Abstract
JASPAR (http://jaspar.genereg.net) is the leading open-access database of matrix profiles describing the DNA-binding patterns of transcription factors (TFs) and other proteins interacting with DNA in a sequence-specific manner. Its fourth major release is the largest expansion of the core database to date: the database now holds 457 non-redundant, curated profiles. The new entries include the first batch of profiles derived from ChIP-seq and ChIP-chip whole-genome binding experiments, and 177 yeast TF binding profiles. The introduction of a yeast division brings the convenience of JASPAR to an active research community. As binding models are refined by newer data, the JASPAR database now uses versioning of matrices: in this release, 12% of the older models were updated to improved versions. Classification of TF families has been improved by adopting a new DNA-binding domain nomenclature. A curated catalog of mammalian TFs is provided, extending the use of the JASPAR profiles to additional TFs belonging to the same structural family. The changes in the database set the system ready for more rapid acquisition of new high-throughput data sources. Additionally, three new special collections provide matrix profile data produced by recent alternative high-throughput approaches.
INTRODUCTION
The wide availability of TF affinity data is becoming essential for an increasing number of research efforts to understand gene regulation in the post-genomic era. The increasing amount of assembled genome sequences, transcriptome data (1), as well as high-throughput studies revealing genome-wide locations of core promoters (2) and enhancer elements (3,4) have resulted in the greatest demand for TF binding site content analyses.
TF binding affinities are typically modeled as position frequency matrices (PFMs, also known as raw count matrices or simply binding profiles), summarizing nucleotide counts in an alignment of active binding sites These can be used to scan genomes for new binding sites (5). Since the first official release of JASPAR in 2004 (6), the research community has embraced it as the leading open-access database of such matrix profiles for TF binding sites. From the beginning, the aim of its core collection has been to provide a non-redundant set of curated, high-quality matrix profiles derived from experimental binding data in the form of position frequency matrices (7); in other words, the goal is to present the best currently available DNA binding model for a given TF, decided by expert curators.
The availability of potentially useful matrices derived by other means (e.g. using a number of genome-wide computational approaches) as well as non-TF binding profiles, prompted the addition of separate JASPAR Collections in the second release (8): the intention was to provide those matrix profiles in the same format and hence usable with the same tools as the core JASPAR database, while keeping the latter reserved for profiles representing experimentally derived data.
While the community has valued the open-access policy and non-redundant nature of JASPAR, a common complaint was that the size of the core collection was small compared to the commercial TransFac database (9), currently the only comprehensive alternative to JASPAR. In this update, our goal was to make this gap smaller by performing a major expansion of the core database, while maintaining the popular non-redundant, curated quality. As a result, this fourth major release introduces a wealth of new and improved matrix profiles and represents the largest expansion of the core database since its inception, with new data coming either from high-throughput methods like Chip-seq, or assembled from TF binding site databases particularly PAZAR (10) described below.
NEW AND IMPROVED MATRIX PROFILES IN JASPAR CORE DATABASE
Profiles from ChIP-seq
Several recent genome-wide studies have revealed thousands of TF binding sites for individual TFs. Compared to the original matrices, the larger number of representative target sequences provides potentially more accurate profiles and brings the added benefit that (unlike in DNA SELEX), all the binding sites come from the actual genome sequence to which the TFs in question are bound in vivo.
To make the derivation of matrices uniform, we extracted the original sets of bound regions from published experiments (11–19). We retrieved 200 bp sequences centered on each peak and performed de novo motif discovery on them using parallelized MEME (20) on a Cray XT4 supercomputing platform, which can handle inputs of many thousands of sequences in manageable time. In most cases, the resulting matrices closely resemble those reported in the original publications, produced using various motif discovery tools. The single exception was the Zfx profile, where our profile obtained with MEME from sites reported in (13) differed reproducibly from the profile reported therein. In this case, we chose to include the newly derived matrix.
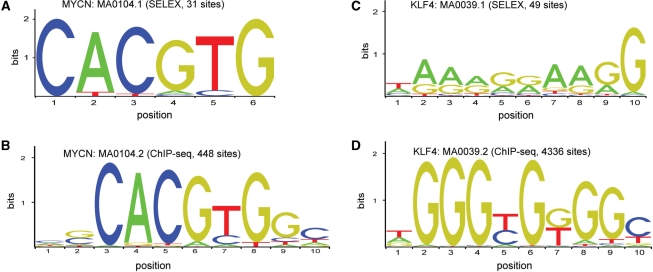
In most cases, the ChIP-seq data resulted in improved matrices with higher information content than the original ones derived from either compiled single promoter assays or from DNA SELEX (Figure 1). This contradicts the widely held view that SELEX is prone to producing over-specified models since many selection rounds are commonly used. Also, somewhat surprisingly, the resulting matrices did not differ much as thresholds were varied for the inclusion of ChIP identified regions (e.g. top 100 highest confidence bound regions versus top 1000).
