Abstract
Citrobacter rodentium (formally Citrobacter freundii biotype 4280) is a highly infectious pathogen that causes colitis and transmissible colonic hyperplasia in mice. In common with enteropathogenic and enterohemorrhagic Escherichia coli (EPEC and EHEC, respectively), C. rodentium exploits a type III secretion system (T3SS) to induce attaching and effacing (A/E) lesions that are essential for virulence. Here, we report the fully annotated genome sequence of the 5.3-Mb chromosome and four plasmids harbored by C. rodentium strain ICC168. The genome sequence revealed key information about the phylogeny of C. rodentium and identified 1,585 C. rodentium-specific (without orthologues in EPEC or EHEC) coding sequences, 10 prophage-like regions, and 17 genomic islands, including the locus for enterocyte effacement (LEE) region, which encodes a T3SS and effector proteins. Among the 29 T3SS effectors found in C. rodentium are all 22 of the core effectors of EPEC strain E2348/69. In addition, we identified a novel C. rodentium effector, named EspS. C. rodentium harbors two type VI secretion systems (T6SS) (CTS1 and CTS2), while EHEC contains only one T6SS (EHS). Our analysis suggests that C. rodentium and EPEC/EHEC have converged on a common host infection strategy through access to a common pool of mobile DNA and that C. rodentium has lost gene functions associated with a previous pathogenic niche.
The mouse has been utilized extensively as a model for studies of infection and immunity (11, 25, 41, 44). Mice can be housed in relatively simple facilities and can be maintained under controlled pathogen- or germ-free conditions. Further, the availability of inbred or genetically manipulated mice greatly enhances the potential of any studies (35, 80). Although murine infection systems are used extensively, relatively few pathogens have been exploited that are naturally virulent for the species.
Spontaneous disease outbreaks in mouse colonies in the United States and Japan in the 1960s and 1970s were associated with infections by the Gram-negative pathogens Escherichia coli (Ex30, also known as murine pathogenic E. coli [MPEC]), Citrobacter freundii ANL, and C. freundii biotype 4280 (3, 9, 25, 43, 64). On the basis of DNA relatedness, these pathogenic strains were subsequently assigned to a separate species, Citrobacter rodentium (54, 84). C. rodentium is highly infectious, causing colitis and transmissible colonic hyperplasia (61). Following ingestion, C. rodentium colonizes the intestines of mice, residing predominantly in the cecum and colon (91), where in some inbred strains it can drive hyperplasia or overgrowth of the epithelium. C. rodentium-mediated colonic hyperplasia is highly infectious in poorly managed mouse facilities, spreading between mice via contaminated feces and environments (93). Some mouse strains are relatively resistant to clinical disease, but in such strains, C. rodentium lives in the intestine as a component of the microbiota; younger mice are generally more susceptible to infection (89).
C. rodentium is genetically related to Escherichia coli and is a member of the Enterobacteriacae. An extracellular pathogen, C. rodentium colonizes the lumen of the mouse gut mucosa by formation of so-called attaching/effacing (A/E) lesions on the apical surfaces of the enterocytes (82; reviewed in references 30 and 61). A/E lesions, which are characterized by effacement of the brush border microvilli and intimate bacterial attachment to pedestal-like structures, are also classically associated with the pathogenesis of enteropathogenic E. coli (EPEC), a leading cause of pediatric diarrhea, and enterohemorrhagic E. coli (EHEC), which causes diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome (HUS) (reviewed in reference 45). In this respect, C. rodentium, EPEC, and EHEC share a common virulence strategy.
A/E lesion formation is dependent on the expression of intimin, a type III secretion system (T3SS) (reviewed in reference 31) and effector proteins encoded on a specific pathogenicity island (PI) known as the locus for enterocyte effacement (LEE) (57). The LEE of C. rodentium, and those of EPEC and EHEC, are horizontally acquired sections of DNA, and although not identical, they are highly related in terms of overall genetic organization and gene function (19, 26, 75). The fact that mice are resistant to EPEC and EHEC infection (60) makes C. rodentium an ideal animal model to study the roles of conserved genes in pathogenesis in vivo.
Aside from the role of the LEE in pathogenesis, a number of other genes have been shown to contribute to C. rodentium disease, including type IV pili (63) and various T3SS effector proteins (33, 39, 46, 55, 62, 86). However, direct comparisons between the virulence gene repertoires of C. rodentium and other enteric pathogens have been limited. Whole-genome sequencing has proved to be a powerful approach to defining the genetic structure of bacteria, providing a genomewide blueprint to guide genetic analysis. A fully annotated genome sequence can guide targeted mutagenesis and transcriptomic or proteomic studies. Here, we report the fully annotated genome sequence of a murine virulent strain of C. rodentium, ICC168, and make comparisons with the genomes of EPEC, EHEC, and other enteric bacteria.
MATERIALS AND METHODS
Bacterial strains and sequencing.
The strain of C. rodentium sequenced, strain ICC168, was obtained in 1993 from S. W. Barthold's original stocks, previously known as Citrobacter freundii biotype 4280 (ATCC 51459) (3, 4), and subjected to minimal passages. The bacterial strains, plasmids, and primers used in the biological studies are shown in Table 1. Ten μg genomic DNA was fragmented by multiple passages through a 30-gauge needle and end repaired with a mixture of Klenow polymerase, T4 DNA polymerase, and polynucleotide kinase before size selection by preparative electrophoresis through a 0.8% agarose gel. The recovered sized fractions were subjected to a second round of end repair and size selection. Two- to 2.2-kb and 2.2- to 4-kb fractions were ligated into pUC19_smaI, and 4- to 5-, 5- to 6-, 6- to 9-, and 9- to 10-kb fractions were ligated into the low-copy-number vector pMAQ1b_smaI. Two bacterial artificial chromosome (BAC) libraries containing 15- to 25- and 25- to 40-kb inserts were constructed as previously described (69). End sequences obtained from 672 clones from each library were used to scaffold the assembly. The whole-genome shotgun was sequenced to approximately 9× coverage on ABI3730 automated sequencers using BigDye terminator chemistry.
TABLE 1.
