulatory regions of metazoan genes due to the relative absence of histones at these genomic loci. Similarly, micrococcal nuclease is typically used to determine nucleosome positions, since this nuclease exhibits a strong preference for linker DNA over nucleosomal DNA. These characteristics have allowed researchers to infer aspects of chromatin structure from broad genomic surveys of nuclease sensitivity. For example, a number of genome-scale studies have measured the locations of DNase I hypersensitive sites in human cell lines (4–6). Here, isolated nuclei are treated with a titration of DNase I, and cleavage sites are recovered and analyzed by microarray or sequencing for the identity of hypersensitive genomic sequences. A similar, but nuclease-independent, technique called FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) is an alternative method that enriches regulatory regions based on differential solubility caused by the differing amounts of protein associated with regulatory vs coding regions (7).
In terms of measurement technology, the DNA microarray was the dominant genomic measurement technology for a decade, but the incredible power of high-depth sequencing has recently spread from dedicated genome centers into wider circulation. As both DNA microarrays and DNA sequencing are relatively well understood, we touch only on advantages and disadvantages of these technologies for chromatin studies.
In a typical DNA microarray study, an isolated nucleic acid population is labeled with a fluorescent dye and hybridized to a microarray. Microarray resolution is limited by probe spacing (and even ultradense tiling does not necessarily achieve single-bp resolution, due to hybridization of sequences with extensive, but incomplete, overlap), and coverage is limited by probe number. Microarrays are relatively cheap, however (250-bp resolution whole-genome yeast microarrays cost roughly $200 each), and two-color hybridization schemes allow relative changes to be sensitively detected for both high- and low-abundance features.
So-called deep sequencing is increasingly used now (particularly in mammalian systems) and offers excellent spatial resolution (single base pair, in principle), and complete genomic coverage. Furthermore, sequencing provides allele-specific information in diploid organisms, whereas single-nucleotide discrimination is nontrivial in microarray studies. The two major sequencing methodologies used to date (more are already available but have not been widely published) have been 454 sequencing, which provides ~100,000 sequences several hundred base pairs in length, and Illumina 1G “Solexa” sequencing, which provides several million shorter (~30–70 bp) sequencing reads. Disadvantages of sequencing are higher cost (~$1000 per run), and the double-edged sword of complete coverage—sequencing mRNA from a mammalian cell will generate huge numbers of reads from housekeeping genes such as actin and GAPDH, meaning that less-abundant genes will yield much lower numbers of reads and higher experimental variability.
YEAST CHROMATIN OVERVIEW
In general, lessons learned from studies of chromatin in the model yeast Saccharomyces cerevisiae hold true in multicellular organisms—mammalian chromatin appears to be more complex than yeast largely as a result of additional histone isoforms and histone modifications, rather than distinctive use of modifications common to all eukaryotes (exceptions, of course, exist). We therefore first discuss genomic studies of chromatin structure in yeast, then turn to additional features found in mammals.
An overriding paradigm emerging from genomic studies of chromatin is that common patterns (which can be conceptualized as motifs) emerge that are widespread though not ubiquitous. These “stereotyped” structures often provide deep insight into the general rules underlying the establishment of chromatin architecture. However, not all promoters (for example) in yeast look like the typical pattern, and the deviations from the average behavior often reveal important regulatory mechanisms at play. We therefore first emphasize common patterns in each section, and then point out examples of genomic loci that depart from the typical pattern.
NUCLEOSOME POSITIONING
Nucleosome occupancy has been studied in yeast using low-resolution DNA microarrays (7, 8), high-resolution tiling oligonucleotide microarrays (9–11), and, most recently, ~4-bp resolution high-throughput sequencing (12–14). In general, each higher-resolution study confirms prior results, with increased resolution additionally allowing appreciation of novel features. A notable surprise from genomic maps of nucleosome positions has been the extent to which nucleosomes are well positioned in the population.
Yeast open reading frames are generally characterized by a strongly nucleosome-depleted region (often called the nucleosome-free region, or NFR, but see below) found upstream, surrounded by two well-positioned nucleosomes. The NFR is the site of the majority of functional transcription factor binding sites, although some transcription factors appear to be able to bind along the DNA wrapped around the −1 nucleosome at locations where the major groove faces away from the octamer core. These results partially explain a longstanding dilemma in the transcription field: Most transcription factors, which bind short (4–10-bp, typically) sequence motifs, only bind a small fraction of their motifs in the genome. Indeed, histone occupancy accounted for a significant subset of sequence motifs bound by purified Leu3 in vitro that are not bound in vivo, supporting a role for nucleosome positioning in transcription factor site accessibility (15).
How Is the Stereotypical 5′ Promoter Architecture Specified?
The 5′ NFR typically contains one or more homopolymeric runs of polyA (or polyT) (10, 16). Poly-dA/dT runs are intrinsically stiff, and the bending required to wrap around the histone octamer is energetically costly for these sequences relative to random sequences (17–19), leading to decreased nucleosome incorporation on model genes in vivo, and, importantly, in vitro (16, 19–21). In addition to constituitive NFRs where nucleosomes are excluded by poly-dA/dT sequences, regulatable NFRs can be generated by the binding of certain proteins, such as Reb1 and Abf1, to their binding sites, which are not nucleosome depleted in in vitro reconstitutions but are strongly nucleosome depleted in midlog yeast cultures (21).
In contrast, AT-rich dinucleotides confer some bend to the DNA duplex, and thus spacing of these dinucleotides with 10-bp periodicity (aligning these bends in the same direction) results in DNA that is thermodynamically favored as a binding site for histones, because less energy must be expended in the bending of the DNA (22, 23). Genome-wide computational studies have recently described the analysis of pronucleosomal sequences in the yeast genome (24–27). One study utilized ~200 nucleosomal sequences to determine a dinucleotide position-specific sequence matrix (PSSM) for nucleosomes (26). Another used conservation of AA/TT periodicity across six Saccharomyces species to define a nucleosome-positioning score (NPS) (24). More recent studies have included discrimination against polyA runs (25, 27). The most notable finding of these studies has been that the first nucleosome in a typical yeast ORF (the +1 nucleosome) is often associated with strong pronucleosomal sequence elements. However, whole-genome data from in vitro reconstitution experiments demonstrate that antinucleosomal sequences explain far more of the in vivo patterning of chromatin than do pronucleosomal sequences (21, see Figure 4a).
Furthermore, while pronucleosomal sequences are most common at the +1 position, microarray studies show that the +2 and +3 nucleosomes are also generally well positioned. In 1988 Kornberg & Stryer proposed a “statistical positioning” model in which nucleosomes on random sequence could appear well positioned in population averages because of constraints imposed by packing many nucleosomes into a short region (28). A useful analogy is that of a can of tennis balls: The can imposes few constraints on the location of a single tennis ball, but three tennis balls will appear uniformly packaged thanks to the limited number of ways the small amount of free space can be distributed. This idea might well account for the surprising amount of order in yeast chromatin, since genes are fairly short (<2 kb average). Consistent with this idea, “fuzzy” or delocalized nucleosomes are typically found distal to NFRs and +1 nucleosomes, which are the two most likely candidates for the borders that constrain a given packaging unit (10, 13). The ideal test of this hypothesis—in vitro reconstitution of histone octamers into long or short shear distributions of genomic DNA, from saturating to limiting octamer: DNA ratios—remains to be reported.
3′ Nucleosome-Free Regions
Long nucleosome-depleted regions do not only occur at 5′ ends of genes—a surprising fraction of gene 3′ ends also exhibit NFRs (13, 14), which also appear to be programmed by antinucleosomal sequences. The function of these has not been explored in any detail, but it should be noted that many antisense transcripts initiate within these 3′ NFRs (13). These sites are often bound by the general transcription factor TFIIB, which has been proposed to contribute to gene looping, in which 5′ and 3′ ends of some yeast genes appear to interact in vivo (29), possibly enabling the local recycling of transcription machinery after each round of transcription.
