Abstract
The Gene Expression Atlas (http://www.ebi.ac.uk/gxa) is an added-value database providing information about gene expression in different cell types, organism parts, developmental stages, disease states, sample treatments and other biological/experimental conditions. The content of this database derives from curation, re-annotation and statistical analysis of selected data from the ArrayExpress Archive of Functional Genomics Data. A simple interface allows the user to query for differential gene expression either (i) by gene names or attributes such as Gene Ontology terms, or (ii) by biological conditions, e.g. diseases, organism parts or cell types. The gene queries return the conditions where expression has been reported, while condition queries return which genes are reported to be expressed in these conditions. A combination of both query types is possible. The query results are ranked using various statistical measures and by how many independent studies in the database show the particular gene-condition association. Currently, the database contains information about more than 200 000 genes from nine species and almost 4500 biological conditions studied in over 30 000 assays from over 1000 independent studies.
INTRODUCTION
In the last decade, genome-wide gene expression assays, mostly employing microarrays and more recently high-throughput sequencing, have become common tools in biomedical and biological research. Most assays are performed to answer specific questions, for instance, to find which genes are differentially expressed in a particular disease state in comparison with healthy condition in a tissue or cell type. Some experiments instead compare a larger number of conditions, such as various tissue or cell types, i.e. the well known and widely used Genomics Institute of the Novartis Research Foundation gene expression atlas dataset for human and mouse (1).
Expression datasets derived from high-throughput experiments have utility beyond answering the specific questions that have been posed in the original experiments generating them. For instance, if a gene expression study has revealed a set of genes differentially expressed in a particular disease, making this information available online may help others working on the same disease, it can help in selecting candidate genes, or prioritizing the existing ones.
In compliance with the MIAME initiative (2), most scientific journals nowadays require publication-related microarray gene expression data to be deposited in public repositories like ArrayExpress (3) or the Gene Expression Omnibus (GEO) (4). Data from over 10 000 independent studies are readily available from these archives in several formats; however, using these deposited data to answer biological questions is not straight-forward. For instance, to find which genes are differentially expressed in a particular disease effectively one would need to download the datasets relevant to this disease and re-analyse them. Secondary databases, such as Oncomine (5) or Genevestigator (6), are doing the work of importing specific datasets from the public archives, re-analysing and making them available through various interfaces. Most of these databases are specific to particular biological domains and are, at least in part, commercial. The GEO profiles service is providing gene-based queries for expression profiles, however, it does not allow searches for genes specific to a particular condition (e.g. a particular disease or tissue), nor for conditions specific to a particular gene.
The European Bioinformatics Institute (EBI) has launched a new database called the Gene Expression Atlas that allows users to query gene expression under various biological conditions, including different cell types, developmental stages, physiological states, phenotypes and disease states. The key questions this new resource can answer can be summarized as:
Under which conditions or where in the organism is a gene of interest differentially expressed?
Which genes are differentially expressed in a condition or site (for instance in a disease, or in an organ)?
Both the questions can also be combined to focus on particular genes and their role in a specific disease, such as identifying members of the Wnt signalling pathway, which are expressed in a specific type of cancer.
The Atlas takes data directly from the ArrayExpress Archive of Functional Genomics Experiments, including data imported from GEO (4). The selected datasets are then systematically curated, genes are mapped to the latest genome builds and the experimental conditions are systematized and mapped to an application ontology, the Experimental Factor Ontology (EFO) (7). Statistical computations are performed, providing P-values linking each gene to each experimental condition in every study. A simple query interface is implemented, and the results are ranked by their P-values and weighted by the number of independent studies linking genes to biological conditions. The advanced interface enables the user to ask more sophisticated questions; tutorial materials are available at http://www.ebi.ac.uk/microarray-srv/tutorials.
The EBI Gene Expression Atlas freely provides its content for online queries and for programmatic access without restriction and without requirement to register. The complete content will be made available for download following the publication of this article.
As of August 2009, the EBI's; Gene Expression Atlas contains data for over 200 000 genes from over 1000 different independent studies, including more than 30 000 samples representing nearly 4500 different biological conditions. Nine different species, including human and model organisms, are included. The database is updated monthly, and is growing constantly. With streamlining of the curation process, we expect its content to double in the next 12 months.