Bacterial strains, plasmids and primers used in this study
| Strain, plasmid, or primer | Description | Reference |
|---|---|---|
| Strains | ||
| ICC169 | Spontaneous Nalr derivative of ICC168 | 92 |
| ICC304 | ΔespA::AphT in ICC169 | This study |
| E2348/69 | Wild-type EPEC O127:H6 | 50 |
| Plasmids | ||
| pCX340 | Expression vector for TEM fusion protein | 14 |
| pICC453 | pCX340-EspS, expressing C. rodentium EspS fused to TEM | This study |
| pICC454 | pCX340-EspH, expressing E2348/69 EspH fused to TEM | This study |
| pACYC184 | Bacterial expression vector | New England Biolabs |
| pICC490 | pACYC184 expressing espA from the tet promoter | This study |
| Primers | ||
| NdeI-EspS-Fw | 5′-AAACTCGAGCATATGTTTAGTATAAAAGGATACCTACC-3′ | |
| EcoRI-EspS-Rv | 5′-CCGGAATTCTCGTCAATATCTGTTGGTTGTGTCC-3′ | |
| NdeI-EspH-Fw | 5′-CCCGAGCATATGCGTTATATAGGGAGGTGTATG-3′ | |
| EcoRI-EspH-Rv | 5′-CGGAATTCCCAACTGTCACACCTGATAAAGAG −3′ | |
| EspA-Fw | 5′-TTATTAGGTGAGGATGATTGGG-3′ | |
| BamHI-EspA-Rv | 5′-CGGGATCCCTGATGTCATAGTTGATGTATCC-3′ | |
| BamHI-EspA-Fw | 5′-CGGGATCCAACTGTTTCAGCAATACCCATTG-3′ | |
| EspA-Rv | 5′-ATCATTATTAAGCACAGTTGTACC-3′ | |
| EcoRV-rbsEspA-Fw | 5′-TTTGATATCAAGAAGGAGATATACCATGGATACATCAACTATGACATCAGTTGC-3′ | |
| BamHI-EspA-Rv | 5′-CGCGGATCCTTATTTGCCAATGGGTATTGCTGAAACAG-3′ |
The sequence was assembled, finished, and annotated as previously described (72). The program Artemis (81) was used to collate data and facilitate annotation. Base 1 of the C. rodentium genome matched that of E. coli K-12 MG1655 to aid whole-genome alignments, making the first gene thrL. The origin of replication is located between positions 4232946 and 4233181.
Pseudogenes were defined as coding sequences (CDS)s that had one or more possible inactivating mutations based on similarity with intact CDSs in related bacteria. Each single frameshift mutation or premature stop codon within a CDS was confirmed by checking the original sequencing data.
In addition, in order to provide a robust whole-genome phylogenetic species tree for Citrobacter, we sequenced the whole genome of Citrobacter freundii ballerup 7851 using 454 FLX pyrosequencing, assembled using the 454/Roche Newbler assembly program into 357 contigs (N50 contig size, 60,237 bp) from 410,057 sequence reads with an average read length of 177 bp to give a total sequence length of 4,904,659 bp.
Bioinformatics analyses.
The following genome sequences were used in genome comparisons with C. rodentium ICC168: E. coli K-12 MG1655 (K-12; accession number U00096), E. coli E2348/69 (EPEC; accession number FM180568), E. coli O157:H7 Sakai (EHEC; accession number BA000007), Salmonella enterica subspecies enterica serovar Typhi CT18 (accession number AL513382), S. enterica subspecies enterica serovar Typhimurium LT2 (accession number AE006468), S. enterica subspecies enterica serovar Enteritidis P125109 (accession number AM933172), Citrobacter koseri ATCC BAA-895 (accession number CP000822), Yersinia enterocolitica 8081 (accession number AM286415), Pectobacterium atrosepticum SCRI1043 (accession number BX950851), Klebsiella pneumoniae 342 (accession number CP000964), Enterobacter sakazakii ATCC BAA-894 (accession number CP000783), and Serratia marcescens Db11 (http://www.sanger.ac.uk/Projects/S_marcescens/).
Pairwise whole-genome comparisons of C. rodentium with a range of enteric bacteria were performed using TBLASTX and visualized using the Artemis Comparison Tool (13). Circular diagrams were made using DNAplotter (12). Average nucleotide identities (ANI) were calculated from BLASTN matches over the entire lengths of the bacterial chromosomes. Orthologues in C. rodentium ICC168 and the three E. coli genomes (K-12, EPEC, and EHEC) were identified using an all-against-all reciprocal FASTA comparison of translated DNA with at least 40% identity over 80% of the length.
Phylogenetic analyses.
The sequences for each of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) were extracted from the whole-genome sequences of several enteric bacteria, individually aligned using MUSCLE (24), and then concatenated. FindModel (77) was used to identify the best phylogenetic model to fit the data. Phylogenetic trees were constructed by the maximum-likelihood method using the General Time Reversible (GTR) plus Gamma model with PhyML (34) and Bayesian likelihood using GTR plus Gamma with 100,000 generations, sampling every 1,000, and a burn-in of 1,000 using MrBayes, and gave consistent results.
Strain and plasmid construction.
C. rodentium strain ICC169 ΔespA (ICC304) was constructed using a modified version of the lambda red-based mutagenesis system (18). The espA gene and flanking regions were PCR amplified from wild-type C. rodentium ICC169 genomic DNA using the primers EspA-Fw/BamHI-EspA-Rv and BamHI-EspA-Fw/EspA-Rv (Table 1). The two fragments were digested with BamHI, ligated to each other, and cloned into pGEMT, and the nonpolar aphT cassette was then inserted into the BamHI site between the two fragments. After verification for correct orientation of the kanamycin resistance gene cassette, the insert was PCR amplified using the EspA-Fw and EspA-Rv primers (Table 1). The PCR product was electroporated into ICC169 containing pKD46 encoding the lambda red recombinase (18). Transformants were selected on kanamycin plates, and the correct insertion of the kanamycin resistance gene cassette was confirmed by PCR.
Full-length espA was amplified using C. rodentium ICC169 genomic DNA as a template and the primer pair EcoRV-rbsEspA-Fw and BamHI-EspA-Rv (Table 1). The PCR product was digested and ligated into the EcoRV/BamHI sites of pACYC184 to produce plasmid pICC490, which constitutively expresses espA from the tetracycline promoter (Table 1). The construction was confirmed by DNA sequencing.
Genes encoding EspS and EspH fusions to TEM-1 β-lactamase were constructed by PCR amplification using ICC169 or E2348/69 genomic DNA and the primer sets NdeI-EspS-Fw/EcoRI-EspS-Rv and Nde-EspH-Fw/EcoRI-EspH-Rv, respectively (Table 1). The plasmids resulting from the ligation of the digested PCR products into pCX340 were called pICC453 and pICC454, respectively.