DEVIATION FROM TYPICAL BEHAVIOR
The features of yeast promoters and 3′ ends described above are widespread, but not universal. For example, the (typical) HIS3 promoter has an NFR that can be recapitulated in vitro using just histones, salts, and DNA (20). Conversely, the PHO5 promoter lacks an NFR in the uninduced state in vivo (30), and its chromatin state can only be approximated in vitro if yeast extracts and ATP are included in the reconstitution reactions (31). This suggests that cellular machinery actively repositions nucleosomes away from the intrinsic locations specified by the sequence cues at PHO5. This repositioning contributes to the regulatory program of PHO5 (32). Below we identify some broad classes of genes apparently subjected to specific regulation by cellular factors.
Tata Versus Non-TATA Yeast Genes
Yeast genes can be grouped into two broad classes, based on the histone lysine acetylase (KAT) involved in Tata-binding protein recruitment to promoters (33–35). SAGA-dominated genes typically have TATA boxes, are stress responsive, are characterized by noisy, or “bursty,” expression, and are regulated by a wide range of chromatin remodeling factors. Conversely, TFIID-dominated genes lack TATA boxes, are expressed during active growth, exhibit little noise in expression levels, and are not affected by deletion of most chromatin-regulatory genes.
These two classes also exhibit differences in promoter chromatin packaging. The majority of yeast genes (~80%) are TFIID dominated, and the above descriptions of NFRs apply to this class in particular. The minority class of TATA-containing stress genes, on the other hand, exhibits more variable promoter architecture. This is true across different genes (i.e., various stress-responsive genes exhibit a range of promoter packaging states) and also appears to be true across individual cells, since these promoters often are associated with delocalized nucleosomes (12, 24, 36). Importantly, transcription factor binding sites at TATA-containing promoters are likely to be occluded by nucleosomes, although rapid exchange of nucleosomes at these promoters (see below) will allow binding sites to be accessed during transient time windows. This competition between nucleosomes and transcription factors might be expected to contribute to cell-to-cell variability (noise) in expression of downstream genes (36, 37).
Isw2
ATP-dependent chromatin remodelers utilize the energy of ATP hydrolysis to distort histone-DNA contacts, with eventual outcomes such as histone sliding, eviction, or replacement. Elegant work by Whitehouse & Tsukiyama showed that the ATPase Isw2p acts to position nucleosomes over unfavorable sequence elements at the POT1 promoter (38). Specifically, nucleosomes in isw2Δ yeast matched positions from in vitro reconstitutions, whereas in wild-type yeast the+1 nucleosome was located further 5′, narrowing the NFR and inhibiting transcription. A subsequent whole-genome study found that +1 nucleosomes or −1 nucleosomes were shifted toward the NFR (often over unfavorable poly-dA/dT tracts) at sites of Isw2 action (9). Promoters with shifted +1 nucleosomes were generally repressed, while repositioning of −1 nucleosomes at other promoters was surprisingly implicated in inhibiting antisense transcription.
Thus, we imagine one could prospectively identify locations where nucleosomes do not follow sequence cues in vivo, such as Isw2-regulated genes or PHO5, and this would reveal sites where cellular factors modulate chromatin architecture to regulate transcription or other processes. We anticipate that advances in computational predictions of thermodynamically favored nucleosome positions, coupled with identification of in vivo nucleosomes that do not match the in vitro/in silico predictions, will provide a rich source of information on the cellular machinery that regulates chromatin structure in vivo.
HISTONE DYNAMICS
Steady-state studies provide an anatomy of chromatin structure in mixed populations, but miss important dynamic behavior such as nucleosome sliding and eviction. In general, histones are among the most stably associated DNA-binding proteins in photobleaching studies (39), but seminal work by Ahmad & Henikoff showed that a fraction of nucleosomes are replaced in a replication-independent (RI) manner (40). In flies, the H3 variant H3.3 marks regions where RIH3replacement has occurred, providing a surrogate to direct dynamic measures.
In yeast, there is no separation between replication-coupled and replication-independent H3 isoforms (yeast H3 most closely resembles H3.3), so replication-independent H3 replacement has been studied using inducible epitope-tagged systems (41–43). Yeast cells are arrested in G1 phase, and tagged H3 is induced after arrest is complete. At varying times after induction, the tag is immunoprecipitated and hybridized to microarrays. Nucleosomes that accumulate tag at early time points are inferred to be replaced rapidly, while slow or undetectable incorporation at a nucleosome implies stable association of histone molecules with the DNA at that genomic location. Since the H3/H4 tetramer forms the core of the histone octamer, H3 replacement is taken as a surrogate for whole-nucleosome exchange.
In yeast, nucleosomes over ORFs in yeast are replaced relatively slowly, while promoter nucleosomes are rapidly replaced. This is surprising given that H3.3 patterns in Drosophila indicate that transcription is a major driving force for histone replacement (40, 44). We believe the yeast result provides an interesting insight into the mechanism for polymerase-dependent histone replacement.
Histone Dynamics: Mechanism
H3 is replaced in yeast only over very highly transcribed genes, but at a given transcription rate, SAGA-dominated genes (34) exchangeH3 more rapidly than do TFIID-dominated genes (41). Why should the mechanism of regulation affect chromatin dynamics over coding regions? One possibility is that SAGA helps recruit histone-displacing factors to genes (45). Alternatively, we note that SAGA-dependent genes exhibit high levels of transcriptional noise (46), ascribed to “bursts” of transcription rather than to evenly spaced initiation events (47). When RNA polymerase moves through a histone octamer, it is believed to result in eviction of a H2A/H2B dimer (48), resulting in a hexamer that will either be repaired or evicted by collision with a second polymerase. Thus, the well-spaced polymerases at TFIID-dominated genes might be less likely to evict nucleosomes than would the bursts of closely spaced polymerases at SAGA-dominated genes: Pol2 density on genes might then account for differences between species in coding region turnover profiles.
At promoters, nucleosomes are generally rapidly replaced. While H3 replacement at several regulated promoters increased upon induction (41, 42), globally promoter H3 replacement was uncorrelated with RNA Pol2. We speculate that rapid promoter replacement could be related to the presence of “antinucleosomal” poly-dAdT sequences at many promoters. ATP-dependent chromatin remodelers are capable of sliding nucleosomes laterally, and in vitro even seemingly well-positioned nucleosomes can make short lateral excursions around their baseline position (49). Even if all nucleosomes in the genome were to make lateral excursions, those adjacent to antinucleosomal sequences could be evicted simply by being pushed onto the adjacent unfavorable sequences (50)—falling off a cliff, as it were. Thus, nucleosome eviction at promoters would result from an interaction between a location-specific intrinsic sequence tendency for histone eviction, and regulatable extrinsic factors such as level of ATP-dependent remodeler present or the extent of competition between nucleosome and transcription factor binding (36).
Histone Dynamics: Consequences
Replication-independent nucleosome replacement is likely to have functional consequences for cellular regulation of transcription, replication, etc. For example, transient DNA exposure by histone replacement may allow access of DNA-binding sites to DNA-binding factors. Indeed, it is entirely possible that nucleosome-“free” regions are in fact very transiently (or very loosely) associated with histones (44, 51), rather than statically naked—the fact that a typical NFR is about one nucleosome wide is consistent with this idea. But even nucleosomes that are highly occupied at steady state, such as the +1 nucleosome, are dynamic, leading to a picture of a nucleosome that is constrained translationally by sequence characteristics, yet which is evicted frequently and replaced rapidly, thereby transiently exposing the underlying DNA to binding factors (or allowing RNA polymerase to pass). Beyond site exposure, histone replacement shows an interesting association with two aspects of heterochromatin in yeast—heterochromatic regions are protected from histone replacement (41, 52), whereas the boundaries of these regions are associated with rapidly replaced nucleosomes (41, 51), suggesting that histone replacement serves to erase laterally spreading chromatin states (53) and thereby insulate chromatin domains from one another.
HISTONE VARIANTS
Metazoans encode a large number of alternative variant histone isoforms, but budding yeast encode only two: the predominantly centromere-localized H3 variant Cse4, and the H2A.Z homolog Htz1. Numerous genomic localization studies have been carried out for Htz1 (12, 54–57). Generally, these studies find Htz1 located at promoters, usually at the +1 nucleosome. One curious observation is that while Htz1 is almost universally found at the+1 nucleosome, Htz1 is also found at the −1 nucleosome at a good fraction of promoters (including many tandemly oriented genes where the −1 is not another gene’s +1), and no published studies have identified features that reliably distinguish +1-type from +1/−1-type promoters.
