Abstract
Plants and animals rely on innate immunity to prevent infections by detection of microbe-associated molecular patterns (MAMPs) through pattern-recognition receptors (PRRs). The plant PRR FLS2, a leucine-rich repeat-receptor kinase, recognizes bacterial flagellin and initiates immune signaling by association with another leucine-rich repeat-receptor-like kinase, BAK1. It remains unknown how the FLS2/BAK1 receptor complex activates intracellular signaling cascades. Here we identified the receptor-like cytoplasmic kinase BIK1 that is rapidly phosphorylated upon flagellin perception, depending on both FLS2 and BAK1. BIK1 associates with FLS2 and BAK1 in vivo and in vitro. BIK1 is phosphorylated by BAK1, and BIK1 also directly phosphorylates BAK1 and FLS2 in vitro. The flagellin phosphorylation site Thr237 of BIK1 is required for its phosphorylation on BAK1 and FLS2, suggesting that BIK1 is likely first phosphorylated upon flagellin perception and subsequently transphosphorylates FLS2/BAK1 to propagate flagellin signaling. Importantly, bik1 mutants are compromised in diverse flagellin-mediated responses and immunity to the nonpathogenic bacterial infection. Thus, BIK1 is an essential component in MAMP signal transduction, which links the MAMP receptor complex to downstream intracellular signaling.
Keywords: pathogen-associated molecular pattern/microbe-associated molecular pattern-triggered immunity, phosphorylation, pattern recognition receptor, BRI1-associated receptor kinase, flagellin sensing 2
Plants and animals live in an environment with a diverse array of microorganisms and have developed the capacity to timely detect potential infectious agents without destroying their own tissues. Innate immunity, the first line of inducible defense, is triggered instantaneously upon the detection of conserved pathogen- or microbe-associated molecular patterns (PAMP/MAMPs) (1–5). In plants, MAMPs are usually perceived by cell-surface pattern-recognition receptors (PRRs) and mount PAMP/MAMP-triggered immunity (PTI). Different MAMPs likely trigger convergent immune signaling events, including changes in cytoplasmic Ca2+ levels, activation of MAP kinase (MAPK) cascades, induction of defense-related genes, production of reactive oxygen species and nitric oxide, deposition of callose to reinforce the cell wall, and stomatal closure to prevent pathogen entry (1–5). PTI is important for plants to thwart off a broad spectrum of potential pathogens.
One of the best-characterized plant MAMP receptors is the leucine-rich repeat receptor kinase (LRR–RK) protein FLS2 that recognizes a conserved 22-amino-acid peptide (flg22) from bacterial flagellin (6). Upon flagellin perception, FLS2 rapidly associates with another LRR–receptor-like kinase (RLK), BAK1, thereby initiating downstream signaling (7, 8). BAK1 was originally identified as a BRI1-associated receptor kinase mediating brassinosteroid signaling (9, 10). Brassinosteroids (BRs), a class of plant hormone with essential roles in plant growth and development, are perceived by LRR–RK BRI1, which is structurally similar to FLS2 (11). Rather than being involved in direct binding of BR to BRI1 and flagellin to FLS2 (7, 12), BAK1 more likely functions as an adaptor or signaling partner for the regulation of FLS2 and BRI1. Furthermore, BAK1 is required for the immune responses triggered by multiple MAMPs other than flagellin, including the bacterial elongation factor EF-Tu, peptidoglycans, lipopolysaccharides, cold-shock protein, and the oomycete elicitor INF1 in Arabidopsis and tobacco (7, 8, 13). Thus, BAK1 appears to associate with multiple PRRs to integrate specific MAMP perception into convergent downstream signaling. However, the substrates of FLS2 and BAK1 kinases have yet to be identified, and how the MAMP signal is transmitted from the BAK1-associated receptor complexes at the plasma membrane to intracellular events remains largely unknown.
The RLK/Pelle/IRAK protein kinase family plays a general role in innate immunity from plants to insects and humans (4, 14). In contrast to animals, plants have expanded a large number of RLK/Pelle/IRAK genes with about 610 members in Arabidopsis, including RLKs and receptor-like cytoplasmic kinases (RLCKs) (15, 16). RLKs are involved in a wide range of biological processes, including plant growth, development, and immunity, by perceiving diverse signals through the extracellular domain. Compared to RLKs, the biological functions of RLCKs are much less understood. Lacking an apparent extracellular domain, RLCKs more likely function in signal transduction rather than ligand perception. Here, we show that one RLCK member, BIK1, plays an important role in mediating early flagellin signaling from the FLS2/BAK1 receptor complex. BIK1 (BOTRYTIS-INDUCED KINASE 1), originally identified as a component in plant defense against necrotrophic fungal pathogens (17), is rapidly phosphorylated at residue Thr237 on flg22 perception in an FLS2- and BAK1-dependent manner. In vivo and in vitro data suggest that BIK1 associates with both FLS2 and BAK1. BIK1 is a substrate of BAK1, whereas BAK1 and FLS2 are also substrates of BIK1, suggesting transphosphorylation events between BIK1 and the FLS2/BAK1 complex. Strikingly, compared to wild-type plants, bik1 mutants display reduced flg22 responses as assayed by flg22-mediated inhibition of seedling growth and immunity to virulent and nonpathogenic bacterial infection. Consistent with the role of BAK1 in multiple MAMP signaling, BIK1 is also phosphorylated by EF-Tu in addition to flg22. The results demonstrate that BIK1 mediates PTI signal transduction from multiple MAMP receptor complexes.
Results
Flg22 Rapidly Induces BIK1 Phosphorylation.