The Role of EspA in vivo.
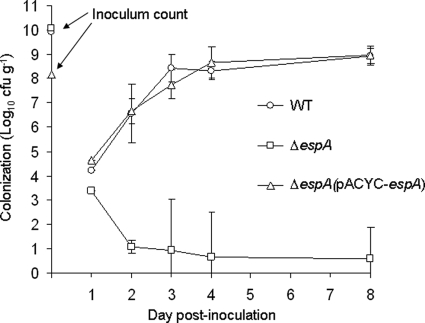
Pathogen-free male C3H/Hej mice, 6 to 8 weeks old, were purchased from Harlan Olac (Bichester, United Kingdom). All animals were housed in individually HEPA-filtered cages with sterile bedding and free access to sterilized food and water. All animal experiments were performed in accordance with the Animals Scientific Procedures (Act 1986) and were approved by the local ethical review committee. Independent single-infection experiments were performed twice using four mice per group. The mice were inoculated by oral gavage with 200 ml of overnight LB-grown C. rodentium suspension in phosphate-buffered saline (PBS) (∼5 × 109 CFU). The number of viable bacteria used as an inoculum was determined by retrospective plating on LB agar containing antibiotics. Stool samples were recovered aseptically at various time points after inoculation, and the number of viable bacteria per gram of stool was determined by plating them on LB agar (93). The experiment was terminated at day 8 postinoculation by sacrificing the mice.
T3SS translocation assay.
The translocation assay is an adaptation of the assay described previously (14). The Swiss 3T3 cell line was grown in Dulbecco's modified Eagle's medium (DMEM) containing 4,500 mg liter glucose−1 supplemented with 10% fetal calf serum and 2 mM glutamine at 37°C in 5% CO2. Two hundred μl of cells was seeded in black-wall/clear-bottom 96-well plates at a density of 2 × 104 cells per well to obtain 100% confluence on the day of infection. C. rodentium strains were grown for 8 h in LB broth and then transferred into fresh, sterile DMEM containing 1,000 mg liter glucose−1 and incubated statically at 37°C in 5% CO2 overnight prior to infection. Each well was infected with 40 μl of the static overnight culture of either C. rodentium ICC169 or ICC304 carrying either pICC453 or pICC454. This was then centrifuged at 1,000 rpm for 5 min at room temperature. The infection was carried out at 37°C in 5% CO2 for 2 h, followed by induction of the TEM-1 β-lactamase fusion protein for 1 h 30 min in the presence of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The cells were washed three times with Hanks' buffered salt solution (HBSS) and incubated for a minimum of 1 h 30 min in the dark at room temperature in 100 μl of 6 mM probenecid in HBSS-20 mM HEPES buffer supplemented with 20 μl of 6× CCF2/AM solution freshly prepared with the CCF2/AM loading kit (Invitrogen). The cells were washed in HBSS, and the fluorescence was quantified on a Fluostar Optima reader with excitation at 410 nm (10-nm band-pass), and emission was detected via 450-nm (blue fluorescence) and 520-nm (green fluorescence) filters. The translocation rate was expressed as the 450/520-nm emission ratio. Experiments were performed in triplicate.
Nucleotide sequence accession numbers.
The annotated genome sequence of C. rodentium ICC168 has been deposited in the public databases under accession numbers FN543502 (chromosome), FN543503 (pCROD1), FN543504 (pCROD2), and FN543505 (pCROD3). The sequence of the fourth plasmid was identical to that of the previously sequenced plasmid pCRP3 (accession number AF311902). The raw sequence data generated for Citrobacter freundii ballerup 7851 has been deposited in the trace archive under accession number ERA000106 (ftp://ftp.era.ebi.ac.uk/vol1/ERA000/).
RESULTS
Genome structure and general features.
The genome of C. rodentium ICC168 consists of a 5.3-Mb circular chromosome and four plasmids (pCROD1 to -3 and pCRP3) that range in size from 54 to 3 kb (Table 2; see Table S1 in the supplemental material). The genome of C. rodentium ICC168 shares significant synteny with those of E. coli K-12 and other members of the Enterobacteriaceae; however, there is evidence of repeated mediated recombination events (see below and data not shown). The chromosome is predicted to contain 4,984 CDSs, including 182 pseudogenes, 86 tRNAs, and 7 rRNA operons. We identified a total of 10 prophage-like regions, including five complete prophages, which were mainly Mu- or P2-like, and four putative lambdoid prophage remnants (Table 3) . Seventeen genomic islands (GIs) were also identified within the chromosome (GI1 to GI16 and the LEE) as regions of aberrant GC content carrying mobility functions or showing evidence of integration, such as being flanked by direct repeats or insertion sequence (IS) elements. The exceptions are GI2, GI6, and GI13, which were identified on the basis of similarity to Salmonella pathogenicity islands 4 and 9. Many of the known and putative virulence factors of C. rodentium are encoded on the GIs (Table 3).
TABLE 2.
Genome features of C. rodentium ICC168
| Parameter | Value |
||||
|---|---|---|---|---|---|
| Chromosome | pCROD1 | pCROD2 | pCROD3 | pCRP3 | |
| Genome size (bp) | 5,346,659 | 54,449 | 39,265 | 3,910 | 3,172 |
| G+C content (%) | 54.7 | 47.2 | 41.8 | 51.0 | 46.8 |
| No. of CDSsa | 4,984 (182) | 60 (5) | 56 (1) | 3 | 8 |
| CDS density (%) | 88.5 | 82.5 | 86.2 | 79.6 | 72.6 |
| No. of tRNAs | 86 | 0 | 0 | 0 | 0 |
| No. of rRNA operons | 7 | 0 | 0 | 0 | 0 |
| No. of miscellaneous RNAs | 56 | 1 | 0 | 0 | 1 |
| No. of prophage regions | 10 | 0 | 0 | 0 | 0 |
| No. of genomic islands | 17 | 0 | 0 | 0 | 0 |
| No. of IS elementsb | 109 (31) | 3 (2) | 1 | 0 | 0 |
The numbers in parentheses indicate the numbers of pseudogenes.
The numbers in parentheses indicate partial/disrupted IS elements.
TABLE 3.