Htz1 levels exhibit a modest anticorrelation with transcription rate—the small fraction (1%–2%) of very highly expressed (>~10 mRNA/hr) genes in yeast are Htz1 depleted (54, 56). These promoters also exhibit extremely fast H3 replacement, and we suspect the absence of Htz1 at these promoters is due to a lag between incorporation of a canonical octamer and subsequent replacement of H2A with Htz1—at very high turnover Htz1 incorporation will not keep up. At the other extreme of transcription rates, Htz1 is found at low levels, diffusely localized in patches throughout heterochromatic regions in yeast (54, 58).
Together, these data are consistent with a model in which a major function for Htz1 is to mark regions where RNA polymerase has initiated. A completely untranscribed gene is devoid of Htz1, but after a round of transcription, Htz1 is assembled at the+1 nucleosome behind RNA polymerase (59). This results in Htz1 being found at most genes in yeast, since most genes are modestly expressed. Of course, the prediction of this model for Htz1 assembly would be that +1/−1 promoters would exhibit bidirectional transcription. The recent isolation and characterization of cryptic unstable transcripts from yeast exosome mutants, which fail to degrade noncoding transcripts such as antisense transcripts (60, 61), will enable a direct test of this hypothesis.
HISTONE MODIFICATIONS
Histones are subject to a bewildering array of covalent modifications, with over 100 different modification sites described to date. This diversity has become the subject of intense research interest, and over the past few years the question of why so many modifications occur has become a focus for much commentary, with a widely cited idea being that of a histone code (62–65) (see below). Many histone modifications are correlated with cellular processes such as transcription, and deletion studies show that elimination of histone-modifying enzymes often affects transcription rates, chromosome stability, or other chromosomal processes. Thus, mapping of histone modifications has become a very popular way to characterize genome activity.
Typical Gene
Broadly speaking, two groups of modification have been mapped in yeast. One group consists of modifications that generally occur over transcribed regions, whose levels correlate with polymerase abundance, and which are possibly deposited by enzymes traveling with RNA polymerase. The other group consists of modifications that either correlate or anticorrelate with rates of replication-independent histone replacement. We hypothesize that their genomic localization patterns likely result from incorporation into the genome of histones drawn from a free pool carrying (or lacking) the modifications in this group. We elaborate on these groups below.
Transcription-Related Marks
A now-classic example of a transcription-related histone mark is H3K4 trimethylation. H3K4 is methylated by Set1p (66), which associates with the Serine 5-phosphorylated initiation form of RNA polymerase (67), and H3K4me3 is found over the 5′ ends of yeast genes at levels that correlate with transcription rate (68, 69). Similarly, H3K36me3 is found over the middle and 3′ ends of coding regions, deposited by a methylase (Set2p) associated with the Serine 2-phosphorylated elongation form of polymerase (70, 71).
A number of histone acetylation states (including H3K9ac, H3K14ac, H4K12ac, and many others) are found at the 5′ ends of coding regions. For these marks, this pattern apparently results from a combination of two effects. First, these residues are acetylated throughout the coding region during transcription. Subsequently, a histone deacetylase complex known as Rpd3S is recruited by the H3K36me3 found over the middle and 3′ end of coding regions, resulting in a shaping of the original coding region pattern to the 5′-biased pattern observed at steady state (70, 71).
Histone Modifications and Regulation of Transcription
Histone modifications often correlate with processes such as transcription, but genetic studies in yeast prove the old saw that correlation is not causation. For example, H3K4me3 occurs universally over the 5′ ends of transcribed genes, yet elimination of all H3K4me3 by deletion of SET1 is well tolerated by yeast, rather than being lethal as expected if all transcriptional initiation ceased. Furthermore, part of the phenotype of set1Δappears to result from Set1 methylation of a nonhistone substrate (72).
It is therefore crucial to emphasize what may be learned from localization studies. Specifically, localization of a mark simply identifies nucleosomes where a modifying enzyme has acted (and in some cases the process resulting in recruitment of the enzyme), not the function of the mark.
Perhaps the clearest example of this comes from studies of H3K36me3. H3K36me3 is deposited during RNA polymerase elongation, and is considered a transcriptional elongation mark, yet deletion of SET2 results in no identifiable effects on elongation per se (73). Instead, H3K36me3 deposition leads to deacetylation of lysines that were acetylated during polymerase passage. In the absence of K36me3 and resultant deacetylation, coding regions remain hyperacetylated, and so-called cryptic internal initiation sites become active (70, 71, 74). Thus, the function of this elongation mark is to reverse perturbations to a gene’s chromatin structure, which presumably aided polymerase passage through the coding region.
Replacement-Related Marks
The second group of histone modifications exhibits genomic localization patterns related to replication-independent histone replacement. Most notably, H4K16ac and H2BK16ac are found over coding regions (except over very highly expressed ORFs), but are depleted from the rapidly replaced nucleosomes flanking the NFR (68). Conversely, H3K56ac, best understood as a mark of newly synthesized histones assembled into chromatin during replication (75), is enriched in nucleosomes subject to replication-independent replacement (43, 76). The H3K56 acetylase Rtt109 can acetylate free histones, but not nucleosomal histones (77). We therefore believe that the localization of replacement-related histone marks results in part from their presence (H3K56ac), or absence (H4K16ac) in the free pool of histones, and subsequent incorporation via histone replacement.
Modulation of histone modification patterns by histone replacement has several interesting potential consequences. First, replacement of a given nucleosome would erase preexisting histone marks at that genomic location. This is of great interest, as it establishes an event horizon beyond which histone modification physiology cannot be discerned. Of course, if an enzyme remains associated with a genomic locus even after histone replacement, it will re-establish its mark—H3K4me3 is apparent at the +1 nucleosome of active genes, despite rapid turnover of +1 nucleosomes (68). However, the enrichment of H3K4me3 is greater at the +2 nucleosome than at the +1, suggesting that some fraction of nucleosomes in the population has been replaced but has not yet been remodified. In any case, marks leading directly to histone eviction might be difficult to find by their nature, requiring mutation of downstream turnover machinery or high temporal resolution kinetic studies of gene activation for their identification.
More speculatively, we note that histone replacement exhibits the potential for positive feedback. We can imagine a first histone replacement event in a given cell cycle, in which a H4K16ac, H3K56deac nucleosome is replaced with a H4K16deac, H3K56ac nucleosome. H3K56ac aids histone replacement, and this appears to be primarily an effect of enhanced eviction rather than incorporation (43, 76, 78). Thus, the first nucleosome evicted at a location should be more difficult to evict than subsequent H3K56ac nucleosomes. The loss of H4K16ac during replacement could have similar consequences via a K16deac->Bdf1->Swr1 pathway of Htz1 incorporation (55, 56, 79–81). Such local positive feedback loops could affect features ranging from gene induction kinetics to expression noise.
Another interesting feature of rapidly replaced nucleosomes is that replacement results in nucleosomes that are expected to have a relatively high affinity for one another. For example, H4K16 interacts with an acidic patch on H2A in an adjacent nucleosome in the nucleosome crystal structure (82), and K16 acetylation inhibits compaction of nucleosome arrays into 30-nm fiber in vitro (83). Thus, the pair of +1/−1 nucleosomes lacking H4K16ac at promoters might be expected to preferentially contact each other, or perhaps H4K16deac nucleosomes at other locations in the genome. Nucleosomes around 3′ NFRs also exchange relatively rapidly (41) and lack H4K16ac, so this phenomenon could contribute to the 5′ to 3′ looping of genes proposed to play a role in recycling of transcriptional machinery (29). Deciphering the influence of these factors on chromosome folding will be of great future interest.
CHROMATIN FEATURES IN GENOMIC ANNOTATION
Given the fairly consistent features of chromatin states from ORF to ORF, one may then seek noncanonical chromatin states as indicators of unusual regulatory mechanisms. For example, as noted above, Htz1 typically marks +1 nucleosomes. Simply browsing publicly available Htz1 mapping data reveals numerous examples of Htz1-containing nucleosomes that appear in the middle of well-characterized genes, often with evidence from RNA maps for associated transcription (84). A cursory glance indicates that many of these are genes specific to developmental programs such as meiosis (e.g., MEI4), possibly indicating a more general role for interfering transcription in regulation of cell-fate programs in yeast (85, 86).
