Abstract
Xenbase (www.xenbase.org), the model organism database for Xenopus laevis and X. (Silurana) tropicalis, is the principal centralized resource of genomic, development data and community information for Xenopus research. Recent improvements include the addition of the literature and interaction tabs to gene catalog pages. New content has been added including a section on gene expression patterns that incorporates image data from the literature, large scale screens and community submissions. Gene expression data are integrated into the gene catalog via an expression tab and is also searchable by multiple criteria using an expression search interface. The gene catalog has grown to contain over 15 000 genes. Collaboration with the European Xenopus Research Center (EXRC) has resulted in a stock center section with data on frog lines supplied by the EXRC. Numerous improvements have also been made to search and navigation. Xenbase is also the source of the Xenopus Anatomical Ontology and the clearinghouse for Xenopus gene nomenclature.
The African frogs Xenopus laevis and X. (Silurana) tropicalis serve as a powerful and widely used model system for exploring gene function in vertebrate development. Both species have unique experimental advantages and combining data from the two synergistically enhances their usefulness. Many of the genes and fundamental mechanism governing early embryonic development were first discovered in the Xenopus system. Xenopus females generate thousands of embryos in response to a simple hormone injection and the large robust embryos are simple to microinject with mRNA or anti-sense reagents. As the embryos develop a full set of differentiated organs within days of fertilization, very rapid high-throughput screens of gene function can be performed. Xenbase (www.xenbase.org) is a free, publicly accessible resource, for Xenopus data, amalgamating data from both of these Xenopus species and linking to both Xenopus and other model organism resources. Xenbase also manages the Xenopus Anatomical Ontology (XAO) (1) and acts as a clearinghouse for Xenopus gene names. Additionally, Xenbase maintains the biological data for the European Xenopus Resource Centre (EXRC).
Xenbase has been growing. Since our last report in 2008 (2), the count of genomic features in Xenbase has grown from 486 000 records to 4 750 000, including ESTs, cDNA clones, mRNA, Ensembl (3) gene models and alignment matches. 26 000 images have been added and Xenopus publications have increased from 35 000 to 38 000.
The initial release of Xenbase included a gene catalog, the GBrowse genome browser (4), a literature compendium, a community directory, a blast search and extensive links to orthologous species gene records at NCBI and other model organism databases. Since then, efforts have focused on integration of the literature and the gene catalog, incorporating gene expression data, expanding the gene catalog, adding support for the EXRC and making numerous improvements to search and usability.
LITERATURE, LINK MATCHING AND INTERACTANTS
The literature section is now much more strongly leveraged in Xenbase. As a result of the development of a link-matching system, it has been possible to integrate the literature section with the gene catalog and mine it for potential interactants. Finally, through agreements with publishers, the literature section has become a key part of Xenbase’s gene expression coverage by incorporating published images that illustrate gene expression patterns.
With 38 000 articles in Pubmed mentioning Xenopus, complete manual curation was impossible. A link-matching system was developed that searches for the presence of gene symbols, synonyms and names in the titles, abstracts and image captions of published papers. These terms are extracted from surrounding punctuation and exclusions are made for common terms that would cause too many false matches (e.g. not). The link matcher searches new articles for gene symbols, names and synonyms on a weekly basis.
Link matching allows us to determine which papers are associated with particular genes. Matched papers are listed in the literature tab of each gene page (Figure 1). Correspondingly, gene symbols, synonyms and names are hyperlinked inside article titles, abstracts and captions. In fact, gene matches are hyperlinked in article titles wherever they appear in Xenbase. This allows easy navigation back and forth between the gene catalog and publications.
Figure 1.
Literature tab. For each gene page the literature tab stores each publication citing the genes name, symbol or synonyms. Records are counted (see tab) and parsed for various key words, such a morpholino, to aid in literature sorting.
The linkages between genes and articles have also been exploited to create an interactants tab in the gene catalog. The interactants tab is generated by using the link-matching system to determine when multiple genes are cited in a paper. Potential interactants are ordered from those with the most co-citations to those with the least and links are provided to the source papers.