Figure 1.
Examples of SELEX-derived matrix profiles replaced by ChIP-seq-derived profiles. (A) The previous MYCN matrix profile (MA0104.1) derived by DNA SELEX. (B) The new MYCN profile (MA0104.2) derived from ChIP-seq binding shows general agreement with the SELEX profile, with additional information derived from hundreds of sites at flanking positions. (C) The previous KLF4 profile (MA0039.1) is an example of SELEX-derived profile that did not correspond well to the handful of individually characterized KLF4 sites. (D) The new KLF profile derived from ChIP-seq data (13) shows a dramatic increase in information content and a good agreement with individually characterized binding sites.
Profiles from ChIP-chip experiments
The ChIP-chip derived TF binding sites, while not providing the resolution of the ChIP-seq data, are a rich source of binding data. Even though they are currently being superseded by ChIP-seq (21), the published sets contain a number of high-quality binding data currently unavailable in the ChIP-seq version. As with ChIP-seq, we use the enriched regions reported by the authors of the study in question, and then apply MEME to find the pattern.
Yeast profiles in core collection
Previous versions of JASPAR did not include any matrix profiles for yeast TFs. Responding to community requests, we have compiled results from several large-scale binding profile projects to produce a non-redundant set of matrix profiles for TFs from Saccharomyces cerevisiae. The sources used, in order of preference, were a recent in vitro binding screen (22), a protein-binding microarray (PBM) experiment (23), the compiled SCPD binding profile database (24), the SwissRegulon computational re-analysis of multiple data collections (25) and a motif discovery-based collection from a widely used ChIP-chip data collection (26). The prioritization of the contributions, as well as the indicated deviations, reflect the curators’ personal perspective. The preferred set, from Badis et al. (22), appeared to offer matrices of consistently high-quality, likely reflecting the curated nature of the effort (new experimental data were compared against existing data for consistency). All matrices were manually curated to remove redundancies and converted to count matrices. In curating the collection, the curators identified a few instances in which profiles were preferred in contradiction with the source priority: GAL4 (SwissRegulon), GCR1 (SwissRegulon), MATALPHA2 (SCPD), PHO4 (UniProbe with the six leftmost and rightmost nucleotides trimmed) and ROX1 (SCPD). The resulting non-redundant set represents a comprehensive open-access compilation of yeast binding profiles, facilitating genome-wide computational studies of yeast regulatory inputs. We are grateful to the commitment of all of the data providers to open information, without which the compilation would have been impossible.
New literature-based profiles from PAZAR
Recently, annotations of hundreds of experimentally validated TF binding sites from published studies have accumulated in the PAZAR database (27), allowing us to produce additional matrices similar in nature to the original JASPAR release (DNA SELEX or compiled from multiple studies on individual binding sites). The PAZAR database was mined to identify TFs with more than 15 annotated binding sites. The resulting data was manually curated, selecting only the results from the most high-quality data collections (i.e. collections manually annotated from the literature by specialists) and discarding any redundant sequences to build the profiles. The resulting set of compiled binding sites for each TF was used as input to the MEME software to obtain a profile. If non-informative positions were obtained on the edges of the matrices, the profiles were trimmed accordingly.
Additional model organism core profiles
For this new release, two major sources of Drosophila melanogaster matrix profiles have been used: DNaseI footprinting data by Bergman et al. (28) and bacterial one-hybrid data by Wolfe and colleagues (29–31). The profiles from these data sets have been curated by the authors to remove redundancies among the results and with the existing profiles in the previous version of JASPAR database. In addition, any profile based on less than 10 sequences has been discarded. This new insect sub-section of JASPAR core includes 123 curated profiles; however, these are heavily dominated by the homeodomain profiles (29). For Caenorhabditis elegans, no large sources of data are currently available. Through literature searches, we identified only five profiles suitable for inclusion in the core database (32–36).
In summary, the JASPAR core database now numbers 457 non-redundant matrix profiles (Table 1). New core profiles are summarized in Supplementary Table S1.
Table 1.