CHROMATIN STRUCTURE IN MAMMALS: OVERVIEW
In broad strokes, the main chromatin features of a gene are conserved from yeast to humans. But the increased complexity of chromatin structure in mammals is evident even within one cell type. There are more distinct histone modifications, and more ways to place, remove, and read each modification. Another major driving force of complexity in mammals is the diversity of cell types. Although each cell in multicellular organisms typically carries the same genome, different genes are activated or silenced in distinct cell types. The chromatin structure in a particular cell reflects both the panel of genes that are active in that cell type, as well as the developmental plasticity of the cell. Furthermore, in mammals specialized chromosomal domains with unique chromatin structures are more numerous than in yeast. These exceptions to the rule typically contain clusters of functionally related genes that are coregulated in unique developmental or biological contexts.
Conserved Features of Chromatin Structure
Many features of chromatin structure are conserved from yeast to mammals. Nucleosome placement appears constrained by some of the same sequence preferences as in yeast, because patterns of dinucleotide repeats identified in yeast nucleosomes and sequences that are depleted in yeast nucleosomes can partially predict nucleosome occupancy in chicken and human chromatin (26, 27). Nucleosome-excluding sequences are widespread but not ubiquitous at transcriptional start sites (TSSs) of mammalian genes, and are enriched at ubiquitously expressed genes (87). Mammalian chromatin also exhibits nucleosome-free regions (NFRs) of approximately 200 bps centered −85 bp upstream of the transcriptional start site and surrounded by positioned nucleosomes; this has been documented indirectly by genome-wide measurement of DNAse hypersensitive sites (DHSs) (6) and by direct measurements of histone occupancy (88–90). The flanking of NFRs by well-positioned nucleosomes is consistent with the idea that NFRs themselves, or the machinery that forms them, have an instructive role in nucleosome positioning.
Next, as in yeast, histone modifications in mammals strongly reflect the anatomy of genes. For example, the start and first ~500 bps of an active gene are typically occupied by H3K4me3; H3K4me2 peaks in the body of the gene with a shallower gradient, and H3K4me1 is weakly localized distally. Also, as in yeast, H3K36me3 is associated with transcriptional elongation in human and mouse, and has been exploited to map the lengths of protein-coding and -noncoding transcripts (91). In both yeast and human cells, genome-scale mapping of asymmetrically methylated H3R2 showed that it is localized to the body of genes but absent from promoters (92, 93), although its level over genes is independent of transcription level. H3R2me2a and H3K4me3 each inhibit placement of the other mark, and this mutual inhibition likely contributes to their distinct localization at 5′ or the body of genes, respectively.
All known histone methylation and acetylation states were mapped in a series of wide-ranging studies from the Zhao group (94, 95). Figure 1b summarizes results for the methylation and acetylation states, aligned by coding regions. Active transcription in human cells is correlated with K4 and K36 methylation, along with additional histone methylations (Figure 1b). In contrast to the complex patterns of histone methylation, analysis of 18 histone acetylations showed that they are all positively correlated with transcription (90, 94–96), particularly at the 5′ ends. Furthermore, as in yeast, histone modifications tend to be found in groups of correlated marks. In human T cells, a common pattern of 17 histone modifications was associated with approximately 25% of promoters, and deviants from this pattern are all individually rare (and much of this deviation is likely to represent an artifact of using thresholding as a computational tool for analysis of genomic localization data).
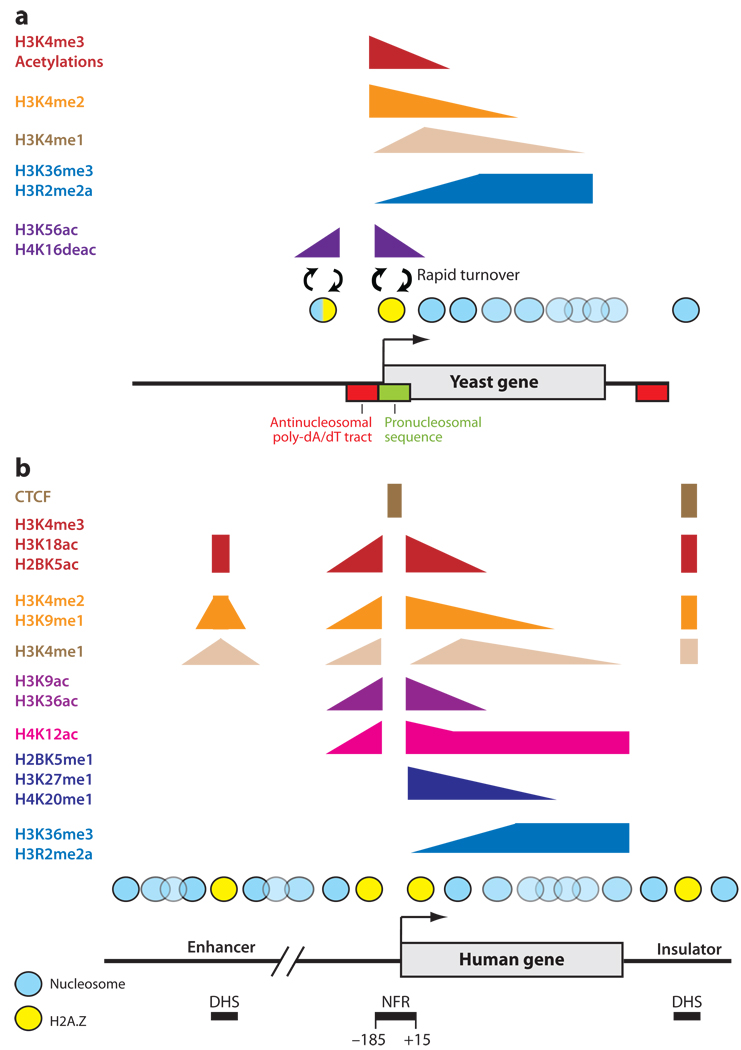
Figure 1.
Chromatin map of typical active genes in yeast and human. (a) Schematic of a typical yeast gene, showing distributions of various histone marks associated with transcription (H3K4 methylation, H3K36 methylation, most acetylation), and associated with histone replacement (H4K16ac, H3K56ac). Boxes indicate prominent sequence rules underlying nucleosome positioning at a typical gene—antinucleosomal poly-dA/dT tracts (red box) are found at many promoters and may specify the abundant nucleosome-free regions, while pronucleosomal sequences (green box) are often found associated with the +1 nucleosome. (b) Schematic of an average gene, enhancer, and insulator in human cells. H3K4me3 is found near the TSS, while H3K4me2, H3K9me1, H2BK5me1, H3K27me1, and H4K20me1 are distributed over the TSS and into the gene body. The me1 occupancy patterns of H3K9 and H3K27 are in striking contrast to the repressive me3 patterns of occupancy, illustrating that the methylation status of a single residue may signal distinct outcomes (94). However, to date no protein has been identified that that specifically recognizes H3K9me1 or H3K27me1. H3K36me3, H3R2me2a, and H4K12ac are distributed over the body of genes. Several histone acetylations, including H2AK9ac, H2BK5ac, H3K9ac, H3K18ac, H3K27ac, H3K36ac, and H4K91ac, are mainly associated with regions around the TSS of active genes, whereas a number of other histone acetylations also extend into the bodies of active genes. While gene-proximal promoters are occupied by H3K4me3, enhancers typically show H3K4me1 and are also occupied by the histone acetyltransferase p300 (90). In a separate study, intergenic DHSs (a subset of which likely includes enhancers) were found to be associated with H2A.Z, H3K4me1, H3K4me2, H3K4me3, H3K9me1, H3K18ac, and H2BK5ac (95). Some of these differences (e.g., H3K4 methylation states) from the two studies suggest that there may be multiple and cell type–specific chromatin signatures of enhancers. Finally, insulators are associated with CTCF binding, strongly phased and positioned nucleosomes that contain H2A.Z, and modifications such as H3K4me3, H3K4me2, H3K9me1, along with other marks typically associated with active TSS.