MATERIALS AND METHODS
Atlas web interface
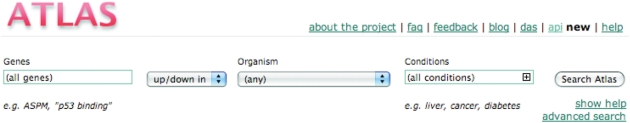
The Gene Expression Atlas interface (Figure 1) allows the user to query for condition-specific gene expression across multiple datasets. There are three basic types of queries: (i) for a gene, or a set of genes, by name or various gene attributes, including synonyms, Ensembl identifiers and Gene Ontology terms; (ii) for a ‘biological condition’, such as, disease name, developmental stage, as well as tissue or cell type; and (iii) for a combination of genes and biological conditions. Biological conditions, also referred to as EFs, are organized using an application ontology called EFO, which is described in more detail in the next section.
Figure 1.
Gene Expression Atlas home page. Querying for gene(s) will identify all genes whose annotation matches your query. The ‘Conditions’ parameter will identify all experiments in which the conditions that match your query appear. Searches can be restricted only to genes belonging to a given organism and also by direction of differential expression.
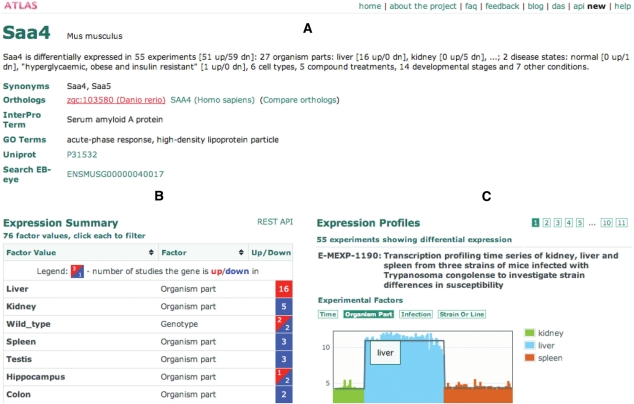
If a query matches one gene uniquely, the ‘Gene page’ for that gene is displayed (Figure 2). This page summarizes the behaviour of the selected gene across all Atlas datasets, providing easy access to both statistical analysis results and expression data. All gene pages can be linked directly, using links of the form http://www.ebi.ac.uk/gxa/gene/IDENTIFIER, where IDENTIFIER is any one of annotated gene attributes (e.g. Ensembl, UniProt and other accessions). Direct links can also be made to Atlas experiments, e.g. http://www.ebi.ac.uk/gxa/experiment/E-AFMX-5. Full details for linking and other Atlas use are available in online Atlas documentation.
Figure 2.
‘Gene page’ for Mus musculus Saa4. The following information is displayed: (A) summary of terms and external databases cross-references, as well as orthologue genes, which allows comparison of orthologues across the Atlas; (B) expression heat map listing all the conditions in which the gene was observed differentially expressed. The heatmap cell colour ranges from red, i.e. up-regulated, to blue, i.e. down-regulated. For each condition, the number of independent studies in which the gene was observed significantly up- or down-regulated is provided. Saa4 is over-expressed in ‘liver’, in 16 independent studies, which is consistent with the notion that liver is the primary site of Saa4 mRNA transcription (8) and (C) thumbnail plots of gene expression profiles for the studies in which the gene was found to be differentially expressed. Saa4 shows the highest significance of differential expression in the experiment E-MEXP-1190, comparing kidney, liver and spleen, each assayed in several replicates. A link to the experimental details in the ArrayExpress Archive of Functional Genomics Data is provided for each experiment.
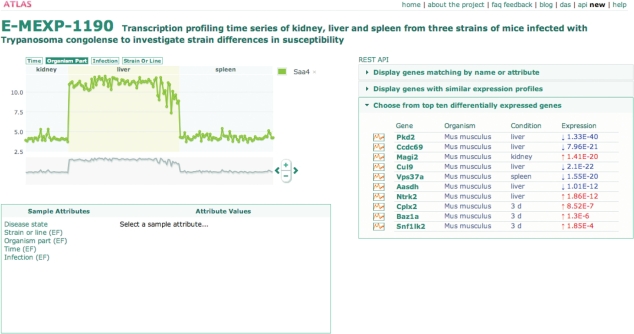
The thumbnail plots provide a direct link to individual experiment pages where the gene expression profile for the selected gene can be viewed in detail (Figure 3). In the experimental page, multiple ‘search’ options allow the user to retrieve genes of interest and add their expression profiles to the main plot (Figure 3, right). The search options available are: (i) search for any gene by name or attribute; (ii) search up to 10 most similar genes, based on Pearson correlation, to any of the genes currently plotted; and (iii) choose any gene from a list of top 10 differentially expressed genes for the selected study. For each gene, a P-value of significance of differential expression is provided.