Protein phosphorylation plays essential roles in diverse MAMP responses from receptor activation to downstream immune response gene expression (18). Although some phosphorylated proteins have been identified in response to MAMP treatment, their exact mode of action in MAMP signaling has remained elusive (19, 20). To identify kinases with a particular interest in RLCKs involved in MAMP signaling in Arabidopsis, we analyzed kinase-encoding transcripts that are induced upon flg22 and other MAMP treatment by searching through microarray data sets (21, 22). One gene identified in this way was BIK1, which was rapidly induced by flg22 and EF-Tu treatments (21, 22). The induction of BIK1 by flg22 was further confirmed by RT–PCR analysis in which BIK1 expression was significantly enhanced 30 min after flg22 treatment in 8-day-old Arabidopsis seedlings (Fig. 1A). Intriguingly, when we expressed HA-epitope-tagged BIK1 in Arabidopsis protoplasts, the flg22 treatment induced a mobility shift of BIK1 as detected by Western blot (Fig. 1B). We examined several other RLCKs that have been shown previously to play roles in plant defense or other signaling: oxidative signal-inducible 1 (OX1) is involved in oxidative-burst-mediated signaling in Arabidopsis (23), brassinosteroid-signaling kinase 1 (BSK1) mediates Arabidopsis BR signaling (24), and Pto induces effector-triggered immunity in tomato (25). The flg22 treatment did not induce the mobility shift of OX1, BSK1, and Pto as detected by Western blot (Fig. 1B).
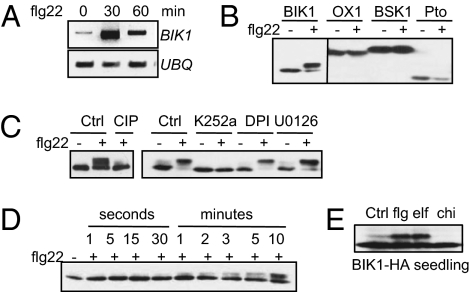
Fig. 1.
flg22 induces BIK1 phosphorylation. (A) flg22 induces BIK1 transcripts. RT–PCR analysis of 8-day-old wild-type (Col-0) seedlings treated with 10 nM flg22. (B) flg22 treatment induces the mobility shift of BIK1, but not of OX1, BSK1, or Pto. Protoplasts were transfected with HA-epitope-tagged BIK1, OX1, BSK1, or Pto for 6 h and treated with 1 μM flg22 for 10 min. (C) flg22-induced BIK1 mobility shift is restored by the treatment of CIP or kinase inhibitor K252a. The control (Ctrl) was nontreatment. (D) BIK1 is phosphorylated by flg22 within the first minutes upon stimulation. The protoplasts were concentrated by a low-speed centrifuge 6 h after transfection and treated with 1 μM flg22 at the indicated time before adding protein sample loading buffer. (E) flg22 and elf18, but not chitin, induce BIK1 phosphorylation in BIK1-HA transgenic plant seedlings. Twelve-day-old BIK1-HA seedlings were treated with H2O (Ctrl), 1 μM flg22, 1 μM elf18, or 50 μg/mL chitin for 10 min.
The mobility of modified BIK1 could be restored to that of the unmodified form by the treatment of calf alkaline intestinal phosphatase (CIP) or a general kinase inhibitor K252a (Fig. 1C). The data indicate that the modification of BIK1 as detected by Western blot was caused by flg22-induced phosphorylation. We further performed a BIK1 phosphorylation time-course assay upon flg22 treatment. BIK1 phosphorylation was evident as early as 1 min upon flg22 treatment and gradually increased within 10 min (Fig. 1D). The rapid phosphorylation of BIK1 upon flg22 treatment demonstrates that BIK1 phosphorylation is one of the earliest events in flagellin signaling. BIK1 phosphorylation in response to flg22 was further confirmed in transgenic plants expressing BIK1 fused with an HA epitope tag (Fig. 1E). Plants could respond to multiple MAMPs and activate the convergent signaling (21). We therefore examined BIK1 phosphorylation upon different MAMP treatments. Significantly, elf18 (the N-terminal 18-amino-acid peptide of EF-Tu), but not chitin, also induced BIK1 phosphorylation in Arabidopsis transgenic plants (Fig. 1E). The results suggest that BIK1 is a convergent component involved in multiple MAMP signaling.
Flg22-Induced BIK1 Phosphorylation Depends on FLS2 and BAK1.
The flg22-induced BIK1 phosphorylation was dependent on the flg22 receptor FLS2 and its signaling partner BAK1 as phosphorylation was absent in the fls2 and bak1 mutant seedlings (Fig. 2A). Importantly, expression of FLS2 in the fls2 mutant protoplasts or of BAK1 in the bak1 mutant protoplasts could restore the BIK1 phosphorylation induced by flg22 (Fig. 2B). However, expression of the corresponding kinase-inactive mutants, FLS2Km or BAK1Km, failed to complement BIK1 phosphorylation, indicating that the kinase activity of FLS2 and BAK1 is required for BIK1 phosphorylation (Fig. 2B). Interestingly, the BAK1 closest homolog, SERK4, is not required for BIK1 phosphorylation as the flg22-induced BIK1 phosphorylation occurred normally in the serk4 mutant (Fig. 2B). The flg22-induced BIK1 phosphorylation could be suppressed by expression of the bacterial effector AvrPto (Fig. 2C), which is consistent with the finding that AvrPto suppresses MAMP signaling by targeting receptor complexes (13, 26). Therefore, the results demonstrate that BIK1 could be quickly phosphorylated by different MAMPs downstream of MAMP receptor complexes.