DIVERSIFICATION OF RULES FOR NUCLEOSOME OCCUPANCY
We have summarized the chromatin state of a typical, transcriptionally active mammalian gene (Figure 1b). Despite the many overlapping characteristics between yeast and metazoan chromatin structure, differences can also be identified. Mammalian genes are associated with an NFR when actively transcribed (88) or when a preinitiation complex has been assembled (89), but not at untranscribed genes, and constitutive sequence-programmed NFRs appear to be uncommon. In mammals and flies, Pol2 and H3K4me3 can be detected at many genes with no appreciable mRNA production, implying the widespread existence of “paused” RNA polymerase complexes poised for transcription at the start of many genes (97–99), unlike yeast (but see Reference 100). The mammalian +1 nucleosome is located at +40 bp relative to the TSS at transcribed genes (88), but at +10 at genes with paused polymerase. Similar differences were also observed in Drosophila (101). Differences in average +1 positioning between species may therefore reflect different transcriptional behavior at a typical gene, or alternatively perhaps the transcriptional machinery of different eukaryotes might engage the +1 nucleosome in fundamentally different ways.
DIVERSIFICATION OF HISTONE MODIFICATIONS
Relative to yeast, mammalian chromatin is characterized by an expanded set of enzymes for the deposition and removal of each modification, increased complexity in the recognition of each modification, and an enlarged repertoire of histone modifications. Here we touch upon three main themes regarding this diversification.
Complexity in Modification Placement and Removal
We examine H3K4 methylation to illustrate this point. At H3K4, me3 and me2 are associated with transcribed genes, me1 is associated with enhancers (see below), and unmethylated H3K4 (H3K4me0) is associated with gene repression. Thus, each methylation state of H3K4 can, in principle, transduce a distinct biological signal. This fine discrimination is achieved by expansion and increased selectivity of protein lysine methyltransferases (KMT), protein lysine demethylases (KDM), and adaptor proteins that recognize these distinct modifications. In the case of H3K4, at least eight KMTs can methylate H3K4me0 all the way to H3K4me3. However, the H3K4 demethylases show distinct specificity: LSD1/KDM1 cannot demethylate H3K4me3 but will efficiently demethylate H3K4me2/1 to H3K4me0. Conversely, at least four KDMs in the KDM5 subfamily can demethylate H3K4me3/me2 to the H3K4me1 state. Some, but not all, Jumonji domain KDMs can also demethylate histone lysine me3 all the way to me0 (102). Thus, there is increased enzymatic specificity in KDMs to distinguish specific residues and individual methylation states. ChIP-chip studies of KMTs and KDMs are an efficient strategy to determine which enzyme is responsible for histone modifications at specific genes (103–107).
Furthermore, while many histone modifications common to yeast and humans are similarly distributed, notable exceptions exist. For example, H3K4me1 in mammals is found over long-distance enhancers (see below). H3K79me3, which blankets yeast coding regions, exhibits a complex pattern of localization in humans; it is found in tight peaks in the TSS of a subset of the highest expressed genes, but otherwise is observed in silent genes.
Complexity in Histone Modification Readers
Second, metazoans have seen a dramatic expansion of the number and diversity of binding domains for the various histone modifications. Distinct H3K4 methylation states are recognized by specific protein binding partners, which couple recognition ofH3K4methylation states to specific gene regulatory events. For instance, H3K4me3 may be linked to transcriptional activation, acute gene silencing, or DNA recombination by the PHD fingers of TAF3 (a basal transcription factor) (108), ING1 (a tumor suppressor) (109), or RAG2 (a recombination factor) (110, 111), respectively. In addition, the expanded “Royal superfamily” of methyl-lysine readers include the Tudor domain, chromodomain, PWWP domains, and Malignant Brain Tumor (MBT) domains (112). These examples reinforce the concept that histone modifications can transduce biological function in a manner independent of their biophysical effect on chromatin fiber; rather, in some cases, recognition of the histone modification by specific readers mediates distinct biological outcomes (63). One possible way to learn how complex chromatin modifications are decoded into distinct biological outcomes in the future is to map the occupancy patterns of modification binders genome wide (113).
Expansion of Histone Modifications
Mammalian chromatin also features more distinct histone modifications than yeast. For instance, gene silencing is enforced not just by histone hypoacetylation, or H3K9me3 (as in Schizosaccharomyces pombe), but is also signaled by H3K27me3 (mediated by the Polycomb repressive complex), by H2A monoubiquitination, or by association with the H2A variant macro-H2A. Curiously, in genomic maps, repressive marks such as H3K27me3 show only modest anticorrelation with transcription (94). Mapping of H3K27me3, H3K9me3, and DNA cytosine methylation in 12 human and mouse cell types revealed that most silent genes are associated with just one of the above modifications (114). This implies that these marks are not used redundantly, but mark silent genes of different functional categories, or genes that are slated for distinct modes of coregulation. Indeed, H3K27me3 occupancy is highly enriched over homeodomain-containing developmental regulators, whereas H3K9me3 and H4K20me3 are highly enriched over zinc finger transcription factors (94, 115), but not globally associated with silent genes. These observations support the idea that histone methylation associated with gene silencing is not merely a consequence of a lack of transcription, but rather reflects active mechanisms that enforce gene silencing toward distinct ends.
CHROMATIN MARK OF ENHANCERS AND INSULATORS
In addition to carrying more histone marks, metazoans also utilize additional chromosomal mechanisms of gene regulation that are scarce in yeast, such as regulation by enhancers, which are regulatory elements that control gene expression from up to hundreds of kilobases away. Enhancers are associated with DNaseI-hypersensitive sites (DHSs), and genome-wide studies have identified thousands of DHSs located distal to genes (and hence unlikely to be promoters) (4, 5, 116). Many distal DHSs have subsequently been confirmed to act as enhancers. Furthermore, genome-scale maps of histone modifications found a distinct chromatin signature of enhancers (90) (Figure 1b). Using the latter criteria, it has been possible to predict the location of enhancers based on their pattern of chromatin modifications, and validate the ability of such sequences to drive gene expression at a distance.
Playing yin to enhancers’ yang is the so-called insulator, a sequence element that prevents communication between enhancer and promoter. Like enhancers, known insulators are associated with DHSs. The best-understood protein component of insulators, the zinc finger factor CTCF, has been mapped across the human genome in human fibroblasts and T cells (94, 117). Surprisingly, binding patterns of CTCF show little variation across cell types. The majority of CTCF binding sites are found in DHSs, and analysis of nucleosome-resolution ChIP-Seq data shows that the nucleosome-free regions at insulators, like those at promoters, are surrounded by well-positioned nucleosomes (118). Phasing can be seen for ~5 nucleosomes on either side of the CTCF binding site, where increasingly distal nucleosomes show decreasingly tight positioning, providing yet more evidence for a “statistical positioning”-type model for nucleosome positioning around NFRs. CTCF binding sites across the genome also tend to have histone modifications typically associated with TSS of active genes, including H3K4me3, H3K4me2, H3K9me1, H2A.Z, and occupancy of Pol II (94). Although the histone modification patterns of enhancers and insulators are useful in predicting novel regulatory elements in the genome, the function of the histone modifications for enhancer or insulator activity has not been directly addressed. Because enhancers and insulators may come into contact with gene promoters via chromosomal looping (119), some of these histone marks may simply reflect the physical proximity of the regulatory elements with the transcriptional machinery.
CHROMATIN AND REPLICATION TIMING
The connection between chromatin states and DNA replication was examined for 1% of the human genome in the ENCODE project (96). The timing of DNA replication genome wide is measured by bromodeoxyuridine (BrdU) incorporation in synchronized cells over multiple time points. Comparison with histone modification data showed a general correlation between active histone modifications (H3 and H4 acetylation, H3K4 methylation) and early replication timing, and between repressive marks (H3K27me3) and late replication timing. These findings are consistent with the notion, obtained previously from single-gene or low-resolution microscopy studies, that euchromatic regions are replicated early whereas heterochromatic regions are late replicating. The difference in replication timing also provides a potential timing-based mechanism for transmission of histone modification states over successive cell generations (120). For chromosomal regions showing replication throughout S phase, many of these encode genes with interallelic differences in expression, suggesting that the replication timing of the two individual alleles tracks with their respective chromatin states.