Figure 3.
Gene expression profile page for experiment E-MEXP-1190 showing the table of genes with similar expression profile to Saa4, identified through similarity search. In the main graph, the horizontal axis shows all samples in this experiment, grouped by EF. The vertical axis shows the expression levels for Saa4 in each sample. The EF ‘organism part’ is selected and, under this condition, Saa4 has notably higher expression values in liver, as expected. Sample attributes can be selected from the ‘Sample attributes’ table and highlighted on the graph.
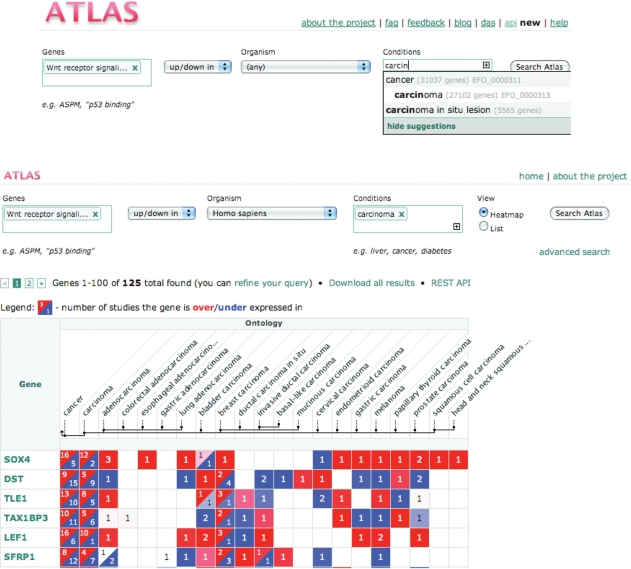
It is also possible to query for such a condition as a particular disease, either over all genes or for those matching specified attributes, such as belonging to a pathway. Figure 4 is an example of a summary view of transcriptional activity among members of the ‘Wnt signalling pathway’ in ‘carcinoma’. Both ‘Genes’ and ‘Conditions’ boxes provide auto complete functionality to help the user formulate queries. Condition queries (Figure 4, top) are expanded using our application ontology (EFO, see next section) to include all child terms available for the original query, so that, for instance, brain queries are expanded to all brain parts. The query results are displayed as a heat map (Figure 4, middle); genes matching the query are listed in the first column and the conditions go across the remaining columns grouped under ‘Ontology’ (when the query matches an ontology term) and ‘Keywords’ (when the query matches a keyword). All conditions mapping to Ontology terms are shown as nodes in the EFO tree (Figure 4, bottom; see next section). As in the ‘gene page’, the red colour corresponds to up-regulation of the selected gene in the selected condition, and the blue corresponds to down-regulation. The numbers in the cell correspond to the number of independent studies where differential expression has been observed, and the colour intensity represents the best P-value of this observation (i.e. the brighter the colour, the more significant the P-value). Results can be downloaded in tab-delimited format. Clicking on a cell in the heat map opens a window, showing information about the expression of the selected gene in the associated experiments including thumbnail plots of the gene expression profile(s) and links to the experimental details in the ArrayExpress Archive of Functional Genomics Data, as shown in Figure 2.
Figure 4.
Query results for human genes matching GO term ‘Wnt signaling pathway’ expressed in condition ‘carcinoma’. The ‘conditions’ auto complete function uses the EFO controlled vocabulary to expand queries, to query synonyms and to suggest query terms (top). In this example, SOX4 gene is over-expressed in adenocarcinoma in three different independent studies. Results are split over several pages and can be downloaded using the link provided (middle). Advanced query functionality is accessed using the ‘advanced search’ link below the ‘Search Atlas’ button.
A query can be refined by using terms enriched in the results (‘refine your query’ link; Figure 4, middle), by adding conditions to the original query (using the ‘advanced search’ link), as well as by adding various filters through the advanced interface, which allows user to formulate complex queries by combining several conditions, gene property and organism filters in one query.