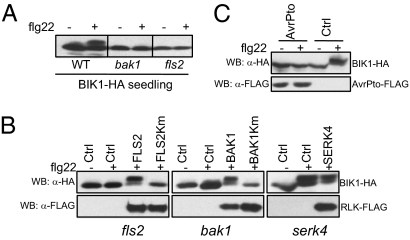
Fig. 2.
FLS2- and BAK1-dependent BIK1 phosphorylation. (A) flg22-induced BIK1 phosphorylation depends on FLS2 and BAK1. BIK1-HA transgenic plant seedlings in a Col-0 [wild type (WT)], bak1, or fls2 background were treated with 1 μM flg22 for 10 min. (B) The kinase activity of FLS2 and BAK1 is required for flg22-mediated BIK1 phosphorylation. Protoplasts were isolated from fls2, bak1, or serk4 mutant plants and cotransfected with HA-epitope-tagged BIK1 and FLAG-epitope-tagged FLS2, FLS2Km, BAK1, BAK1Km, or SERK4. The control (Ctrl) was a vector control. (C) AvrPto blocks flg22-induced BIK1 phosphorylation. Protoplasts were cotransfected with a control vector or AvrPto-FLAG and treated with 1 μM flg22 for 10 min.
BIK1 Associates with FLS2 and BAK1.
Our data suggest that BIK1 is a component in the early MAMP signaling. Consistent with this notion, expression of BIK1 in Arabidopsis protoplasts moderately, but significantly, activated FRK1-LUC and WRKY29-LUC, two MAMP-responsive genes fused to a luciferase reporter (Fig. S1A). MAPK activation is an immediately early event upon MAMP perception (27). To determine the relationship of BIK1 phosphorylation and MAPK activation, we transiently coexpressed BIK1 with constitutively active MKK5, MEKK1, or full-length MEKK1 that has been shown to be activated in flg22 signaling (27). The MAPK components did not induce the phosphorylation of BIK1 (Fig. S1B), suggesting that BIK1 functions either upstream of the MAPK cascade or independently of MAPK signaling.
BIK1 is a plasma membrane localized protein with a putative myristoylation motif (17). To test whether BIK1 could associate with FLS2 and BAK1, we performed a coimmunoprecipitation (Co-IP) assay with coexpressing HA-epitope-tagged BIK1 and FLAG-epitope-tagged FLS2 or BAK1 in protoplasts. Clearly, BIK1 immunoprecipitated both FLS2 and BAK1 in vivo (Fig. 3 A and B). Interestingly, the association of BIK1 with FLS2 or BAK1 appears to be reduced upon flg22 treatment (Fig. 3 A and B). This result suggests that BIK1 might be released from the receptor complex upon phosphorylation by flagellin. The kinase activity of BIK1, BAK1, and FLS2 seems not to be required for the association because their kinase-inactive mutants still associate normally by Co-IP assay (Fig. S2 A and B). The BIK1-FLS2 association is independent of BAK1, and BIK1–BAK1 association is independent of FLS2 because bak1 and fls2 mutants did not affect the association of BIK1 with FLS2 or BAK1 (Fig. S3 A and B). To confirm this interaction, we further performed a pull-down assay with an N-terminal GST–BIK1 fusion protein immobilized on glutathione beads as bait against lysates of Arabidopsis protoplasts expressing HA-epitope-tagged FLS2 or BAK1. Both FLS2 and BAK1 could be pulled down strongly by GST-BIK1, but not by GST itself (Fig. 3C). The treatment of flg22 seems not to significantly affect FLS2–BIK1 and BAK1–BIK1 interaction with this assay in which BIK1 was not exposed to flg22 treatment. BIK1 is a membrane-associated intracellular cytoplasmic kinase. We next tested whether FLS2 and BAK1 associate with BIK1 through their kinase domains. Using a GST pull-down assay, BIK1 could be clearly pulled down by GST-BAK1K and GST-FLS2K, the kinase domain of BAK1 (BAK1K) or FLS2 (FLS2K) fused to an N-terminal GST (Fig. 3D). This was further confirmed by a Co-IP assay, in which BIK1 coimmunoprecipitated FLS2K (Fig. 3E) and BAK1K (Fig. 3F). Taken together, the data demonstrate that BIK1 functions in MAMP signaling by interacting with the MAMP receptor complex.
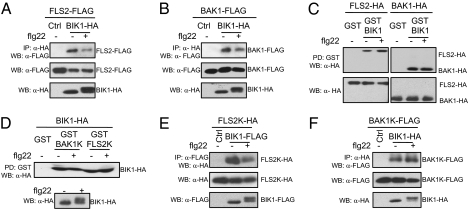
Fig. 3.
BIK1 interacts with FLS2 and BAK1. (A) BIK1 associates with FLS2 in vivo. The protoplasts were coexpressed with FLS2-FLAG and BIK1-HA or a control vector. Co-IP was carried out with an anti-HA antibody (IP: α-HA), and the proteins were analyzed using Western blot with an anti-FLAG antibody (WB: α-FLAG). (Top) BIK1 coimmunoprecipitates with FLS2. (Middle and bottom) The expression of FLS2 and BIK1 proteins. Protoplasts were stimulated with 1 μM flg22 for 10 min. (B) BIK1 associates with BAK1 in vivo. Co-IP was performed with protoplasts coexpressing BAK1-FLAG and BIK1-HA or a control vector. (C) GST-BIK1 pulls down both FLS2 and BAK1. Protoplasts were transfected with FLS2-HA or BAK1-HA and stimulated with or without 1 μM flg22 for 10 min. The cell extracts were incubated with GST or GST-BIK1 beads (PD: GST). (Top) GST-BIK1 beads pull down FLS2 and BAK1. (Bottom) Protein expression of FLS2 and BAK1. (D) Kinase domains of BAK1 and FLS2 pull down BIK1. Protoplasts were transfected with BIK1-HA and stimulated with or without 1 μM flg22 for 10 min (bottom: protein expression). The cell extracts were incubated with GST, GST-BAK1K, or GST-FLS2K beads. The proteins were detected using Western blot with an anti-HA antibody (top). (E) The kinase domain of FLS2 associates with BIK1 with Co-IP assay. (F) The kinase domain of BAK1 associates with BIK1 with Co-IP assay. The control (Ctrl) was a vector control.