Prophages and genomic islands of C. rodentium ICC168
| Name | Start position | End position | Length (bp) | Insertion site | Flanking repeat (bp)a | Type | Virulence-related and other notable factors encoded |
|---|---|---|---|---|---|---|---|
| Prophages | |||||||
| CRP28 | 285,573 | 325,997 | 40,425 | lfiG-lfiI | ND | Mu-like prophage; intact | Dam |
| CRP99 | 990,424 | 1,027,608 | 37,185 | ROD_08971 | ND | Mu-like prophage; intact | Dam |
| CRPr11 | 1,143,137 | 1,151,111 | 7,975 | ROD_t18 (tRNASer) | 70 DR | Putative lambdoid prophage; remnant | |
| CRPr13 | 1,304,879 | 1,315,433 | 10,555 | potC-potB | 12 DR | Putative lambdoid prophage; remnant | Three T3SS effectors (EspX7, EspN1c, EspK) |
| CRPr17 | 1,762,632 | 1,769,107 | 6,476 | uspF-ttcA | 25 DR (2 MM) | putative lambdoid prophage; remnant | Three T3SS effectors (NleC, NleB2c, NleG1) |
| CRPr20 | 2,096,166 | 2,115,165 | 19,000 | fliC | 8 DR | P2-like prophage; remnant | Dam |
| phiNP | 2,730,430 | 2,775,336 | 44,907 | ssrA (tmRNA) | 25 DR | Untypeable prophage; intact | Five DNA methyltransferases (3 Dam, 1 Dcm, 1 unknown), DinI |
| CRPr33 | 1,304,879 | 1,315,433 | 3,912 | ROD_31701-ROD_31781 | ND | Putative lambdoid prophage; remnant | Three T3SS effectors (NleG2c, NleGc, EspM3) |
| CRP38 | 3,838,308 | 3,875,066 | 36,759 | ROD_36451 | ND | P2-like prophage; intact | HokA |
| CRP49 | 4,945,776 | 4,986,231 | 40,460 | gatD | 28 IR (8 MM) 4 DR | Mu-like prophage; intact | tRNAArg |
| Genomic islands | |||||||
| GI1 | 363,963 | 378,377 | 14,415 | ROD_t04(tRNAThr)-ROD_03491 | 40 IR (2 MM) | Genomic island; no integrase; previously IS flanked | Two T3SS effectors (EspS, NleA/EspI), Aap-like protein, tRNAThr |
| GI2 | 981,787 | 1,036,635 | 17,664 | ROD_t17(tRNASer) | ND | Putative genomic island; no integrase | T1SS and large repetitive proteinc |
| GI3 | 1,151,112 | 1,154,180 | 3,069 | ROD_t18(tRNASer) | 70 DR (1 D, 3 MM) | Genomic island; integrasec | |
| GI4 | 1,185,005 | 1,194,883 | 9,879 | ROD_t19(tRNASer) | 18 DR (1 MM) | Genomic island; integrasec | Three T3SS effectors (EspL2, NleB1 and NleE) |
| GI5 | 2,147,048 | 2,164,244 | 17,197 | ROD_t28(tRNASer) | 19 DR (1 MM) | Genomic island; integrase | Two H-NS |
| GI6 | 2,712,024 | 2,730,042 | 18,019 | smpB_ssrA | ND | Putative genomic island; no integrase | T1SS and large repetitive protein |
| GI7 | 2,775,891 | 2,794,475 | 18,585 | ROD_26351-ROD_26491 | ND | Genomic island; no integrase; IS flanked | LifA |
| GI8 | 3,124,334 | 3,142,596 | 18,263 | ROD_29461-ROD_29651 | ND | Genomic island; no integrase; IS flanked | CroR/I, T1SS, and large repetitive protein |
| LEE | 3,142,637 | 3,183,242 | 40,606 | ROD_29641-ROD_30191 | ND | Genomic island; no integrase; IS flanked | T3SS, intimin, seven T3SS effectors (EspG, EspF, EspB, Tir, Map, EspH, EspZ) |
| GI9 | 3,340,622 | 3,370,076 | 29,455 | ROD_t53(tRNAPhe) | ND | Genomic island; integrasec | LifA-like protein, one T3SS effector (EspN2-1c) |
| GI10 | 3,517,032 | 3,557,418 | 40,387 | ROD_t56(tRNALeu) | 51 DR (1MM) | Degenerate ICE element (ICECr1); integrasec | T1 RM system (M+,S+,R−) |
| GI11 | 4,316,630 | 4,333,192 | 16,563 | ROD_40761-ROD_41011 | ND | Genomic island; no integrase; IS flanked | Intimin-like proteinc, five T3SS effectors (EspM1c, EspO, EspT, NleG8. and EspM2) |
| GI12 | 5,017,131 | 5,037,202 | 20,072 | ROD_47721-ROD_47851 | ND | Genomic island; no integrase; IS flanked | LifA-like protein, one T3SS effector (EspN2-2) |
| GI13 | 5,064,524 | 5,087,409 | 22,886 | ROD_48131-ROD_48201 | ND | Putative genomic island, no integrase | T1SS and large repetitive proteinc |
| GI14 | 5,146,469 | 5,158,968 | 12,500 | yjjG-prfC | 22 DR (4 MM) | Genomic island; integrasec | Four T3SS effectors (NleH, NleF, NleG7, and EspJ) |
| GI15 | 5,207,322 | 5,212,668 | 5,347 | ROD_t85(tRNA-gly) | 21 DR | Genomic island; integrase | |
| GI16 | 5,218,554 | 5,271,734 | 53,181 | ROD_t86(tRNAPhe) | 17 DR | Genomic islandb; integrase | AIDA-like protein, hemolysin, PagC-like protein |
ND, none detected; DR, direct repeat; IR, inverted repeat; MM, mismatch; D, deletion.
Also known as GIpheV-CRICC168 (32).
Pseudogene.
A total of 113 IS elements comprising 29 different types were found within the genome of C. rodentium ICC168, six of which (ISCro1 to -6) were newly identified in this study. The most abundant element, ISCro1, which had 22 intact copies in the genome sequence, was previously referred to as IS679 (19) but is actually a novel IS element. ISCro1 was found to be very similar to the single IS679 element in C. rodentium (59%, 97%, and 98% amino acid identity to transposases A, B, and C, respectively), and it is possible, therefore, that one is a derivative of the other. It has been suggested that IS679-like elements may be involved in the horizontal transfer of virulence determinants (32), and it is notable that both GI7, which encodes the virulence factor LifA (47), and the LEE are flanked by intact copies of ISEc14 at one end and ISCro1 at the other end.
Relationship between C. rodentium and other Enterobacteriaceae.