Biological conditions—the concepts of EFs and their values
High-throughput gene expression experiments are typically used to compare gene expression in different biological conditions. Our approach to describe biological conditions is based on the concept of EF and EF value (EFV). An EF is defined as the experimental variable that is tested for gene expression variation, and EFVs are the values of this variable. For instance, in an experiment that compares gene expression in leukemic to normal blood, EF is a disease state, which has two values, leukemia and normal.
For a study to be included in the Atlas, it must have at least one defined EF with at least two different EFVs, and each EFV should be tested in at least two replicates. Experiments can have multiple EFs, for instance normal and leukemic gene expression can be profiled in peripheral blood or bone marrow. Such an experiment has two EFs, disease state and organism part.
Using free text keywords to describe EFs and their values limits their utility. For instance, to identify genes differentially expressed in ‘cancer’, we would also like to find genes that are studied in an experiment profiling ‘leukemia’. One way to achieve this is to use a disease ontology linking ‘leukemia’ to ‘cancer’ as a type of cancer, and then expand the query to all cancer types. Since in Gene Expression Atlas we are dealing with EFs mapping to a wide range of biological concepts such as organism parts, diseases or treatments, we require multiple source ontologies to describe these conditions. Initial mapping to existing ontologies identified the NCI Thesaurus (9) as providing best coverage due to the large amount of gene expression data performed on cancer samples. However, as no external ontology covered all our EFs and EFVs, we developed our own application ontology called EFO (7).
Mappings from EFO to external ontologies are maintained as identifiers from the external resource into a ‘denition citation’ annotation property in EFO. Equivalent classes are thus mapped from EFO into multiple other ontologies, for example a ‘neoplasia’ in EFO maps to ‘neoplasia’ in the NCI Thesaurus (9). This appears in the EFO ‘denition citation’ property, which has the value ‘NCI Thesaurus:C3262’. Typically, there are multiple maps to external ontologies as many biomedical ontologies and controlled vocabularies are non-orthogonal. EFO is released synchronously with the Atlas, new terms being added to each release where needed to describe Atlas data. EFO is mapped to 14 external ontologies including the NCI Thesaurus, the Foundational Model of Anatomy (10) and Chemical Entities of Biological Interest (ChEBI) (11) (for full list see http://www.ebi.ac.uk/efo/overview). As illustrated in the previous section, EFO is used in the Atlas to expand queries as outlined above, as well as a means to browse the Atlas content.
Where datasets are submitted directly to ArrayExpress, the submission tools guide the users through annotation of their EF and EFV, which later are checked and curated by the ArrayExpress staff. For data imported into ArrayExpress from GEO, we use text mining tools using EFO as the dictionary to identify the potential EFs and EFVs, and then introduce them via curation.
Statistical computations to rank query results
The meta-analysis approach taken in the construction of the Atlas can be outlined as follows. For each experiment:
identify differentially expressed genes for each EF;
for each gene found, identify EFVs where the gene's; mean expression level is significantly different from its overall mean across all factor values; and
perform multiple comparisons correction and aggregate the identified Gene-EFV associations for storage and retrieval.
If an experiment has several factors, i.e. a multi-factorial experiment design, we treat each factor independently from all others, and identify Gene–EFV associations separately for each EF in an experiment. This represents a simplification, and assumes that there are no interactions between EFs (see ‘Discussion’ section). We observed that in most cases (data not shown) of multi-factorial experiment designs, even if the factors are treated independently, once one statistically significant factor is identified for a gene, secondary factors’ effects become readily visible to the investigator when visualizing the data. For this reason, to construct a basic platform for further data analysis, we initially limited statistical analysis to the factor independence scenario.
The detailed description of the data analysis procedure outline is below:
If ygj are expression values for genes g = 1,…,G and arrays j = 1,…,J, pre-processed, background-corrected and normalized, we describe systematic expression effects for each gene by a linear model E(yg) = Xβg, where yg = (yg1}, …,ygJ)T is the vector of expression values for gene g, X is a known design matrix with full column rank K and βg = (βg1,…,βgK)T is a gene-specific vector of regression coefficients. The design matrix depends on the experimental design and choice of parameterization, and the regression coefficients represent comparisons of interest between RNA sources in the experiment. These coefficients are estimated with a least squares linear model fitting procedure in the Bioconductor package limma (12) and tested for differential expression (i.e. testing any particular βgk equal to 0) with moderated Student's; t-statistic via the empirical Bayesian statistics developed by Smyth (12). For each EF in the experiment, we set up a complete pairwise-comparisons contrast matrix (equivalent to one-way Analysis of Variance) as an omnibus test of differential expression across all factor values in the selected factor. We then can accept or reject the ‘equal means for all groups’ null hypothesis on the basis of P-values computed for the omnibus F-statistic via limma as described above, at a specified significance level. These P-values, with appropriate multiple testing adjustment to control the False Discovery Rate (FDR) at 5% (13), allow us to identify differentially expressed genes.