Summary of the content and growth of the JASPAR database
| JASPAR | Brief description | Subset | Number of profiles in JASPAR 3.0 | New profiles in JASPAR 4.0 | Updated profiles | Removed profiles | Total profiles (including all versions) | Total profiles (non- redundant) |
|---|---|---|---|---|---|---|---|---|
| Core | ||||||||
| Non-redundant, literature-derived, curated models | Vertebrates | 101 | 29 | 16 | 1 | 145 | 130 | |
| Plants | 21 | – | – | 21 | 21 | |||
| Insects | 14 | 109 | 1 | – | 124 | 123 | ||
| Nematoda | – | 5 | – | – | 5 | 5 | ||
| Fungi | – | 177 | – | – | 177 | 177 | ||
| Urochordata | 1 | – | – | – | 1 | 1 | ||
| Total core | 137 | 321 | 17 | 1 | 474 | 457 | ||
| Collections | ||||||||
| POLII | Core promoter element profiles | – | 13 | – | – | 13 | 13 | |
| FAM | Familial ‘consensus’ profiles for major structural families of transcription factors | – | 11 | – | – | – | 11 | 11 |
| CNE | Profiles overrepresented in vertebrate highly conserved non-coding elements | – | 233 | – | – | – | 233 | 233 |
| PHYLOFACTS | Evolutionary conserved profiles in 5′ promoter regions | – | 174 | – | – | 174 | 174 | |
| SPLICE | Splice sites | – | 6 | – | – | – | 6 | 6 |
| PBM | Protein binding microarray profiles | – | – | 208 | – | – | 208 | 208 |
| PBM_HOMEO | Protein binding microarray profiles focused on homeodomain TFs | – | – | 176 | – | – | 176 | 176 |
| PBM_BHLH | Protein binding microarray profiles focused on bHLH domain TFs | – | – | 19 | – | – | 19 | 19 |
| Total collections | 437 | 403 | – | – | 840 | 840 |
NEW COLLECTIONS
In addition to the expansion of the core database, we remain committed to providing other collections of matrix profiles within JASPAR.
Recently, the PBM technology has emerged as a new in vitro method for the characterization of TF binding affinities (37). The UniPROBE database hosts the PBM datasets and makes the derived matrix profiles available to the community (38). We have selected three of these new datasets as new collections in JASPAR:
PBM, the set derived by (39) from binding preferences of 104 mouse TFs. For each TF, both the primary and secondary motifs identified in the study were incorporated.
PBM_HOMEO, the set derived by (40) includes 176 profiles from mouse homeodomains. From the original 168 TFs analyzed, two were discarded because they could not be identified (Dobox4 and Dobox5) and ten have two alternative profiles.
PBM_HLH, the set derived from binding preferences of dimers of C. elegans bHLH TFs, including nine homodimers and ten heterodimers (41).
With these additions, JASPAR now holds 840 profiles within collections outside of the core database.
GENERAL ORGANIZATIONAL CHANGES
Version control and taxonomic catagories
In line with our goal of presenting the best currently available binding model for any TF, we updated some previous JASPAR entries motivated by new available data. Seventeen entries of the previous release were updated. The replacement of existing matrices with the new ones led us to the introduction of version numbers in matrix IDs, in a manner equivalent to the management of sequence versions in GenBank. For example, the old GATA1 profile MA0035 is replaced with a new one, and the full identifier of the new matrix is MA0035.2, while the old one becomes MA0035.1. By default, the latest version of non-redundant database includes the latest version of each profile. A search for ‘MA0035’ also retrieves the newest version, with an option to view older versions. Older versions can also be downloaded from the JASPAR web site.
The addition of 177 yeast matrices to the core collection means that the JASPAR matrices now span the entire eukaryote crown group. Even before that, a typical user scenario included the selection of only a subset of matrices derived from a particular taxonomic category of organisms, across which the TFs are strictly orthologous and their binding activities largely unchanged (e.g. vertebrates). For that reason, both the JASPAR web interface and the download section now present the database content split into major taxonomic categories—vertebrates, insects, nematodes, (higher) plants and fungi—within which most of the binding sites are transferable across species. The option to search with and download the entire core collection is still available and behaves as before.
A standardized TF classification
Up to now, JASPAR used an ad hoc structural class annotation for the TFs associated with each matrix profile. In this release, we have updated the structural class annotation using our recently published catalog for mouse and human TFs (42) in which DNA binding proteins are associated with a structural classification system. We adopted the two-level classification described by Luscombe et al. (43) and extended it to accommodate additional binding domain structures. For the TFs from other species, we extrapolated the structural class and family based on the PFAM annotation of the DNA-binding domains. This addition to JASPAR provides a standardized system for the classification of TFs and allows a better grouping into families (or sub-families) with potentially similar binding preferences. A curated list of putative mouse/human DNA-binding proteins is provided at the JASPAR web site. It is also possible to browse the catalog by structure, to see what profiles that are available within the web interface.
Changes in the underlying database structure and interface
The underlying database schema was updated to accommodate matrix versions and to allow multiple species and TF accession numbers, as well to allow the storage of multiple collections in the same sql database. A Perl API (JASPAR5) for the new schema is available as part of the open-source TFBS Perl framework (44).