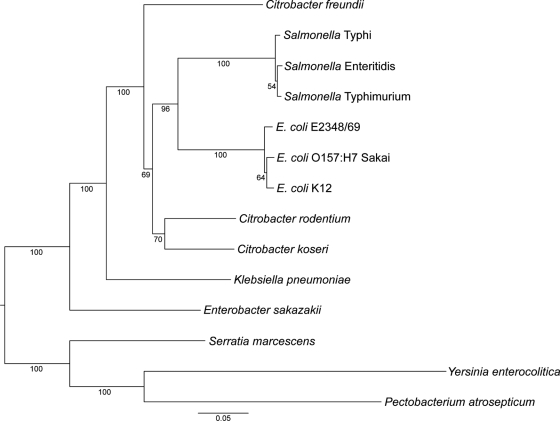
To determine the relationship between C. rodentium, E. coli, and other enteric bacteria, a phylogenetic tree was constructed, based on the DNA sequences of seven conserved backbone genes located in core regions of the genome (Fig. 1). Figure 1 shows that C. rodentium clusters within the tree between other members of the Enterobacteriaceae. However, even though C. rodentium and C. koseri appear to cluster, the branch lengths separating them are almost as long as those separating E. coli and Salmonella, and C. freundii is more distantly related. These data indicate that Citrobacter as a genus is polyphyletic. This is consistent with the whole-genome ANI shared between C. rodentium and C. koseri (92.2%), E. coli K-12 (90.7%), Salmonella Typhi CT18 (90.7%), and C. freundii (88.9%).
FIG. 1.
Phylogeny of C. rodentium showing the phylogenetic relationship of C. rodentium to various enteric bacteria based on the nucleotide sequences of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA). The tree shows bootstrap values (percentages of 1,000 replicates) below the branches and was rooted using an outgroup comprising Yersinia, Serratia, and Pectobacterium. The posterior probability for each node was 1 in every case and thus is not shown on the tree. The scale bar represents the number of substitutions per site.
Genome comparison with EPEC E2348/69 and EHEC Sakai.
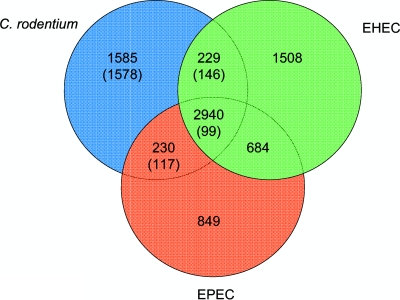
Reciprocal FASTA comparisons showed that the number of genes shared between C. rodentium, EPEC O127:H6 E2348/69 (referred to here as EPEC) (42), and EHEC O157:H7 Sakai (referred to here as EHEC) (37) is 2,940 (Fig. 2). This compares to 3,305 shared between E. coli strains K-12 MG1655 (referred to here as K-12) (6), EPEC, and EHEC.
FIG. 2.
Orthologous CDSs in C. rodentium ICC168, E. coli E2348/69 (EPEC), and E. coli O157:H7 Sakai (EHEC). The Venn diagram shows the number of genes that are unique to one strain, or shared between two or three of the strains, based on the results of reciprocal FASTA analysis with a minimum similarity of 40% identity over 80% of the CDSs. The numbers in parentheses indicate the numbers of C. rodentium genes in that category that have no orthologue in E. coli K-12 strain MG1655.
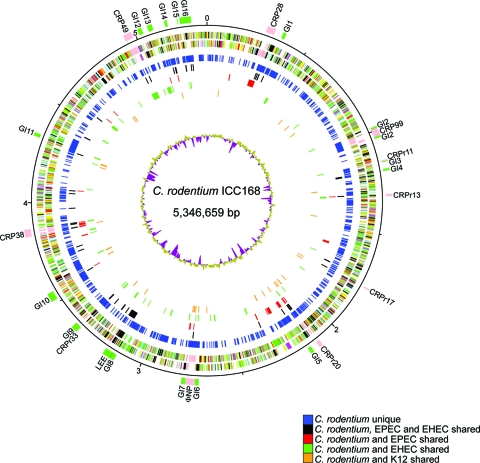
The numbers of CDSs unique to C. rodentium and those shared exclusively between C. rodentium and either EPEC or EHEC were also high: 1,585, 230, and 229 CDSs, respectively (Fig. 2). We analyzed these three CDS groups in more detail. We subtracted from the groups those genes that were also shared with K-12 (Fig. 2). This comparison has been used previously to differentiate core E. coli genes from pathotype-specific functions (42). Almost half of the CDSs C. rodentium shares with either EPEC (75/117 CDSs) or EHEC (67/146 CDSs), but without an orthologue in K-12, were found to be clustered and located on GIs or prophages (Fig. 3). Of the genes shared with either EPEC or EHEC that were in the “backbone” of C. rodentium, many had putative functions associated with metabolism and sugar transport and utilization. Of particular note, C. rodentium carries all of the genes in the propanediol utilization operon, which are also found in EPEC but are absent in many other E. coli strains, including EHEC and K-12 (42). This supports the suggestion that the cob-pdu locus originated in a common ancestor of the Enterobacteriaceae and has been lost repeatedly by different lineages (42, 79). Also, among the genes shared only with EHEC was a urease gene cluster. The lack of urease production in EHEC has been linked to a premature stop codon in ureD (38, 65); however, C. rodentium encodes the full-length UreD and can produce urease (84). Interestingly, also among the EHEC and C. rodentium shared genes were those encoding the clustered regularly interspaced short palindromic repeats (CRISPR)-associated proteins, which were absent from EPEC and divergent from those found in K-12. The C. rodentium genome sequence also contains two CRISPRs, located in genomic locations equivalent to those in EHEC but which contain different numbers of repeats and spacers. No CRISPRs were apparent within the genome of EPEC. Figure 3 shows that C. rodentium-specific genes were located throughout the chromosome, although 414 of the 1,578 unique genes (genes not shared with EPEC, EHEC, or K-12) were located on genomic islands or prophages and are likely to have been acquired by recent horizontal gene transfer.
FIG. 3.