BIK1 Transphosphorylates FLS2 and BAK1.
BIK1 is predicted to encode a Ser/Thr protein kinase (17). To determine the amino acid residues that are essential for flg22-induced phosphorylation and gain insight into how BIK1 functions, we searched the potential BIK1 phosphorylation sites in The Arabidopsis Protein Phosphorylation Site Database (http://phosphat.mpimp-golm.mpg.de/app.html) and aligned the activation domain of BIK1 with several related kinases (Fig. 4A). We individually substituted several putative Ser and Thr phosphorylation sites in the BIK1 activation domain with Ala and tested their effect on flg22-induced BIK1 phosphorylation. Significantly, a Thr237 → Ala237 (T237A) mutation completely eliminated the BIK1 phosphorylation by flg22 whereas Ser233, Ser236, and Thr242 seem to be dispensable for flg22-induced BIK1 phosphorylation (Fig. 4B). The data suggest that Thr237 is a major phosphorylation site of BIK1 in response to flg22. However, additional phosphorylation sites that are not detected by Western blot might be present in BIK1 (19). BIK1T237A did not reduce its interaction with FLS2 and BAK1 as detected by an in vivo Co-IP assay (Fig. S2).
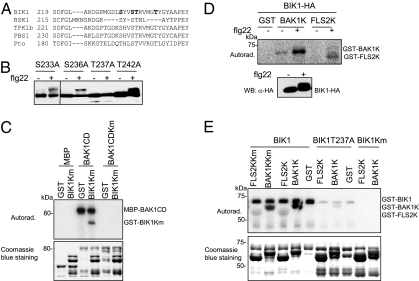
Fig. 4.
Transphosphorylation between BIK1 and the FLS2-BAK1 complex. (A) Alignment of the activation domain of BIK1 with several related kinases, including BSK1, TPK1b, PBS1, and Pto. The residues of BIK1 individually mutated to alanine are indicated in bold. (B) BIK1T237A mutation completely eliminates the flg22-induced BIK1 phosphorylation detected by Western blot. (C) BAK1 phosphorylates BIK1. An in vitro kinase assay was performed by incubating MBP, MBP-BAK1CD, or MBP-BAK1CDKm with GST or GST-BIK1Km. Proteins were separated with SDS/PAGE and analyzed by autoradiography (Upper), and the protein loading control was shown by Coomassie blue staining (Lower). (D) BIK1 phosphorylates BAK1 and FLS2 with an immunocomplex kinase assay. HA-epitope-tagged BIK1 was immunoprecipitated with an anti-HA antibody and subjected to an in vitro kinase assay with GST, GST-BAK1K, or GST-FLS2K as substrate. (E) BIK1 phosphorylates BAK1 and FLS2 in vitro. An in vitro kinase assay was performed by incubating GST-FLS2K, GST-BAK1K, or their kinase mutants with GST-BIK1 or its mutants. Proteins were separated with SDS/PAGE and analyzed by autoradiography (Upper). (Upper) Autophosphorylated GST-BIK1, phosphorylated GST-BAK1K, and phosphorylated GST-FLS2K. The protein loading control was shown by Coomassie blue staining (Lower).
Our findings on BIK1 interaction with FLS2/BAK1 and rapid flg22-induced BIK1 phosphorylation suggest that BIK1 might be a substrate of BAK1 and/or FLS2. BIK1 exhibited strong autophosphorylation activity with an in vitro kinase assay (Fig. S4A). This autophosphorylation was eliminated in BIK1Km, which carries a mutation in the ATP-binding site (Fig. S4A). Thus, GST-BIK1Km was used as a substrate for the in vitro kinase assay. We purified the cytosolic domains of BAK1 (BAK1CD) and FLS2 (FLS2CD) as MBP or GST fusion proteins in Escherichia coli. The cytosolic domain includes both the intracellular kinase domain and the juxtamembrane domain. Significantly, BAK1CD directly phosphorylated BIK1Km in vitro in the presence of [32P]-γ-ATP (Fig. 4C). The phosphorylation depends on the kinase activity of BAK1 because BAK1CDKm completely eliminated its phosphorylation on BIK1 (Fig. 4C). A similar result was obtained with an immunocomplex kinase assay in which FLAG-epitope-tagged BAK1 was expressed in protoplasts with flg22 treatment for an additional 10 min. BAK1 was pulled down with FLAG antibody and subjected to an in vitro kinase assay using GST-BIK1Km as a substrate. The immunoprecipitated BAK1, not kinase-inactive BAK1Km, phosphorylated BIK1Km (Fig. S5). The BIK1 phosphorylation seems to be enhanced on flg22 treatment (Fig. S5). We did not observe the in vitro phosphorylation of BIK1Km by FLS2CD. However, the autophosphorylation activity of FLS2CD was weaker than that of BAK1CD under our in vitro assay condition (Fig. S4B). The data indicate that BAK1 could directly phosphorylate BIK1 and that BIK1 is a substrate of BAK1.