Finally, we have expanded our use of the image scraping system that allows curators to automatically extract images and captions from the online version of journals. Cell, Current Biology, Development, Developmental Biology, Developmental Cell, Developmental Dynamics, Mechanisms of Development and Proceedings of the National Academy of Sciences allow Xenbase to display images depicting gene expression.
GENE EXPRESSION
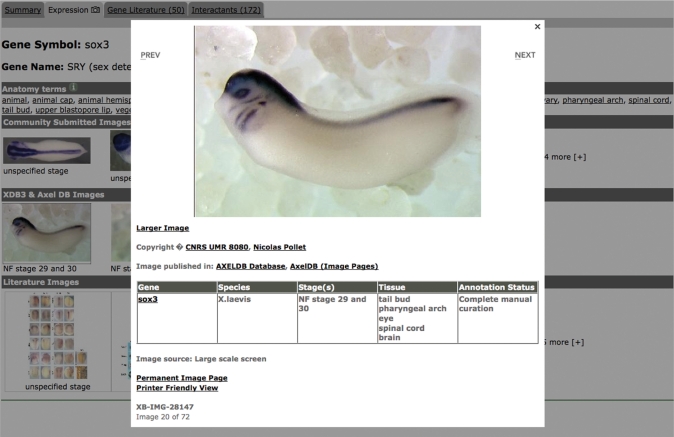
Support for gene expression data is the largest expansion of Xenbase. Gene expression data can be accessed in two ways: via the expression tab of the gene catalog and a new expression search. The expression tab lists the anatomy terms and expression stages where evidence of expression exists for each gene. Any expression thumbnail images are organized into categories by their source and are ordered from the latest to the earliest development stage. Clicking on an expression thumbnail will display a modal window with the standard-sized version of the image (Figure 2). This window also contains a chart summarizing any associated annotations such as the development stages (5) when expression occurs and XAO terms for tissues where expression occurs. Additional information may include a caption, copyright information, a link to a large image and a permanently linkable page. Indeed, clicking on an image anywhere in Xenbase will pull up a modal image box with this information. Users can navigate directly to the previous or next image in the series from the expression tab by clicking on the right or left side of the modal dialog box.
Figure 2.
Modal dialog of gene expression information. When a thumbprint image is selected by the user, a modal dialog box is launched illustrating an enlarged view and additional data.
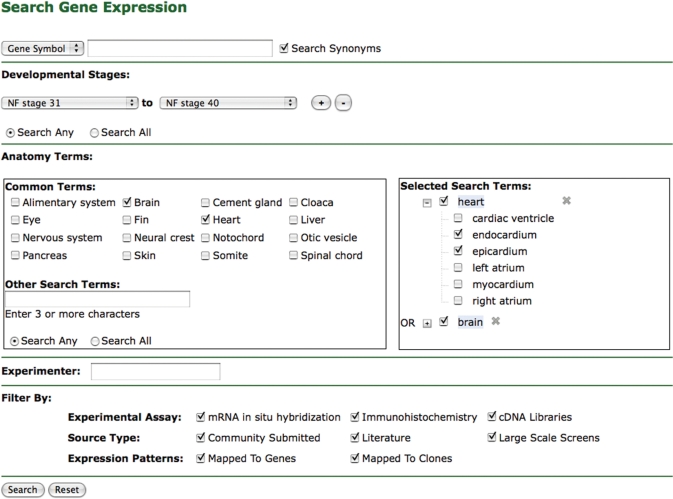
An expression search feature complements the expression tab by allowing users to search for expression patterns using a broad criteria and find expression patterns for clones not yet mapped to a gene. Users can search expression by a combination of gene name, development stages, XAO anatomy terms, data source type, experimenter and assay type or source type (Figure 3).
Figure 3.
Expression search interface. Over 500 000 gene expression objects can be searched using a variety of criteria.
To enhance usability, users can select XAO search terms by clicking checkboxes of commonly searched terms or by using a suggestion box that allows users to search for tissues by their XAO term or common synonyms. A suggestion box lists possible match terms as the user types. When the user selects an anatomy term, it appears in a list to the right of the search box. Users can drill-down to more specific terms by clicking the plus sign to the left of a tissue and examine parts of the tissue. In this manner, the user can find specific search terms without strong knowledge of the anatomy ontology.