Circular map of the C. rodentium ICC168 chromosome. From the outside in, the first circle shows the positions of genomic islands and prophages (detailed in Table 3). The second circle shows the genomic positions in Mbp. The third and fourth circles show the CDSs transcribed clockwise and counterclockwise, respectively (color coded according to the predicted functions of their gene products: black, pathogenicity or adaptation; gray, energy metabolism; red, information transfer; green, membrane or surface structure; yellow, central or intermediary metabolism; cyan, degradation of macromolecules; cerise, degradation of small molecules; pale blue, regulator; pink, prophage or IS element; orange, conserved hypothetical; pale green, unknown; and brown, pseudogene). The fifth circle shows C. rodentium CDSs (dark blue) that lack orthologues (by reciprocal FASTA analysis) in EPEC E2348/69, EHEC Sakai, or K-12 MG1655. The sixth circle shows C. rodentium CDSs (black) that have orthologues (by reciprocal FASTA analysis) in both EPEC and EHEC but not K-12. Circles 7, 8, and 9 show the positions of C. rodentium CDSs that have orthologues (by reciprocal FASTA analysis) in EPEC (red), EHEC (green), or K-12 (orange) (excluding those CDSs that also had orthologues in one or both of the other E. coli strains). The innermost circle shows a plot of G+C content.
Of the 2,940 core genes present in C. rodentium, EPEC, and EHEC, only 99 had no orthologues in K-12 (see Table S2 in the supplemental material), and of these 76 were located on GIs and prophages. Over half (49) of these 76 genes were associated with virulence and included those found on the LEE, as well as other non-LEE-encoded T3SS effectors. Of note, among the remaining shared genes was a cluster with a putative role in the production of 4-hydroxybenzoate decarboxylase, which is found sporadically in other members of the Enterobacteriaceae, as well as elsewhere, and may be involved in metabolism of aromatic compounds under anaerobic conditions (20, 53, 56).
Patterns of functional gene loss.
The genome of C. rodentium ICC168 has 188 pseudogenes, 43% of which are found in prophages, GIs, or plasmids (summarized in Table S3 in the supplemental material). Almost a quarter of the pseudogenes (42/188 CDSs) have been caused by prophage- or IS-mediated insertional inactivation, including disruption of flagellum biogenesis genes and other core and accessory functions (see Table S3 in the supplemental material). The remainder of the pseudogenes are truncated by frameshift mutations or premature stop codons or are gene remnants identified by similarity to full-length genes in other bacteria. Fifty-six of 188 pseudogenes had only a single frameshift mutation or premature stop codon, indicating that these inactivating mutations were relatively recent events. There are a number of sugar uptake and energy generation pseudogenes (see Table S3 in the supplemental material), which may be indicative of metabolic streamlining. Also, a number of pseudogenes are associated with virulence factors, including several T3SS effector proteins and essential functions within three distinct fimbrial operons (Table 4; see Tables S3 and S4 in the supplemental material).
TABLE 4.
C. rodentium T3SS effectors and their homologues in EPEC and EHEC
| Effector | No. ina: |
|||
|---|---|---|---|---|
| C. rodentium ICC168 | O127:H6 E2348/69 | O111:NM B171 | O157:H7 Sakai | |
| Cif | 0 | 1 (1) | 1 | 0 |
| EspB | 1 | 1 | 1 | 1 |
| EspF | 1 | 1 | 1 | 1 |
| EspG | 1 | 2 | 1 | 1 |
| EspH | 1 | 1 | 1 | 1 |
| EspJ | 1 | 1 | 0 | 1 |
| EspK | 1 | 0 | 0 | 1 |
| EspL | 1 | 3 (2) | 3 (2) | 4 (1) |
| EspM | 3 (1) | 0 | 3 (1) | 2 |
| EspN | 3 (2) | 0 | 1 | 1 |
| EspO | 1 | 1 (1) | 0 | 2 |
| EspR | 0 | 0 | 0 | 4 (1) |
| EspS | 1 | 0 | 0 | 0 |
| EspT | 1 | 0 | 0 | 0 |
| EspV | 1 | 0 | 1 (1) | 1 (1) |
| EspW | 0 | 0 | 1 | 1 |
| EspX | 1 | 0 | 0 | 6 (1) |
| EspY | 0 | 0 | 0 | 5 (1) |
| EspZ | 1 | 1 | 1 | 1 |
| Map | 1 | 1 | 1 | 1 |
| NleA | 1 | 1 | 1 | 1 |
| NleB | 2 (1) | 3 (1) | 3 (2) | 3 (1) |
| NleC | 1 | 1 | 2 (2) | 1 |
| NleD | 2 | 1 | 0 | 1 |
| NleE | 1 | 2 | 1 | 1 |
| NleF | 1 | 1 | 1 | 1 |
| NleG | 5 (2) | 1 | 6 | 14 (5) |
| NleH | 1 | 3 (1) | 2 (1) | 2 |
| Ibe | 0 | 0 | 2 (1) | 0 |
| TccP | 0 | 0 | 1 | 2 (1) |
| Tir | 1 | 1 | 1 | 1 |
| Total | 35 (6) | 27 (6) | 40 (12) | 62 (12) |
The numbers in parentheses indicate pseudogenes.
Virulence determinants.
Through targeted investigations and comparison with EPEC and EHEC, a number of factors have previously been shown to be important for C. rodentium virulence. Here, we describe the full repertoire of the known and potential virulence-associated determinants that were identified in the genome sequence of ICC168 (summarized in Table S4 in the supplemental material).
T2SS.
C. rodentium has a type II secretion system (T2SS) encoded in a single locus (ROD_44871 to ROD_45001). Nine putative substrates for this secretion system were also identified, including four likely polysaccharide-degrading enzymes with similarity to chitinases encoded adjacent to the T2SS itself and four additional similar enzymes (including one encoded by a pseudogene) encoded elsewhere in the C. rodentium genome. C. rodentium also carries an orthologue of yodA, the product of which has been shown to be secreted by the pO157-encoded T2SS and to promote virulence in EHEC (40).
LEE-encoded T3SS.
The LEE pathogenicity island, along with its associated T3SS, is conserved among the genomes of EPEC, EHEC, and C. rodentium, but with some notable differences. While the LEEs of EPEC and EHEC are inserted into the SelC tRNA locus, the C. rodentium LEE is in a nonsynonymous location flanked by IS elements, adjacent to another genomic island (GI8). Moreover, in relation to EPEC and EHEC, the rorf1 and espG genes of C. rodentium are located at the opposite end of the LEE (19).
The C. rodentium T3SS translocated-protein repertoire.