Surprisingly, BIK1 could also phosphorylate FLS2 and BAK1. In an immunocomplex kinase assay, the immunoprecipitated BIK1 could phosphorylate GST-BAK1K and GST-FLS2K, the kinase domains of BAK1 and FLS2 fused with GST (Fig. 4D). GST-BAK1K and GST-FLS2K did not exhibit detectable autophosphorylation activity (Fig. S4A). Apparently, the phosphorylation was significantly enhanced with the BIK1 that was activated upon flg22 treatment (Fig. 4D). This unexpected finding was substantiated by an in vitro kinase assay in which GST-BIK1 directly phosphorylated GST-FLS2K and GST-BAK1K (Fig. 4E and Fig. S6A). The specificity of this phosphorylation was demonstrated by the fact that BIK1Km did not phosphorylate FLS2K and BAK1K (Fig. 4E and Fig. S6A). To rule out the possibility that phosphorylation of GST-BAK1K and GST-FLS2K might result from the enhanced autophosphorylation of BAK1K or FLS2K in the presence of BIK1, we generated the catalytically inactive kinase mutants BAK1KKm and FLS2KKm. GST-BIK1 also phosphorylated GST-BAK1KKm and GST-FLS2KKm in vitro (Fig. 4E). Similarly, BIK1, but not BIK1Km, could phosphorylate BAK1CDKm (Fig. S6B). Our data suggest the transphosphorylation of BIK1 with FLS2/BAK1 in the flagellin receptor complex. This phosphorylation was significantly reduced by BIK1T237A, although not completely eliminated (Fig. 4E). BIK1T237A also significantly reduced its autophosphorylation activity (Fig. 4E). The results suggest that BIK1 Thr237, the flg22-mediated phosphorylation site, is required for its phosphorylation on FLS2 and BAK1. Importantly, expression of BIK1T237A significantly reduced the activation of the MAMP-responsive genes FRK1-LUC and WRKY29-LUC compared to wild-type BIK1 (Fig. S7), suggesting that this residue is essential for BIK1 to function in MAMP signaling. The functional importance of Thr237 in plant defense gains further support from the recent study concluding that the corresponding residue in the tomato BIK1 ortholog TPK1b is required for mediating Botrytis cinerea resistance in Arabidopsis (28).
BIK1 Is Required in Flagellin-Triggered Immunity.
To further verify the functional significance of flg22-induced BIK1 phosphorylation, we isolated an insertional mutant of the BIK1 gene from the Salk T-DNA insertion collection (Salk_005291) (29). RT–PCR analysis confirmed that bik1 is a null allele with undetectable transcript [Fig. S8 A and B (17)]. The bik1 mutant did not affect flg22-induced FLS2 and BAK1 association (Fig. S3C), suggesting that BIK1 is not required for flg22 binding to FLS2. We tested the bik1 sensitivity to flg22 with seedling growth assay. The bik1 mutant showed a clear reduction of sensitivity to flg22 compared to wild-type plants although to a lesser extent than the bak1 mutant (Fig. 5A). We next performed a pathogen infection assay with wild-type and bik1 mutant seedlings and adult plants. Significantly, bik1 mutant seedlings lost flg22-induced resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 infection. Flg22-pretreated wild-type seedlings were more resistant than H2O-pretreated seedlings as detected by bacterial multiplication 3 and 5 days after infection (Fig. 5B). However, the bacterial growth did not differ significantly in flg22- and H2O-pretreated bik1 seedlings (Fig. 5B). Although bik1 mutants are more resistant to virulent Pst DC3000 infection than wild-type plants because of the elevated salicylic acid (SA) level (17), the enhanced resistance of plate-grown seedlings was not as pronounced as soil-grown plants probably because high humidity in plates could reduce the high SA level in the bik1 mutant (Fig. 5B and Fig. S8C). The flg22-induced resistance was also not observed in 4-week-old bik1 mutant plants inoculated with Pst DC3000 (Fig. S8C). Furthermore, when we infected wild-type and bik1 mutant seedlings with the nonpathogenic Pst DC3000 type III secretion mutant hrcC, bik1 mutant seedlings were significantly immunocompromised (Fig. 5C and Fig. S8D). Bacterial growth assay indicated that the bacterial number was about five- to-eightfold higher in the bik1 mutant than in wild-type seedlings 3 days after infection (Fig. 5C). Six days after infection with hrcC, bik1 mutant seedlings exhibited severe necrotic symptoms, whereas the wild-type seedlings were still green at this time point (Fig. S8D). The bik1 mutant complemented with the BIK1 gene completely restored plant immunity to hrcC to the wild-type level (Fig. 5C and Fig. S8D). Plant immunity to nonpathogenic Pst DC3000 hrcC infection is likely attributed by the action of multiple MAMPs. The data strongly support that BIK1 is an important signaling component in flagellin and likely in other MAMP signaling.
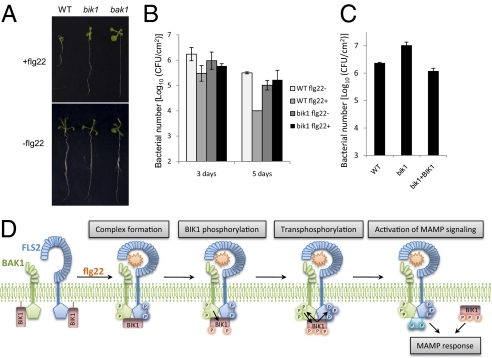
Fig. 5.