Given a set of expression values for a gene under k different conditions, hence k group means μg1,…,μgk and a significant F-statistic, we look at k differences μ − μi, of each group mean to the global mean, and seek to identify which ones are significant, and in what direction. The problem of multiple comparisons with the overall mean is known in statistical literature as an Multiple Comparisons with the Mean procedure. We follow Hsu (14) to make direct inference on multiple comparisons of the means. For each gene we compute k simultaneous confidence intervals governed by a multivariate t-distribution and computed from quantiles of the Studentized maximum modulus statistic and look at their directionality.
Steps (i) and (ii) for each factor in a given experiment produce a matrix of up/down calls (−1, 0 or 1) according to the directionality of the confidence interval and respective P-values: one call/P-value for each Gene–EFV combination. Multiple testing adjustment is performed on these P-values to control the global (across genes and contrasts, i.e. EFVs) FDR at 5%, following recommendations by Smyth in limma (12). Taking advantage of the robustness of the tests performed, we use the P-values significance-based calls to aggregate differential expression results into ‘votes’: each time a gene has been observed as differentially expressed in a particular EFV, we use that as a vote for up/down activity for that Gene–EFV combination. These are the numbers displayed in the heatmap view of the Gene Expression Atlas. All the P-values and computed statistics as well as the aggregated votes are stored in a database and indexed for fast retrieval in the interface.
Currently, all data included in the Atlas are based on microarray assays. The statistical method and the data integration framework developed are generic and can be extended to many other technologies, such as RNA-seq and in situ hybridizations. The R-code for this procedure is available as Supplementary Data and will be released as a separate Bioconductor package (in preparation).
Gene expression data
Data for the Atlas are selected from ArrayExpress Archive and selection is based on various criteria outlined earlier. As currently we are using only microarray data, our first consideration is whether sufficient array annotation is given to enable us to map the array design elements to existing gene identifiers. We use two routes for this mapping: we preferentially map array probe sequences to Ensembl genomes (15) or we attempt to map the design element annotation identifiers to gene annotation in UniProt database (16). Where re-annotation fails, experiments that are performed on such arrays cannot be included in the Atlas. The array re-annotation pipeline will be released as a software package, described and published separately (Sarkans et al., in preparation).
Experiments in ArrayExpress Archive that are performed on well-annotated arrays, which have high MIAME scores (2,17), where the EF/EFV annotation and sufficient replication criteria (as well as some other technical criteria not described here), and where normalized data are present, are annotated as ‘suitable for Atlas’. When all basic criteria are satisfied, experiment selection for the Atlas is motivated by the quality of annotation, use of standard platforms and large sample sizes, without any preference for any biological conditions. Recently, we started to produce themed Atlas data releases, e.g. species oriented or addressing a specific research domain, or by curating user-requested studies. Experiments selected for Atlas are then exported from the Archive. The submitter's; normalized data are used, hence we do not perform any renormalization. Prior to loading into the Atlas, annotations are harmonized, experimental descriptions checked for consistency and non-standard terms are standardized. Maps to EFO are added where the term required is present in the ontology. If terms are not in EFO, we examine source ontologies and provide a term name, definition and maps to external ontologies. The term is then placed in the EFO hierarchy that is optimized for the Atlas visualization.
Once data are loaded, statistical computations, as described in the previous section, are performed and for each new experiment, for each EF and EFV, for each gene the P-value is computed.
Currently, the Atlas contains data from nine species. Table 1 shows the number of assays and the number of studies (experiments) included from each. The experiments included in the Atlas together have more than 40 different EFs, covering over 4500 different EFVs. The distribution of the number of assays for the most frequently studied (at least 50 experiments for each factor) EFs and EFVs are given in Table 2.
Table 1.
Number of studies and assays for each species in the Atlas
| Species | Assays | Studies |
|---|---|---|
| Homo sapiens | 13 703 | 410 |
| Mus musculus | 7539 | 373 |
| Rattus norvegicus | 4858 | 133 |
| Arabidopsis thaliana | 1607 | 88 |
| Saccharomyces cerevisiae | 813 | 43 |
| Drosophila melanogaster | 790 | 40 |
| Schizosaccharomyces pombe | 458 | 19 |
| Danio rerio | 214 | 13 |
| Caenorhabditis elegans | 166 | 5 |
| Total | 30 148 | 1124 |
Table 2.