After executing the search, the user is presented with a list of expression pattern results organized by the gene (or clone, if unmapped). The results include which stages and XAO terms were matched for each gene or clone. Searching for a particular tissue will return items annotated to parts of that tissue. For example, a search for pronephric kidney will also return results annotated to the glomerulus. From the initial search results, the user can drill-down to experiments matching their search criteria and then detailed information on each experiment.
Xenbase gene expression evidence is drawn from expressed sequence tags (ESTs), in situ hybridization and immunohistochemistry assays. For EST evidence, genes are aligned to ESTs from particular tissues at specific development stages. Gene expression images come from three sources: literature, large scale screens and community submissions. Images extracted from papers with the permission of publishers are manually curated and associated with genes, tissues and development stages based on information in the image caption and article content.
For older literature that we have scraped images from, we have performed an automated first pass of curation. Using the link-matching system, we identify gene names and synonyms in the captions. If a single gene is mentioned in the caption we also search for the use of XAO tissue terms or their synonyms. We then infer that the single gene mentioned must be expressed in all of the identified tissues. Development stage descriptions vary too much to extract them using automated means. Therefore, stages for these expression patterns are set to ‘unspecified’. While imperfect, this process allows initial associations between uncurated literature gene expression images and genes (and possibly XAO tissues). These literature images will be manually curated as time permits.
The largest block of expression images come from two large in situ screens AxelDB (6) and XDB3 (Naoto Ueno, NIBB, Okazaki, Japan), consisting of 2600 and 18 600 images, respectively. The Axeldb images are annotated by the development stage and often contain tissue annotation. The XDB3 images were only annotated with development stages. For both of these screens the gene involved has been determined by aligning the sequences for the probes used with mRNA for Xenbase gene catalog entries.
The last source of images is community-submitted images, of which there are currently over 2000. These images are submitted with at least curation of stage and accessions for the sequences used to generate probes. These clone sequences are aligned with mRNAs in our gene catalog to create gene associations. We would like to encourage users to submit their gene expression image data. To this end, there is a submit data button on each Xenbase page where users can upload data files—this includes an optional template spreadsheet for entering image annotation.
The alignment threshold used by Xenbase to align clone sequences from expression experiments (e.g. ESTs) to gene catalog mRNAs is a blast hit with a maximum e-value of 1e−20, a minimum 90% identity and 65% alignment. This methodology does leave the possibility that a single probe may be incorrectly associated with two genes with similar sequences. In these rare cases, a curator chooses the correct assignment.
GENE CATALOG
The gene catalog remains at the core of Xenbase. In addition to the new literature, interactants and expression tabs described above, several improvements have been made to the gene page summary tab itself. Most significantly, the contents of the gene pages have been greatly expanded. New links to additional related data sources include: Ensembl, UniProtKB, Affymetrix and GEO (7). The literature summary section also provides links to papers that specifically discuss morpholino loss of function data. There are also links to several clone suppliers. Finally, the gene page summary tab now supports laevis gene duplicates (e.g. sox17a). However, so far, only a small number have been identified.
The most important expansion of the gene catalog has been through the growth in the number of gene pages that has grown from an initial set of 1934 human-annotated gene pages to 4705 human-annotated gene pages and 10 833 machine-annotated gene pages. Of the 4705 human-annotated genes, there are 4159 with synteny to other organisms determined via Metazome, our primary method for establishing orthology. Another 546 human-annotated genes have sequence similarity with another organism but no synteny evidence or are novel genes waiting for Human Genome Nomenclature Committee approval. (Xenbase uses human nomenclature to facilitate accessibility to the broader research community.) The 10 833 machine-annotated gene catalog entries are marked as ‘provisional- machine annotated only’. Of these, 9264 gene catalog entries were added based on sequence similarity to a human gene based on a maximum e-value of 1e−10 with a minimum 55% identity and 65% coverage. Their symbols and names have the stipulation that they were machine-predicted matches using Entrez gene. The remaining 1558 genes were added based on synteny evidence to some other organism. Their symbols are set as unnamed and their names are set as the Metazome homology description.
STOCK CENTER
Xenbase has entered into an agreement to provide bioinformatics support for the EXRC. At the current time, Xenbase supports a catalog of Xenopus lines carried by the EXRC.