C. rodentium possesses a repertoire of 35 genes (including six pseudogenes) (Table 4) encoding proteins that show significant sequence homology to known T3SS effector proteins in other systems or that we have shown to be translocated by the Citrobacter T3SS. In comparison, EPEC E2348/69 encodes 27 T3SS effectors (42), EPEC B171 has 40 (42), and EHEC Sakai has 62 effector proteins (88) (Table 4). All of the C. rodentium effectors are encoded in regions thought to have been laterally acquired. Of the 29 intact C. rodentium effectors, seven are encoded on the LEE; five are present on the prophage remnants denoted CPRr13, CPRr17, and CPRr33; and 14 effector proteins are encoded on five GIs (GI1, GI2, GI4, GI11, and GI14) (Table 3). The three remaining effectors are located within regions with low GC content. The three prophage-like elements, CRPr13, CRPr17, and CRPr33, each consist of two genes encoding putative phage-related functions, along with three different T3SS effectors each. It is notable that these regions share similarity with and are integrated at the same sites as lambdoid prophages identified within the genomes of EPEC and EHEC (Table 3) (37, 42). CRPr13 encodes the effectors EspX7, EspN1, and EspK, which are homologues of effectors encoded on EHEC prophage Sp6. CRPr17 encodes an Sp10-like integrase and homologues of the effectors NleC, NleB, and NleG, which are found encoded on different lambdoid prophages in EPEC and EHEC, and CRPr33 shares similarities with prophage Sp17 and encodes homologues of three of the four effectors found on this prophage. It therefore seems likely that these three regions are remnants of lambdoid prophages and that the nine associated effector proteins have been carried into the C. rodentium genome on these phages by specialized transduction.
T3SS effector families.
C. rodentium genes encode at least one intact member of each of the core set of effector families, consisting of the seven LEE-encoded effectors plus the non-LEE-encoded NleA/EspI, NleB, NleE, NleF, NleG, NleH, and EspL, previously suggested to represent the minimum set required to facilitate infection (42), as they are common to all LEE-encoding species sequenced to date. All the effector families that are not represented in the C. rodentium genome are also absent from at least one of the other sequenced A/E pathogens, E2348/69, B171, and Sakai (Table 4).
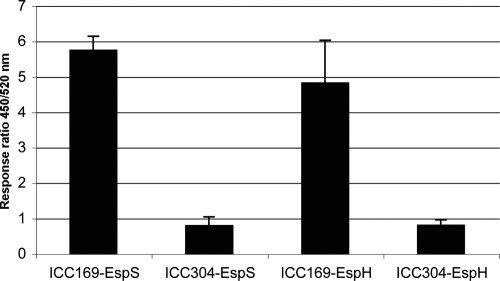
In addition to homologues of EPEC and EHEC effectors, we identified a new T3SS translocated protein in C. rodentium, named EspS, which has some similarity to OspB of Shigella. Using a β-lactamase fusion, we showed that EspS is translocated in an EspA-dependent manner (Fig. 4). Mouse inoculation showed that a ΔespA mutant was severely attenuated in virulence (Fig. 5). C. rodentium also encodes the effector EspT, found only rarely in EPEC (1), which subverts the eukaryotic cytoskeleton by activating the small Rho GTPases Rac1 and Cdc42 (10), and an intact copy of EspV, which is considered to be a pseudogene in EHEC Sakai due to an N-terminal truncation (88). Interestingly, two identical copies of NleD are encoded within the genome of C. rodentium adjacent to identical transposases, and three EspN family members are present (though two of these are encoded by pseudogenes), whereas E2348/69, B171, and Sakai have only one or no representatives from these four families (Table 4). This indicates that there has been an expansion of the NleD-like gene repertoire in C. rodentium.
FIG. 4.
EspS is a T3SS translocated protein. Translocation of EspS-TEM-1 and the control EspH-TEM-1 from wild-type (ICC169) and ΔespA (ICC304) C. rodentium was quantified using a Fluostar Optima reader with excitation at 410 nm (10-nm band-pass). Emission was detected via 450-nm (blue fluorescence) and 520-nm (green fluorescence) filters. The translocation rate was expressed as the 450/520-nm emission ratio. EspS and EspH were translocated from wild-type C. rodentium, but not from the ΔespA mutant. The error bars represent mean standard deviations (SD).
FIG. 5.
Colonization dynamic of wild-type, ΔespA (ICC304), and ΔespA(pACYC-espA) C. rodentium strains after oral inoculation of C3H/Hej mice. Mice inoculated with wild-type (WT) and ΔespA(pACYC-espA) C. rodentium exhibited similar colonization dynamics, whereas ΔespA C. rodentium was unable to colonize. The inoculum count is the number of viable bacteria in 200 ml used to inoculate mice by oral gavage. The data are represented as means ± SD.
T6SS.
A new class of secretion system, the type VI secretion system (T6SS), has recently been described in a variety of bacterial species. T6SS gene clusters commonly consist of between 15 and 25 genes and were initially identified based on the presence of a gene encoding a homologue of the Legionella pneumophila type IV secretion system (T4SS) protein IcmF (17). A contribution to virulence was shown for T6SSs of several pathogenic bacteria (28). The T6SS apparatus is not well characterized yet, but recent studies indicate that several core components resemble bacteriophage tail proteins (49). The Hcp and VgrG proteins have been demonstrated to be secreted in several organisms; however, a possible dual role of these proteins as a structural part of the T6SS and secreted effector molecules is becoming evident (78).
There are two distinct T6SS gene clusters in the genome of C. rodentium, which we have designated Citrobacter rodentiumtype six secretion system cluster 1 (CTS1) and 2 (CTS2). CTS1 is composed of 39 genes, 18 of which are conserved components of the T6SS (Fig. 6). CTS1 is almost identical to a T6SS gene cluster carried by Enterobacter cancerogenus and also shows limited similarity to components of SPI-6 (also known as Salmonella enterica centisome 7 genomic island [sci]) (29). Like the T6SS cluster encoded by E. cancerogenus and the sci locus, CTS1 also carries a chaperone-usher fimbrial biogenesis operon (29). CTS1 contains all of the components that have been determined to be necessary for a functional T6SS in other bacteria (95). Interestingly, the icmF homologue in CTS1 has a frameshift at a region containing a polyadenosine tract, which may allow regulation of the gene via transcriptional slippage in an manner analogous to that of mixE of Shigella or the pyrBI locus in E. coli (52, 74).