Most frequently used EFs and the number of EFVs and studies for each factor
| EFs | EFVs | Studies |
|---|---|---|
| Genotype | 389 | 211 |
| Compound treatment | 425 | 196 |
| Disease state | 214 | 137 |
| Organism part | 267 | 98 |
| Cell type | 164 | 61 |
| Growth condition | 122 | 61 |
| Strain or line | 227 | 51 |
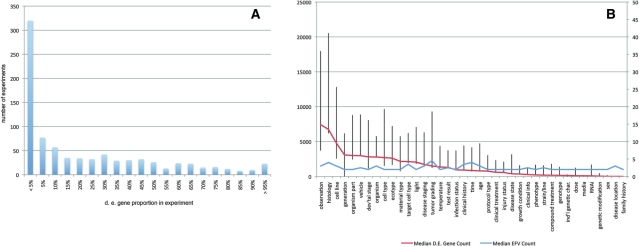
The method used in Gene Expression Atlas analytics allows us to examine trends in differential gene expression across all Atlas data. Figure 5A shows the distribution of proportions of differentially expressed genes across all experiments. There are approximately 400 experiments (from over 1000) with fewer than 10% of all genes showing differential expression; the mean proportion of genes differentially expressed in an experiment, according to our FDR criteria, is 25%. Further, when we examine the number of differentially expressed genes per factor (Figure 5B), we observe that the numbers are highest in the factors ‘observation’, ‘histology’, ‘cell line’, ‘generation’ and ‘organism part’. It appears that, broadly, across species, transcriptional activity is strongly driven by its context: by tissue (‘histology’, ‘organism part’ and, by extension, ‘cell line’), followed by developmental stage and then cell type, while the main extrinsic drivers of transcriptional activity such as xenobiotic responses (‘compound treatment’) and disease states contribute to differential expression to a smaller extent. We can also observe that the number of differentially expressed genes is largely independent of the number of EFVs (the median factor value count is around 3 EFVs).
Figure 5.
Distributions of differentially expressed genes over (A) experiments and (B) EFs. Error bars in (B) mark the 25% and 75% quantiles in the differentially expressed gene count for each EF.
Programmatic access
REST (Representational State Transfer) is a simple technology that allows users to retrieve data in an easy-to-parse format by going directly to a web address Universal Resource Identifier (URI). For instance, all information available on gene matching ‘aspm’ anywhere in gene property fields can be obtained by entering a URI: http://www.ebi.ac.uk/gxa/api?geneIs=aspm.
This will produce a simple output either in JSON or XML format (the latter will be used if ‘&format=xml’ is appended to the URI). Example programmatic queries are provided in Table 3 below.
Table 3.
Examples of gene expression search API links
| Description | Query |
|---|---|
| Retrieve all Atlas data on genes with InterPro ID IPR003822 | http://www.ebi.ac.uk/gxa/api?geneInterProIdIs=IPR003822 |
| Retrieve all Atlas data on genes with GO IDs GO:0003006 or GO:0005000 | http://www.ebi.ac.uk/gxa/api?geneGOIdIs=GO:0003006+GO:0005000 |
| Retrieve data on a specific experiment in Atlas | http://www.ebi.ac.uk/gxa/api?experiment=E-GEOD-2487 |
| Retrieve complete differential gene expression analysis output for a specified experiment and a list of genes | http://www.ebi.ac.uk/gxa/api?experiment=E-MEXP-1190&gene=ENSMUSG00000040017, &gene=ENSMUSG00000055923 |
These APIs allow advanced users to search and retrieve complete information on any gene or experiment from the Atlas, including all gene and sample attributes, details of experimental design, meta-analysis statistics and gene expression values. Additionally, the Atlas provides a gene view-based Distributed Annotation System (18) track at http://www.ebi.ac.uk/gxa/das that can be viewed with any Distributed Annotation System client such as the Ensembl genome browser.
Examples of output for these queries are available from the Atlas documentation at http://www.ebi.ac.uk/gxa/help/AtlasApis.