FUTURE DEVELOPMENTS
In the forthcoming months and years, a large amount of whole-genome binding data from ChIP-seq and related techniques will become available. We have created the first steps towards a standardized way of including this new data into JASPAR, which is expected to expand significantly with the concomitant increase in the quality of matrix data. At the same time, JASPAR collections outside the core will continue to include interesting matrix sets derived by other means.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EU Framework Programme 6 integrated project EuTRACC (to S.T.); YFF grant 180435 from the Norwegian Research Council (NRF), and by Bergen Research Foundation (BFS) (to B.L.). Novo Nordisk Foundation to the Bioinformatics Centre (to X.Z., E.V. and A.S.); The European Research Council under the EU 7th Framework Programme (FP7/2007-2013)/ERC grant agreement 204135 (to A.S.); Scholar of the Michael Smith Foundation for Health Research (to W.W.); Canadian Institutes for Health Research, GenomeCanada (via the Pleiades Promoter Project), GenomeBritishColumbia and the Canada Foundation for Innovation (to W.W. research laboratory). Funding for open access charge: Norwegian Research Council (NFR) (project no. 180435).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Frank Grosveld and Eric Soler for permission to include into JASPAR of ChIP-seq derived profiles prior to publication. We thank the laboratories of Yair Benita, Martha Bulyk, Richard Gronostajski, Steven Jones, and Zhiping Weng for suggestions and/or contributions of data reviewed for inclusion in the new release. We are grateful to Debra Fulton for her efforts to make the TFCat catalog available to the community.
REFERENCES
- 1.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 2.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 3.Heintzman ND, Heintzman ND, Hon GC, Hawkins RD, Hawkins RD, Kheradpour P, Kheradpour P, Stark A, Stark A, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- 6.Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandelin A, Wasserman W. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 8.Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Portales-Casamar E, Arenillas D, Lim J, Swanson MI, Jiang S, Mccallum A, Kirov S, Wasserman WW. The PAZAR database of gene regulatory information coupled to the ORCA toolkit for the study of regulatory sequences. Nucleic Acids Res. 2009;37:D54–D60. doi: 10.1093/nar/gkn783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuteja G, White P, Schug J, Kaestner K. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 16.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Guillon N, Tirode F, Boeva V, Zynovyev A, Barillot E, Delattre O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE. 2009;4:e4932. doi: 10.1371/journal.pone.0004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Børgesen M, Francoijs K-J, Mandrup S, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welboren W-J, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey T, Boden M, Buske F, Frith M, Grant C, Clementi L, Ren J, Li W, Noble W. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shendure J. The beginning of the end for microarrays? Nat. Methods. 2008;5:585–587. doi: 10.1038/nmeth0708-585. [DOI] [PubMed] [Google Scholar]
- 22.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu C, Byers K, McCord R, Shi Z, Berger M, Newburger D, Saulrieta K, Smith Z, Shah M, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J, Zhang MQ. SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics. 1999;15:607–611. doi: 10.1093/bioinformatics/15.7.607. [DOI] [PubMed] [Google Scholar]
- 25.Pachkov M, Erb I, Molina N, van Nimwegen E. SwissRegulon: a database of genome-wide annotations of regulatory sites. Nucleic Acids Res. 2007;35:D127–D131. doi: 10.1093/nar/gkl857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Portales-Casamar E, Kirov S, Lim J, Lithwick S, Swanson MI, Ticoll A, Snoddy J, Wasserman WW. PAZAR: a framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007;8:R207. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergman CM, Carlson JW, Celniker SE. Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- 29.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng X, Brodsky MH, Wolfe SA. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 2005;23:988–994. doi: 10.1038/nbt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development. 1999;126:873–881. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]
- 34.Hristova M, Birse D, Hong Y, Ambros V. The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol. Cell Biol. 2005;25:11059–11072. doi: 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berger MF, Bulyk ML. Protein binding microarrays (PBMs) for rapid, high-throughput characterization of the sequence specificities of DNA binding proteins. Methods Mol. Biol. 2006;338:245–260. doi: 10.1385/1-59745-097-9:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Peña-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJM. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fulton DL, Sundararajan S, Badis G, Hughes TR, Wasserman WW, Roach JC, Sladek R. TFCat: the curated catalog of mouse and human transcription factors. Genome Biol. 2009;10:R29. doi: 10.1186/gb-2009-10-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-reviews001. REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lenhard B, Wasserman WW. TFBS: computational framework for transcription factor binding site analysis. Bioinformatics. 2002;18:1135–1136. doi: 10.1093/bioinformatics/18.8.1135. [DOI] [PubMed] [Google Scholar]