CELL TYPE–SPECIFIC CHROMATIN STATES
A major contributor to genome-wide chromatin structure in mammalian cells is their developmental states. The differentiation of each of the hundreds of cell types in the human body is associated with, and mediated by, the transcriptional activation and repression of thousands of genes. Genome-scale maps of chromatin state therefore reflect the changing status of gene activities. In one such study, DHS in 1% of the genome across six cell types showed cell type–specific patterns in putative enhancers, whereas ubiquitous distal DHSs were mostly insulators (116). Moreover, comparison of genome-scale chromatin maps of different mammalian cell types reveals the lineage potential of the cells—in other words, their possible future trajectories.
Embryonic stem cell (ESC) differentiation provides an excellent example of the role of chromatin changes in development. ESCs can be differentiated to any cell type in the body. Conversely, differentiated somatic cells may also be reprogrammed back to an ESC-like state by the enforced expression of certain transcription factors (so-called induced pluripotent stem cells, or iPS cells), which provides a model to examine mechanisms involved in cell fate maintenance (121).
In ESCs, many lineage-specific developmental master regulators (often transcription factors) are not transcribed, and thus are described as being poised for transcription in differentiated progenitors. The regulatory regions of these developmental regulators are occupied by “bivalent domains”: broad domains of both H3K27me3 and H3K4me3, modifications normally associated with gene silencing and activation, respectively (122). When ESCs differentiate into lineage-specific progenitors, many (but not all) bivalent domains are resolved in a lineage-specific fashion (91). For example, in neural precursor cells, the bivalent domain over neural-specific regulators resolves into a K4me3-only domain; conversely, regulators of other lineages, such as muscle or liver, become occupied by H3K27me3 but not H3K4me3. Successful reprogramming of somatic cells into iPS results in reconfiguration of chromatin modification patterns to that of authentic ESCs (123, 124), whereas partially reprogrammed iPS cells possess intermediate patterns. The functional importance of bivalent domains has been called into question, however, by the recent finding that the K27 methylase PRC2 may be dispensable for ES cell pluripotency (125).
Global change in chromatin states is not only associated with long-term lineage commitment but also occurs during homeostatic differentiation (126, 127). For example, keratinocytes differentiate and turn over every 28 days. Differentiation genes are silenced in basal keratinocytes by PcG and H3K27me3, and differentiation is signaled by the recruitment of the H3K27 demethylase JMJD3, eviction of PRC2, and removal of H3K27me3 from a subset of activated epidermal differentiation genes (128). Depletion of JMJD3 prevents epidermal differentiation, whereas ectopic expression of JMJD3 triggers precocious epidermal differentiation (without activation of genes indicative of other lineages). Thus, chromatin states appear to have major roles in the developmental plasticity of mammalian cells, and are likely to underlie many cell fate decisions.
SPECIALIZED GENE LOCI
Mammalian genomes encompass many specialized gene loci with distinct chromatin structures that likely play roles in the unusual expression behaviors of genes in these loci (see below). Common features of these specialized gene loci include (a) large chromosomal domains of histone modifications that are exceptions to the typical chromatin structure of a gene (see above), (b) transcription of long non-coding RNAs (ncRNAs) that regulate histone modifications, and (c) binding of the insulator protein CTCF, which can organize chromosomal looping to regulate accessibility to enhancer elements.
X Chromosome Inactivation
A canonical example of a large-scale, specialized chromatin domain is the X chromosome in female mammalian cells. Here, one of the two X chromosomes is transcriptionally silenced by a process termed X chromosome inactivation (XCI) (reviewed in Reference 129). The inactive X is a prototype of constitutive heterochromatin: It is transcriptionally silent, enriched in H3K27me3 and H3K9me3, heavily DNA methylated, late replicating, and cytologically condensed during interphase. Initial choice of XCI among female somatic cells is random—either X can be inactivated. The choice of XCI is dictated by the transcription of a ~15 kilobase ncRNA termed XIST from the future inactive X. XIST binds to and spreads over the inactive X, and initiates H3K27 methylation and silencing of the XCI. Ectopic expression of XIST on a human autosome is sufficient to silence a large contiguous portion, but not the entirety, of the autosome (130). The choice of which X chromosome to inactivate involves interaction between XIST and competing overlapping antisense ncRNAs, mediated by CTCF.
The mechanism of spreading of the X silencing complex is not well understood, but genomic mapping studies in Drosophila and Caenorhabditis elegans have yielded insights into this process (although dosage compensation in these organisms differs in detail from that in mammals). In Drosophila, the MSL dosage compensation complex is recruited to the X chromosome in male cells by the X-specific roX noncoding RNAs (131), where it binds actively transcribed genes and up-regulates their transcription by approximately twofold (potentially because of its activity as a H4K16 acetylase). MSL targeting to actively transcribed genes depends on the binding of subunit MSL3 to the elongation mark H3K36me3, and depletion of H3K36me3 reduces MSL binding to X (132). In C. elegans, dosage compensation occurs by halving transcription across each X chromosome in XX cells, leading to the same transcriptional output as the XO male. The worm dosage compensation complex (DCC) binds discrete foci across the X; these binding foci are distinguished by clustering of a specific DNA sequence motif, but also by proximity to upstream regions of transcriptionally active genes (133). The positive correlation between the level of DCC binding and transcription rates of the neighbor gene suggests the potential use of transcription to tune the level of DCC binding and spreading. In sum, despite the use of distinct molecular machinery, dosage compensation in several species involves the gender-selective targeting and chromatin modification-mediated discontinuous spreading over one or more sex chromosomes.
Allele-Specific Gene Expression
Mammalian somatic cells are diploid; each gene is present in two copies (alleles) on homologous chromosomes, one inherited from each parent. For a subset of genes, termed imprinted genes, only the paternal or maternal allele is active, and each allele is associated with a specialized chromatin state (134). Nonimprinted genes can also be expressed in an allele-specific manner: Specialized genes, such olfactory receptors or immunoglobulin genes, possess intricate mechanisms to ensure monoallelic exclusion, but recent evidence suggests that perhaps ~1000 human genes can demonstrate monoallelic expression (135). If single-nucleotide polymorphisms can be identified between the two relevant alleles, then either microarrays or sequencing can be used along with ChIP to identify allele-specific chromatin states. In one such analysis, 4% of RNA polymerase II occupancy sites in the genome of diploid fibroblasts (>450 genes) showed allele-specific bias (136). Although allele specificity in genome-wide chromatin maps has yet to be fully explored, we can anticipate rich information based on the known importance of chromatin modifications in imprinted genes.
The HOX Loci
All bilaterians have a segmented body plan, in which the HOX homeodomain-containing transcription factors play a major role. In mammals, 39 HOX genes are clustered on four chromosomal loci, and are expressed in a nested segmental fashion along the anterior-posterior and proximal-distal axes of the body, with each additional HOX gene being expressed in increasingly posterior or distal anatomic sites. The pattern of HOX gene expression along the anterior-posterior axis is mirrored by their physical location on the chromosomes, with 3′ HOX genes expressed more anteriorly and 5′ HOX genes more posteriorly. HOX expression is maintained through adulthood—in the skin, dermal fibroblasts from different positional identities maintain features of the embryonic patterns of HOX expression (137, 138), and this adult HOX code is required to drive the site-specific gene expression programs of these cells (139). That cells maintain their positional memory over time—in humans, over decades—predicts the existence of a robust epigenetic system to ensure the faithful transmission of transcriptional memory.