IMPROVED SEARCH, NAVIGATION AND BLAST
To improve users’ experience we have made numerous improvements to Xenbase search and navigation. Suggestions have been added to the mini search bar. Terms matching database records are provided as the user types their search term. An advanced gene search now allows users to search the gene catalog by any combination of gene symbol, names, synonyms, ortholog symbols, function and OMIM, GO, Affymetrix or Xenbase IDs. We have also introduced a beta ‘search all’ to the minibar that allows users to search all dynamic and static content from one search box. Finally, the Xenbase gene catalog is now indexed and can be searched via Google.
In addition to added search capability, we have added navigation controls to search results, allowing users to page forward and backward through query search results. We have also made extensive efforts to enhance the speed of Xenbase queries. We perform constant database tuning and make use of techniques such as materialized views to ensure that database queries are sufficiently fast. We currently target under 5 s for page load times. The current average onsite gene search time and gene page load time are 4 and 2.5 s, respectively. Our dedicated blast search has been placed on faster servers and provides almost instant searches of Xenopus sequence data.
XENBASE AND THE COMMUNITY
Xenbase provides stewardship over the XAO and acts as a clearinghouse for Xenopus nomenclature. As a community-directed database, we take feedback from a variety of community sources: an advisory board, the Xenopus steering committee, community surveys, feedback at community meetings and requests sent to us via email or our feedback and ‘submit data’ links. Xenbase annotations are used as inputs into NCBI Entrez gene and Uniprot Xenopus records. Xenbase also participates in the Phenoscape project, Amphibanat, Open Biological Ontologies and Generic Model Organism Database (GMOD).
FUTURE DIRECTION
Our primary future direction will place a stronger emphasis on manual curation. This curation will focus on gene expression, literature and phenotypes. We will also start generating A and B versions of laevis genes by using both published gene pair sets (8,9) and our own pair predicting system. Also, the ability to search gene expression data by sequence (10) will be added to the expression search. To improve community participation, a Xenopus Wiki will be integrated with Xenbase. Further work will focus on developing support for phenotypes, microarrays NextGen seqeunce data and ultimately gene regulation networks.
FUNDING
National Institute of Child Health and Human Development (R01HD045776 for Xenbase); Wellcome Trust and the Biotechnology and Biological Sciences Research Council via collaboration with the EXRC (BB/F020627/1) for stock center section. Funding for open access charge: University of Calgary Libraries and Cultural Resources.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are indebted to the University of Calgary Department of Computer Science for systems support, space, infrastructure and user of servers for operating our Blast search. Large scale in situ data was generously provided by Nicolas Pollet and Naoto Ueno.
REFERENCES
- 1.Segerdell E, Bowes JB, Pollet N, Vize PD. An ontology for xenopus anatomy and development. BMC Dev. Biol. 2008;8:92. doi: 10.1186/1471-213X-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowes JB, Snyder KA, Segerdell E, Gibb R, Jarabek C, Noumen E, Pollet N, Vize PD. Xenbase: a xenopus biology and genomics resource. Nucleic Acids Res. 2008;36:D761–D767. doi: 10.1093/nar/gkm826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin) New York: Garland; 1994. [Google Scholar]
- 6.Pollet N, Schmidt HA, Gawantka V, Vingron M, Niehrs C. Axeldb: a Xenopus laevis database focusing on gene expression. Nucleic Acids Res. 2000;28:139–140. doi: 10.1093/nar/28.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chain FJ, Ilieva D, Evans BJ. Duplicate gene evolution and expression in the wake of vertebrate allopolyploidization. BMC Evol. Biol. 2008;8:43. doi: 10.1186/1471-2148-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellsten U, Khokha MK, Grammer TC, Harland RM, Richardson P, Rokhsar DS. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilchrist MJ, Christensen MB, Harland R, Pollet N, Smith JC, Ueno N, Papalopulu N. Evading the annotation bottleneck: using sequence similarity to search non-sequence gene data. BMC Bioinform. 2008;9:442. doi: 10.1186/1471-2105-9-442. [DOI] [PMC free article] [PubMed] [Google Scholar]