BIK1 is required in flg22-mediated immunity. (A) bik1 mutants show reduced sensitivity to flg22 in seedling growth assay. Wild-type (Col-0, WT), bak1, and bik1 seedlings were grown for 10 days in the presence of 200 nM flg22 (Upper) or in the absence of flg22 (Lower). (B) bik1 mutants are compromised in flg22-mediated immunity to Pst DC3000 infection. Twelve-day-old WT and bik1 seedlings were pretreated with or without flg22 and then infected with Pst DC3000. The bacterial growth assays were performed 3 and 5 days after infection. (C) bik1 mutants are compromised in plant immunity to Pst DC3000 hrcC. Results of a bacterial growth assay of WT, bik1 mutant, and BIK1 complementation (bik1+BIK1) seedlings 3 days after infection are shown. (D) A model of BIK1 in flagellin signaling. In the absence of flagellin (flg22), BIK1 associates with FLS2 and BAK1 in an inactive state. On flagellin binding to FLS2, flg22 induces FLS2 and BAK1 association and probably phosphorylation. The activated BAK1 phosphorylates BIK1, which in turn transphosphorylates the FLS2–BAK1 complex. The fully active FLS2–BAK1 may further phosphorylate BIK1 and other substrates (blue configuration), and then the active BIK1 is likely released from the FLS2–BAK1 complex to activate downstream intracellular signaling.
Discussion
Plants and animals respond to an array of MAMPs from both pathogenic and nonpathogenic microbes and activate convergent immune signaling (1, 4). However, little is known about the molecular and biochemical mechanisms of MAMP receptor activation and signal transduction to intracellular signaling upon MAMP perception. We provide compelling evidence that a cytoplasmic kinase BIK1 associates with the flagellin receptor complex FLS2/BAK1 and functions in MAMP signaling. BIK1 is rapidly phosphorylated by flg22 within the first minutes after stimulation (Fig. 1D), which may happen instantaneously with the formation of the FLS2/BAK1 complex. BIK1 appears to function downstream of FLS2/BAK1 complex formation and phosphorylation because BIK1 phosphorylation requires not only the presence of both FLS2 and BAK1, but also their kinase activity (Fig. 2 A and B). BIK1 is directly phosphorylated by BAK1; however, BIK1 could also phosphorylate BAK1 and FLS2 (Fig. 4 C–E; Fig. S5 and S6), which may result in enhanced phosphorylation on BIK1 and other substrates by BAK1/FLS2. This is supported by the fact that BIK1 phosphorylation was gradually enhanced upon flg22 treatment within 10 min (Fig. 1D). The association of BIK1 with FLS2 and BAK1 appears to be reduced upon flagellin perception (Fig. 3 A and B), suggesting that the fully activated BIK1 is likely released from the MAMP receptor complex to propagate MAMP signaling (Fig. 5D). BIK1 was originally identified as an important component in Arabidopsis resistance to necrotrophic fungi (17). The bik1 mutant was much more susceptible to B. cinerea and Alternaria brassicicola than wild-type plants with the attenuated jasmonate- and ethylene-regulated defense response. It has been suggested that BIK1 may function as an early component in plant defense signaling. However, bik1 mutants display enhanced resistance to the virulent bacterial pathogen Pst DC3000. The seemingly contradictory disease responses are largely attributable to the enhanced SA level in bik1 mutants because bik1NahG plants, in which the elevated SA was removed by expressing the NahG gene that degrades SA, were more susceptible to DC3000 infection, suggesting a positive role of BIK1 in plant immunity that is independent of the SA pathway (17). flg22-induced resistance appears to be independent of SA signaling (22). Consistent with this notion, we found that bik1 mutants lost flg22-mediated immunity to pathogenic bacteria and were significantly immunocompromised to nonpathogenic bacterial infection. Taken together, the results point to the functional importance of BIK1 in plant PTI.
In line with our molecular, biochemical, and genetic analyses, existing evidence supports the functional association of BAK1 and BIK1 in several aspects. Both bak1 and bik1 mutants are more susceptible to the necrotrophic fungi B. cinerea and A. brassicicola with enhanced H2O2 production upon fungal challenge (17, 30). It has been documented that bik1 and bak1 mutants are more resistant to some biotrophic pathogens, such as bik1 to Pst DC3000 and bak1 to oomycete Hyaloperonospora parasitica (17, 30). In addition to defense, both bak1 and bik1 mutant plants show a certain similarity in growth defects, such as reduced leaf growth (17, 30). The growth defect of bak1 is largely attributable to a deficiency in BR signaling. It is not known how BIK1 is involved in normal plant growth and development. Importantly, in contrast to the defense responses, bik1 growth defects are not restored to wild-type levels by expression of NahG (17), suggesting that BIK1 controls immunity and development with distinct signaling mechanisms. This assumption gains further support from the studies of the heterogeneous expression of the BIK1 tomato ortholog TPK1b in the Arabidopsis bik1 mutant (28). TPK1bT238, which is equivalent to BIK1T237 and identified as a major flg22 phosphorylation site in our study, is required for its function in disease resistance, but not in growth. Similarly, BAK1T450 is essential in flg22 signaling, whereas it is dispensable in BR-mediated growth (31). The question remains whether the distinct signaling output mediated by BAK1 and BIK1 in the control of plant immunity and development is specified by the unique phosphorylation sites upon different stimuli.