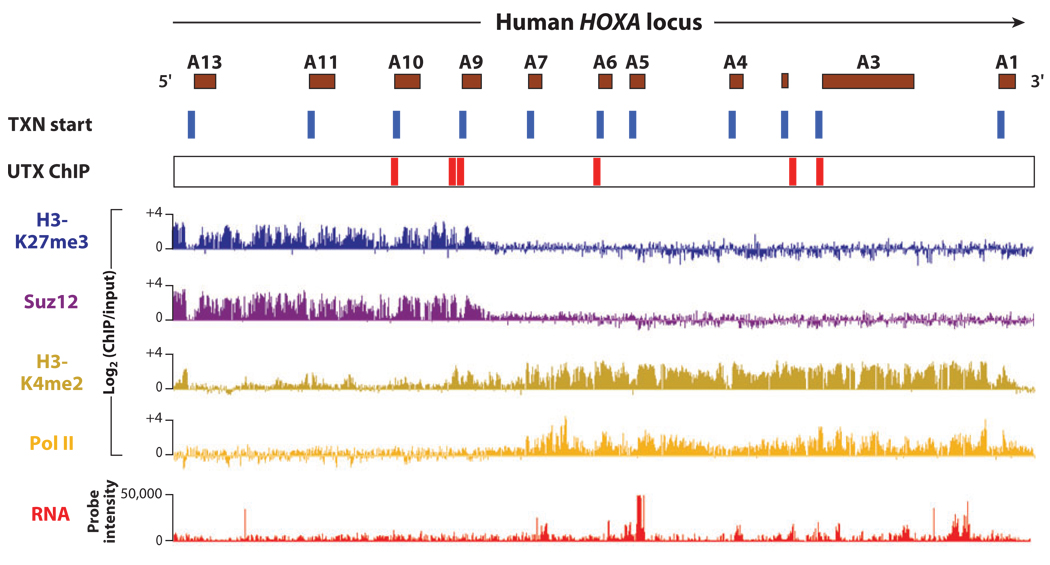
Early genetic studies of homeosis identified mutations that do not affect initial pattern formation, but are required for pattern maintenance (140). The Trithorax (Trx) family of genes are required for continued activation of HOX genes, and encode H3K4 KMT complexes and H3K4me3 binding proteins. Conversely, Polycomb group genes (PcG) are required for continued silence of HOX and other developmental genes and encode at least two main complexes. The Polycomb Repressive Complex 2 (PRC2) is an H3K27 KMT, whereas the Polycomb Repressive Complex 1 (PRC1) recognizes H3K27me3 and mediates H2A ubiquitination and nucleosome compaction. Unlike the chromatin organization at most gene loci, active HOX genes are marked by broad H3K4me3/2 that spans multipleHOX genes and their intergenic regions; these regions are also broadly bound by the Trx protein MLL1 (103). Conversely, the silent HOX genes are occupied by large continuous blocks of H3K27me3 and Polycomb group proteins (104, 105). Further, occupancy of H3K27me3 KDM UTX constitutes an additional, independent layer of regulation by targeting the beginning of HOX genes (Figure 2).
Figure 2.
Chromatin map of the human HOXA locus. The HOX loci are prominent examples of H3K4me3/H3K27me3 bivalent domains in ESCs that resolve upon differentiation. This chromatin map of ~100 kb of HOXA from differentiated lung fibroblasts shows that activated HOX genes and their surrounding ncRNAs and intergenic regions are broadly occupied by H3K4me3/2; the silent HOX genes are broadly occupied by H3K27me3. The resolution of bivalent domains implies that the histone methylation in ESCs must be erased upon differentiation, enzymatically or via histone turnover. Of course, even enzymatic erasure could occur via general (global erasure of all of some mark at a given developmental stage) or specific schemes. The H3K27me3 demethylase UTX does not occupy the HOX in undifferentiated ESCs. Upon differentiation, UTX is focally targeted to the first ~500 bases of multiple HOX genes concomitant with loss of occupancy of PcG proteins and H3K27me3 (106, 153, 154), but this can occur in the midst of broad H3K27me3 domains or H3K4me3/2 domains (106). Reprinted with permission from Reference 106.
How are these long tracks of chromatin modifications established and maintained? In Drosophila, binding elements for Polycomb and Tithorax proteins have been identified (termed PREs), and PcG and Trx proteins are apparently restricted to these sites even though their cognate histone marks, H3K27me3 and H3K4me3, can spread for kilobases around them (140). In contrast, mammalian PREs have yet to be identified, and PcG and Trx proteins broadly occupy the HOX loci. The original genetic studies in flies by Lewis and Hogness identified homeotic mutations that mapped to nonprotein coding regions of the HOX loci, many of which later turned out to generate long noncoding RNAs and microRNAs (see sidebar titled Noncoding RNAs and Long-Distance Targeting of Chromatin Regulators). Drosophila PREs can also be transcribed, which may alter their accessibility and favor Trx binding over PcG binding (141–143). In humans, the four HOX loci harbor a surprisingly large number of noncoding transcripts; there are 39 HOX genes, as compared to 231 transcribed noncoding regions, many of them highly conserved (144). These noncoding RNAs are also expressed in an anatomic position-specific manner, and, importantly, the maintenance of appropriate site-specific HOX chromatin modification and gene expression require the action of HOX ncRNAs. A 2.2-kB HOX ncRNA termed HOTAIR encoded in the HOXC locus binds to the PRC2 complex and is required for PRC2 occupancy, H3K7me3 occupancy, and transcriptional silencing over dozens of kilobases of the HOXD locus on a different chromosome (144). Thus, HOTAIR RNA may guide PRC2 to the HOXD locus to mediate H3K27me3 and gene silencing. However, physical occupancy of HOTAIR on HOXD locus has not yet been shown, and indirect models of PcG positioning by RNA are also possible.
THE HISTONE CODE
The influential phrase histone code embodies three main ideas that cover many of the most interesting features of chromatin structure. First, the original idea that a histone modification could act via recruitment of a modification-regulated binding protein, as proposed by Turner, has been emphatically confirmed over the years (63). Indeed, a universe of modification-regulated histone-binding proteins has been identified, and more are reported almost every month.
Second, Strahl & Allis reused the code metaphor to emphasize that the large number of histone modifications implies a massive number of combinations, each to be recognized by specific readers for unique biological outcomes (62). The combinatorial complexity of histone modifications has been an extremely influential idea; however, the bulk of genomics studies go against it. Virtually all modification-mapping studies reveal extensive correlation between histone modifications, such that only a small subset of the massive number of possible combinations actually occurs in the cell. Even when combinations occur, we may ask whether the combination signals something specific to the cell. Consider H3K4me3 and H3K36me3—these modifications occur at the 5′ end, and the middle/3′ end, of coding regions. So at the +2 or +3 nucleosome of many genes, these patterns overlap, but to date there is no evidence for specialized function of these combinatorially marked K4/K36 nucleosomes. It is our belief that the purpose of histone modification cross talk described by many (148–150) will be best determined using the tools of systems biology and network biology.
Finally, Turner has revisited the code metaphor to argue (as have many others) for heritability of chromatin states (151). As noted in the Introduction, this idea is widely believed to be true but remains unproven (3). While many chromatin regulators are required for inheritance of some gene expression states (140), in many instances it may be other molecules, such as transcription factors, that are the heritable substrate, which re-establish functional chromatin states after each cell division. Whether chromatin structure per se is heritable or not is a question of practical as well as intellectual significance, since the appeal of chemotherapeutics that specifically target epigenetic defects in cancer (152) is that they would only need to be used transiently to reset the heritable information carrier; hence, knowing exactly what is inherited is therefore of paramount interest.
While much of this review has focused on basic types of insight generated by the first generation of chromatin state maps that provide static pictures of cell populations, we anticipate the next generation maps will reveal the even richer dynamics of histone modifications in different scales of space and time.
NONCODING RNAs AND LONG-DISTANCE TARGETING OF CHROMATIN REGULATORS
Transcription of noncoding RNAs is increasingly implicated in alteration of chromatin structure. In S. cerevisiae, examples include cases of direct repression via action of Pol2 in cis (86), repression via RNA-mediated recruitment of Hda1 in cis (145), and silencing of retrotransposons via histone modification in trans (146). In S. pombe, transcription of pericentromeric dg and dh repeats is key to the establishment of silencing, although these RNAs are apparently restricted to function in cis via action of the siRNA exonuclease Eri1 (147).
In mammals, the role of HOTAIR is reminiscent of XIST in initiating the XCI chromatin state. Studies in Drosophila have also suggested specific HOX ncRNAs that support or prevent Trx action in cis (143). However, HOTAIR is unlike XIST in that HOTAIR can work in trans on distantly located genes, suggesting the existence of new mechanisms for genomic targeting of ncRNAs and potentially much broader roles of long ncRNAs in gene regulation. Indeed, recent genomic studies documenting the presence of thousands of long ncRNAs in the genome suggest that they may be major elements shaping the chromatin landscape.