DISCUSSION
More than 60% of the experiments in the Atlas have two or three EFs; our current assumption that they are independent may not be valid. One approach to deal with potential EF interactions is to introduce a single ‘meta-factor’, with values made of all occurring combinations of the individual factor values in the comprising factors. We are currently investigating this and related methods.
In the presented implementation, the experiments are ranked by a simple ‘vote-counting’ method, first described by Light and Smith (19). It has several known deficiencies, for instance, it does not incorporate the sample size into the vote, is imprecise, and occasionally has low statistical power. We are working on employing a more statistically robust procedure for meta-analysis of P-values derived from individual differential expression tests. Using earlier work by Hedges and Olkin (20) and making use of the semantic enrichment provided by EFO curation, we have developed a new effect-size estimation-based method for data integration, which will be incorporated in the future Atlas releases.
Currently the Atlas provides information on expression of only protein-coding genes. In the near future we will also plan to include data on known micro-RNA expression. It is possible to deal with the expression of alternative splice variants, or with expression at the exonic level using the same methodology. At the moment, all data included in the Atlas are derived from microarray-based assays. In the future, as ultra high-throughput sequencing becomes widespread and we plan to include RNA-seq and related data types. Presently, the number of RNA-seq experiments that focus on assaying expression of known genes and for which the processed data are available is still relatively small.
Among the new features under development are graphical gene expression query and display based on anatomograms. Another improvement will be a possibility to query and visualize the results by ontology terms of the user's; choice, not just EFO. We are also building an Atlas of ‘normal gene expression’, i.e. the gene expression in different organism parts under ‘normal’, non-diseased conditions.
The code will be released as open source with installation and data-loading procedures, allowing the users to run the Atlas locally and use it with their own data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EMBL (Core Funding); and the European Commision (FELICS, EMERALD). Funding for open access charge: EMBL (Core Funding).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
In addition to the invaluable help from the entire Microarrays Team, we would like to thank especially the work of Richard Coulson, Ekaterina Pilicheva, Niran Abeygunawardena and Ugis Sarkans on the gene re-annotation pipeline; the work of Ugis Sarkans, Sergio Contrino and Hugo Berube on the earlier ArrayExpress Warehouse software served as a platform and motivation for the Atlas; Nikolay Kolesnikov, Andrew Tikhonov, Miroslaw Dylag and Roby Mani for contributing with components and work on maintenance of the infrastructure on which the Atlas service runs; and Wolfgang Huber for the invaluable discussions.
REFERENCES
- 1.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Gene. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 3.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update–from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37(Suppl. 1):D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008:1–5. doi: 10.1155/2008/420747. doi:10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malone J, Zheng-Bradley HJ, Rayner T, Parkinson H. Developing an application focused experimental factor ontology: embracing the OBO community. In: Lord P, Shah N, Sansone S, Cockerill M, editors. BioOntologies Meeting. Canada: ISMB, Toronto; 2008. pp. 21–24. [Google Scholar]
- 8.deBeer MC, Yuan T, Kindy MS, Asztalos BF, Roheim PS, deBeer FC. Characterization of constitutive human serum amyloid a protein (SAA4) as an apolipoprotein. J. Lipid Res. 1995;36:526–534. [PubMed] [Google Scholar]
- 9.Fragoso G, deCoronado S, Haber M, Hartel F, Wright L. Overview and utilization of the NCI thesaurus. Comp. Funct. Genomics. 2004;5:648–654. doi: 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosse C, Mejino JLV. A reference ontology for biomedical informatics: the foundational model of anatomy. J. Biomed. Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Degtyarenko K, deMatos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcntara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smyth GK. Limma: Linear Models for Microarray Data. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate – a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 14.Hsu JC. Multiple Comparisons: Theory and Methods. Florida: CRC Press, Bota Raton; 1996. [Google Scholar]
- 15.Hubbard TJP, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37(Suppl. 1):D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.UniProt Consortium. The universal protein resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brazma A, Parkinson H. ArrayExpress service for reviewers/editors of DNA microarray papers. Nat. Biotechnol. 2006;24:1321–1322. doi: 10.1038/nbt1106-1321. [DOI] [PubMed] [Google Scholar]
- 18.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Light RJ, Smith PV. Accumulating evidence: procedures for resolving contradictions among different research studies. Harvard Educational Rev. 1971;41:429–471. [Google Scholar]
- 20.Hedges LV, Olkin I. Statistical Methods for Meta-analysis. Florida: Academic Press, Orlando; 1985. [Google Scholar]