Recently, BSKs, members of RLCKs, were identified as signaling components in transducing early BR signaling (24). However, BSKs differ from BIK1 in that BSKs associate only with the BR receptor BRI1 and are phosphorylated by BRI1, not by BAK1 (24). In contrast, BIK1 associates with both the flagellin receptor FLS2 and its signaling partner BAK1 and is phosphorylated by BAK1 (Figs. 3 and 4C and Fig. S5). Furthermore, BIK1 transphosphorylates BAK1 and FLS2 (Fig. 4 D and E and Fig. S6). Significantly, the flg22 phosphorylation site Thr237 is essential for this transphosphorylation (Fig. 4E), suggesting that BIK1 is likely first phosphorylated upon flagellin perception and then subsequently phosphorylates BAK1–FLS2. This is consistent with the fact that flagellin-induced BIK1 phosphorylation requires the FLS2–BAK1 receptor complex (Fig. 2 A and B). Our finding of a transphosphorylation event in the MAMP receptor kinase complex is also distinct from a finding that, in the BR receptor complex, transphosphorylation occurs only between BRI1 and BAK1 (31). In conclusion, our results point to a unique model of plant innate immune signaling via a receptor kinase complex in which flg22-stimulated FLS2–BAK1 complex formation induces BIK1 phosphorylation. Subsequently, BIK1 transphosphorylates the FLS2–BAK1 complex to enhance the flg22 signaling by further phosphorylating BIK1 and other possible substrates, and the phosphorylated BIK1 is likely released from the FLS2–BAK1 complex to activate downstream intracellular signaling (Fig. 5D).
Experimental Procedures
Plant Material and Growth Conditions.
Wild-type (Col-0), bik1, fls2, bak1-4, and serk4 mutant Arabidopsis plants were grown in a growth room at 23°C, 60% relative humidity, 70 μE light with a 12-h photoperiod for 30 days before protoplast isolation or bacterial inoculation. The fls2, bak1-4, and serk4 mutants and bik1 complementation lines were reported previously (13, 17). The bik1 mutant (Salk_005291) was obtained from the Arabidopsis Biological Resource Center and confirmed by PCR and RT–PCR analyses. Seedlings were grown on a 1/2 Murashige and Skoog medium (MS) plate with 1% sucrose and 0.9% agar at 23°C and 70 μE light with a 12-h photoperiod for 12 days. Seedlings were transferred to 2 mL H2O in the six-well tissue culture plates 1 day before flg22 treatment for 10 min. RT–PCR analysis and flg22-mediated inhibition of seedling growth were carried out as described (13, 32). All experiments were repeated three to four times with reproducible results.
Plasmid Constructs, Protoplast Transient Assay, and Generation of Transgenic Plants.
Arabidopsis BAK1 and FLS2 constructs were reported previously (13). BIK1, OX1, and BSK1 genes were amplified by PCR from Col-0 cDNA and introduced into a plant expression vector with an HA or FLAG epitope tag at the C terminus. BIK1 point mutations were generated by a site-specific mutagenesis kit (Stratagene). The primer sequences for different kinases and BIK1 point mutations are listed in the SI Experimental Procedures. Full-length BIK1, a cytosolic domain, or the kinase domain of BAK1 and FLS2 were subcloned into the modified GST fusion protein expression vector pGEX4T-1 (Pharmacia) or pMAL-c2 (New England Biolabs) with BamHI and StuI digestion. Protoplast transient assay was carried out as described (13, 27). For BIK1 phosphorylation assays, 0.1-mL protoplasts at a density of 2 × 105/mL were transfected with 20 μg of plasmid DNA. For Co-IP assays, 1-mL protoplasts were transfected with 200 μg of DNA. For GST pull-down assays, 0.5-mL protoplasts were transfected with 100 μg of DNA. The BIK1 transgenic plants in Col-0, fls2, and bak1-4 were generated by Agrobacterium-mediated transformation with the BIK1 construct under the control of a constitutive cauliflower mosaic virus 35S promoter with an HA epitope tag.
Elicitor and Chemical Inhibitor Treatments.
The flagellin peptide flg22 (32) and the EF-Tu peptide elf18 (21) were used in a concentration of 1 μM if not otherwise stated. Chitin fragments (33) were used at a final concentration of 50 μg/mL. Chemical inhibitors K-252a, diphenylene iodonium (DPI), and U0126 were purchased from A. G. Scientific, prepared as a stock solution of 1 mM, and used at a final concentration of 1 μM for K-252a, 5 μM for DPI, and 1 μM for U0126. Different chemical inhibitors were added 1 h before the 1-μM flg22 treatment. CIP was purchased from New England BioLabs, and the treatment was carried out following the instruction.
Coimmunoprecipitation and GST Pull-Down Assays.
Protoplasts were lysed with 0.5 mL of extraction buffer (10 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100, and a protease inhibitor mixture from Roche). After vortexing vigorously for 30 s, the samples were centrifuged at 12,470 ×g for 10 min at 4°C. For the Co-IP assay, the supernatant was incubated with anti-HA or anti-FLAG antibody for 2 h and then with protein-G-agarose beads for another 2 h at 4°C with gentle shaking. For the GST pull-down assay, the supernatant was incubated with prewashed GST, GST-BIK1, GST-BAK1K, or GST-FLS2K beads for 2 h at 4°C with gentle shaking. The beads were collected and washed three times with washing buffer (10 mM Hepes, pH7.5, 100 mM NaCl, 1 mM EDTA, 10% glycerol, and 0.1% Triton X-100) and once with 50 mM Tris·HCl, pH7.5. The immunoprecipitated proteins were analyzed by Western blot with an anti-HA or anti-FLAG antibody. The protein bands with appropriate molecular weight are shown.