SUMMARY POINTS
Genomic mapping studies are providing unprecedented insight into the structure of chromatin in eukaryotes.
In general, mapping of histone marks yields insight into the mechanism of deposition of the mark, not necessarily the function of the mark.
Mapping studies often reveal stereotypic motifs, which help suggest the mechanism for chromatin state assembly, while deviation from these motifs indicates active regulation.
In yeast, sequence rules and RNA polymerase together account for much of the chromatin structure observed in midlog cultures.
Patterns of histone modifications reveal correlations between many individual modifications, arguing against models for chromatin function that invoke information-rich combinatorial encoding.
Many aspects of chromatin structure are conserved from yeast to human, but human cells exhibit diversification of almost all classes of chromatin regulating machine.
Specialized domains in mammals are more numerous, and often are related to key aspects of multicellularity such as epigenetic inheritance of cell state.
FUTURE ISSUES
Breathtaking advances in genomics technologies will of course continue, such that this review will be partly out of date before it is even published. One key advance will be the development of techniques to measure chromatin structure across the entire genomes in small numbers of cells (such as, for example, oocytes). Another will be the further exploration of genomic methods to determine folding of the “beads on a string,” particularly secondary structure characteristics such as the 30-nm fiber. Intellectually, the function served by having correlated groups of histone modification will be a fruitful area for investigation for some time. Finally, understanding how and where (and if) chromatin states are in fact the substrate for epigenetic inheritance will be of great mechanistic interest and may help guide cancer therapies of the future.
ACKNOWLEDGMENTS
We apologize to authors whose work was not cited due to space constraints. O.J.R. is supported in part by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. This research was supported by grants to O.J.R. and H.Y.C. from the National Institutes of Health, and to H.Y.C. from the American Cancer Society, California Institute for Regenerative Medicine, Emerald Foundation, and Damon Runyon Cancer Research Foundation.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 2.Olins AL, Olins DE. Spheroid chromatin units (v bodies) Science. 1974;183:330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 3.Ptashne M. On the use of the word ‘epigenetic’. Curr. Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 4.Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 5.Crawford GE, Davis S, Scacheri PC, Renaud G, Halawi MJ, et al. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nat. Methods. 2006;3:503–509. doi: 10.1038/NMETH888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- 10.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 11.Lee W, Tillo D, Bray N, Morse RH, Davis RW, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 12.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 13.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–1528. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson JD, Widom J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. Mol. Cell. Biol. 2001;21:3830–3839. doi: 10.1128/MCB.21.11.3830-3839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson HC, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 19.Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl. Acad. Sci. USA. 1985;82:8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2008;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottesfeld JM, Bloomer LS. Nonrandom alignment of nucleosomes on 5S RNA genes of X. laevis. Cell. 1980;21:751–760. doi: 10.1016/0092-8674(80)90438-9. [DOI] [PubMed] [Google Scholar]
- 23.Young D, Carroll D. Regular arrangement of nucleosomes on 5S rRNA genes in Xenopus laevis. Mol. Cell. Biol. 1983;3:720–730. doi: 10.1128/mcb.3.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nat. Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 25.Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, et al. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput. Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, et al. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 30.Almer A, Horz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korber P, Horz W. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J. Biol. Chem. 2004;279:35113–35120. doi: 10.1074/jbc.M405446200. [DOI] [PubMed] [Google Scholar]
- 32.Lam FH, Steger DJ, O’Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- 34.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 35.Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 36.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat. Struct. Mol. Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 39.Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 41.Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 42.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol. Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 44.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 45.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol. Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 47.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, et al. Noise in protein expression scales with natural protein abundance. Nat. Genet. 2006;38:636–643. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 48.Kulaeva OI, Gaykalova DA, Studitsky VM. Transcription through chromatin by RNA polymerase II: histone displacement and exchange. Mutat. Res. 2007;618:116–129. doi: 10.1016/j.mrfmmm.2006.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 50.Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr. Opin. Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 52.Choi ES, Shin JA, Kim HS, Jang YK. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res. 2005;33:7102–7110. doi: 10.1093/nar/gki1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 54.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, et al. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shia WJ, Li B, Workman JL. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 2006;20:2507–2512. doi: 10.1101/gad.1439206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, et al. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:974–975. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 62.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 63.Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. [PubMed] [Google Scholar]
- 64.Turner BM. Histone acetylation and an epigenetic code. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 65.Turner BM. Defining an epigenetic code. Nat. Cell Biol. 2007;9:2–6. doi: 10.1038/ncb0107-2. [DOI] [PubMed] [Google Scholar]
- 66.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are trimethylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 67.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 68.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 70.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 71.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 72.Zhang K, Lin W, Latham JA, Riefler GM, Schumacher JM, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 74.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 75.Masumoto H, Hawke D, Kobayashi R, Verreault A. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature. 2005;436:294–298. doi: 10.1038/nature03714. [DOI] [PubMed] [Google Scholar]
- 76.Kaplan T, Liu CL, Erkmann JA, Holik J, Grunstein M, et al. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4:e1000270. doi: 10.1371/journal.pgen.1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsubota T, Berndsen CE, Erkmann JA, Smith CL, Yang L, et al. Histone H3-K56 acetylation is catalyzed by histone chaperone-dependent complexes. Mol. Cell. 2007;25:703–712. doi: 10.1016/j.molcel.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc. Natl. Acad. Sci. USA. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 81.Kurdistai SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 82.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 83.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 84.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, et al. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 86.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 87.Radwan A, Younis A, Luykx P, Khuri S. Prediction and analysis of nucleosome exclusion regions in the human genome. BMC Genomics. 2008;9:186. doi: 10.1186/1471-2164-9-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 90.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 91.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 93.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 95.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, et al. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 101.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 103.Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc. Natl. Acad. Sci. USA. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 106.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 107.Wang J, Scully K, Zhu X, Cai L, Zhang J, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 108.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 109.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ramon-Maiques S, Kuo AJ, Carney D, Matthews AG, Oettinger MA, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl. Acad. Sci. USA. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 113.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Komashko VM, Acevedo LG, Squazzo SL, Iyengar SS, Rabinovich A, et al. Using ChIP-chip technology to reveal common principles of transcriptional repression in normal and cancer cells. Genome Res. 2008;18:521–532. doi: 10.1101/gr.074609.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.O’Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, et al. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fu Y, Sinha M, Peterson CL, Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000138. e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 120.Zhang J, Xu F, Hashimshony T, Keshet I, Cedar H. Establishment of transcriptional competence in early and late S phase. Nature. 2002;420:198–202. doi: 10.1038/nature01150. [DOI] [PubMed] [Google Scholar]
- 121.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 122.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 123.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 125.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 127.Attema JL, Papathanasiou P, Forsberg EC, Xu J, Smale ST, Weissman IL. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc. Natl. Acad. Sci. USA. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22:1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huynh KD, Lee JT. X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nat. Rev. Genet. 2005;6:410–418. doi: 10.1038/nrg1604. [DOI] [PubMed] [Google Scholar]
- 130.Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl. Acad. Sci. USA. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oh H, Park Y, Kuroda MI. Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev. 2003;17:1334–1339. doi: 10.1101/gad.1082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol. Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 133.Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat. Genet. 2007;39:403–408. doi: 10.1038/ng1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell. 2007;27:353–366. doi: 10.1016/j.molcel.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 136.Maynard ND, Chen J, Stuart RK, Fan JB, Ren B. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat. Methods. 2008;5:307–309. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- 137.Chang HY, Chi J-T, Dudoit S, Bondre C, van de Rijn M, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. USA. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22:303–307. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 141.Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 143.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, et al. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 146.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional transsilencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 148.Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 149.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nelson CJ, Santos-Rosa H, Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- 151.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 152.Brown R, Strathdee G. Epigenomics and epigenetic therapy of cancer. Trends Mol. Med. 2002;8:S43–S48. doi: 10.1016/s1471-4914(02)02314-6. [DOI] [PubMed] [Google Scholar]
- 153.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 154.Lee MG, Villa R, Trojer P, Norman J, Yan KP, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]