In Vitro Phosphorylation and Immunocomplex Kinase Assays.
Expression of the GST and MBP fusion proteins and affinity purification were performed as standard protocol. The protein concentration was determined with the BIO-RAD Quick Start Bradford Dye Reagent and confirmed by the NanoDrop ND-1000 Spectrophotometer. For in vitro kinase assay, kinase reactions were performed in 30 μl of kinase buffer (20 mM Tris·HCl, pH 7.5, 10 mM MgCl2, 5 mM EGTA, 100 mM NaCl, and 1 mM DTT) containing 10 μg of fusion proteins with 0.1 mM cold ATP and 5 μCi of [32P]-γ-ATP at room temperature for 3 h with gentle shaking. The reactions were stopped by adding 4× SDS loading buffer. The phosphorylation of fusion proteins was analyzed by autoradiography after separation with SDS/PAGE. For immunocomplex kinase assays, protoplasts were lysed with 0.5 mL of IP buffer (50 mM Tris·HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM DTT, 2 mM NaF, 2 mM Na3VO3, 1% Triton, and a protease inhibitor mixture from Roche). After centrifugation at 12,470 ×g for 10 min at 4°C, the supernatant was incubated with anti-HA or anti-FLAG antibody for 2 h and then with protein-G–agarose beads for another 2 h at 4°C with gentle shaking. The beads were collected and washed once with IP buffer and once with kinase buffer (20 mM Tris·HCl, pH 7.5, 20 mM MgCl2, 5 mM EDTA, and 1 mM DTT). The kinase reactions were performed in 20 μl of kinase buffer with 2 μg of GST fusion proteins, 0.1 mM cold ATP, and 5 μCi of [32P]-γ-ATP at room temperature for 1 h with gentle shaking. The phosphorylation of GST fusion proteins was analyzed by 10% SDS/PAGE.
Pathogen Infection Assays.
P. syringae tomato DC3000 and hrcC strains were grown overnight at 28°C in King’s B medium with 50 μg/mL rifampicin. Bacteria were collected, washed, and diluted to the desired density with H2O. For the 4-week-old plant flg22 protection assay, leaves were preinoculated with 200 nM flg22 or an H2O control for 24 h and then infiltrated with Pst DC3000 at the concentration of 5 × 105 cfu/mL using a needleless syringe (22). For seedling infection assay, seedlings were grown in 1/2 MS medium for 10 days in a 12-well tissue culture plate. Bacteria were added at the concentration of 2 × 108 cfu/mL in each well. Bacterial counting was performed from six leaves as three repeats by surface sterilization with 70% ethanol (34, 35).
Supplementary Material
Acknowledgments
We are grateful to Dr. Tesfaye Mengiste for generously sharing the mutant and transgenic plant seeds and Arabidopsis Biological Resource Center for the Arabidopsis T-DNA insertion lines. We thank Drs. Martin Dickman and Jen Sheen for critical reading of the manuscript. We appreciate Drs. Mehdi Kabbage, Brett Williams, Xiuren Zhang, Tim Devarenne, Keyan Zhu-Salzman, and Martin Dickman for various assistance during the lab setup. This work was supported by Texas A&M University start-up funds to L.S. and P.H.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909705107/DCSupplemental.
References
- 1.Boller T, He SY. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 5.Bent AF, Mackey D. Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu Rev Phytopathol. 2007;45:399–436. doi: 10.1146/annurev.phyto.45.062806.094427. [DOI] [PubMed] [Google Scholar]
- 6.Gomez-Gomez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 7.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 8.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104:12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 10.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 11.Belkhadir Y, Wang X, Chory J. Arabidopsis brassinosteroid signaling pathway. Sci STKE. 2006;364:cm5. doi: 10.1126/stke.3642006cm4. [DOI] [PubMed] [Google Scholar]
- 12.Kinoshita T, et al. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 13.Shan L, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veronese P, et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18:257–273. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peck SC. Early phosphorylation events in biotic stress. Curr Opin Plant Biol. 2003;6:334–338. doi: 10.1016/s1369-5266(03)00056-6. [DOI] [PubMed] [Google Scholar]
- 19.Benschop JJ, et al. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics. 2007;6:1198–1214. doi: 10.1074/mcp.M600429-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Nuhse TS, Bottrill AR, Jones AM, Peck SC. Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 2007;51:931–940. doi: 10.1111/j.1365-313X.2007.03192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 22.Zipfel C, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 23.Rentel MC, et al. OXI1 kinase is necessary for oxidative burst-mediated signalling in Arabidopsis. Nature. 2004;427:858–861. doi: 10.1038/nature02353. [DOI] [PubMed] [Google Scholar]
- 24.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin GB, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 26.Xiang T, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 27.Asai T, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 28.Abuqamar S, Chai MF, Luo H, Song F, Mengiste T. Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell. 2008;20:1964–1983. doi: 10.1105/tpc.108.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 30.Kemmerling B, et al. The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol. 2007;17:1116–1122. doi: 10.1016/j.cub.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 33.Wan J, et al. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber K, Ckurshumova W, Peek J, Desveaux D. A high-throughput chemical screen for resistance to Pseudomonas syringae in Arabidopsis. Plant J. 2008;54:522–531. doi: 10.1111/j.1365-313X.2008.03425.x. [DOI] [PubMed] [Google Scholar]
- 35.Songnuan W, et al. A seedling assay for MAMP signaling and infection studies. In: Lorito M, Loo SL, Scala F, editors. Biology of Plant-Microbe Interactions. St. Paul, MN: International Society for Molecular Plant-Microbe Interactions; 2007. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.