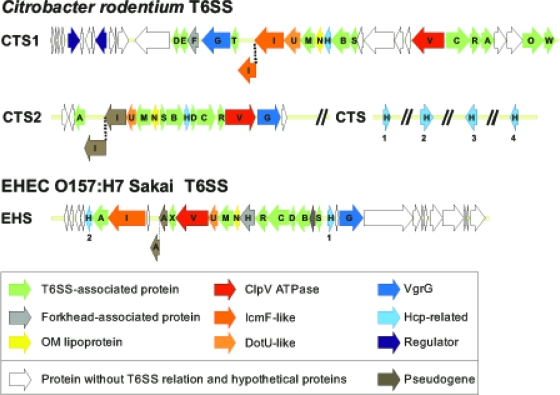
FIG. 6.
T6SSs in C. rodentium and EHEC. Shown is a schematic representation of the C. rodentium ICC168 T6SS gene clusters CTS1 and CTS2 and the EHEC O157:H7 Sakai T6SS cluster EHS. Conserved T6SS components are highlighted. The differing architectures and lack of homology between CTS1, CTS2, and EHS suggest that these T6SS clusters have distinct evolutionary histories.
CTS2 consists of 16 genes, 13 of which are conserved within other T6SS clusters (Fig. 6). Although CTS2 encodes all the components required for assembly of a functional T6SS, the icmF homologue in this cluster has a bona fide frameshift, which makes it unlikely that CTS2 is sufficient to produce a functional T6SS. CTS2 is highly similar to a putative T6SS in C. freundii, which has conserved gene order and an intact icmF homologue in a single frame (data not shown). CTS2 also shares limited similarity with components of the T6SS clusters in Pseudomonas species (59, 85).
The differing architectures and lack of homology between CTS1 and CTS2 suggest that these two T6SS clusters have distinct evolutionary histories. In addition to CTS1 and CTS2, which each encode one Hcp family protein, there are four other Hcp homologues encoded in the C. rodentium chromosome, which share little homology to each other and thus may also have been acquired independently.
EHEC also encodes a T6SS, which we have designated the enterohemorrhagic E. coli type six secretion system cluster (EHS), which is encoded in O island 7 and has high homology to the Shigella sonnei T6SS (85). EHS is distinct from both C. rodentium T6SSs and consists of 33 genes, 18 of which are conserved components of the T6SS (Fig. 6). Unlike in CTS1 and CTS2, the icmF gene in the EHS operon is not interrupted by a frameshift. Additionally, downstream of ehsG there is a region including genes that are associated with Rhs proteins (51, 90). EHEC encodes only two Hcp homologues, EhsH1 and EhsH2, which are both encoded in the EHS cluster and have very limited similarity to each other. In contrast to the T3SS, which is conserved in all A/E pathogens and is essential for virulence, the overall lack of homology between EHS, CTS1, and CTS2 suggests that the T6SS may facilitate more subtle strain-specific adaptations to divergent host or environmental niches.
Other secretion systems.
Type I secretion systems (T1SS) are involved in the transport of a wide range of substrates, and in common with many other enteric bacteria, C. rodentium encodes multiple T1SSs. Of particular note are four genomic islands (GI2, GI6, GI8, and GI13) (Table 3) that are predicted to each encode T1SS apparatus, as well as a presumptive secreted substrate, a large (1,637- to 5,979-amino-acid [aa]) repetitive protein (Table 3). The gene products forming the T1SSs share ∼50% amino acid identity with each other. The associated repetitive proteins show higher variation in protein length and lower overall sequence conservation, with ∼25% amino acid identity between ROD_08971, ROD_25701, ROD_29581, and ROD_48171.
The arrangement, nature, and proposed functions of CDSs seen in the four C. rodentium T1SS genomic islands appear to suggest that they are functional modules that have been laterally acquired. Wider comparisons also showed that these GIs are similar in gene content, arrangement, and, in some instances, sequence to GIs previously seen in other enteric bacteria. Comparisons with these other loci showed that the large repetitive protein encoded by GI6 shares 47% identity over 3,790 aa with the large repetitive protein of S. Typhi encoded on SPI-9. Both GIs are inserted in the same intergenic region between ssrA and smpB.
GI2, GI8, and GI13 also have some similarity to SPI-9 and the related SPI-4; however, GI13 has a high degree of similarity to O island 28 in EHEC EDL933 (51% identity over 5,392 aa to the large repetitive protein), and the large repetitive protein of GI2 is similar to a putative hemagglutinin/hemolysin-related protein in Ralstonia solanacearum. SPI-9 has been shown to be required for intestinal colonization in mice and in vitro biofilm formation in S. Enteritidis (48); therefore, GI2, GI6, and GI13 may encode factors that play roles in the virulence of C. rodentium. However, transcription of the large repetitive protein of GI2 is unlikely to be due to the insertion of prophage CRP99 in the middle of the CDS. In addition, the CDS for the large repetitive protein on GI13 has been truncated by a premature stop codon; however, as it is located close to the C terminus, it is possible that a 5,884-aa protein may be expressed as opposed to the 5,979-aa full-length protein.
There are 20 putative type V secretion systems, or autotransporters, encoded in the C. rodentium genome. Fourteen of these, including one encoded by a pseudogene, were identified as putative autotransporter adhesins and were related to AIDA-I, TibA, or pertactin, which are involved in virulence in EPEC, enterotoxigenic E. coli (ETEC), and Bordetella pertussis, respectively (5, 8, 27). Two other autotransporters were predicted to belong to the serine protease autotransporter of the Enterobacteriaceae (SPATE) family of proteins, members of which have been shown to contribute to virulence in other pathogens (7, 23, 70). The majority of the autotransporters are located in the chromosomal backbone; however, one, AdcA, which was shown to function as an adhesin but had no obvious role in colonization of mice (36), is encoded on a genomic island. In addition, the two SPATE autotransporters are encoded by genes on the large plasmid pCROD1, along with a TibA-like autotransporter adhesin and its associated glycosyltransferase.
Adhesins.
C. rodentium carries a total of 19 fimbrial biogenesis operons, including four incomplete operons (Table 5). Of the intact operons, the product of the Kfc chaperone-usher fimbrial operon and CFC type IV fimbriae were previously determined to be involved in gastrointestinal colonization (36, 63), whereas long polar fimbriae had no discernible role in the virulence of C. rodentium (87).
TABLE 5.
Fimbrial biosythesis operons identified in C. rodentium
| Operon | CDS ID | Genes | Descriptionc | EPEC orthologue | Sakai orthologue |
|---|---|---|---|---|---|
| 1 | ROD_01101-01121 | hofCB-ppdD | Type IV fimbriae | E2348_C_0109-0111 | ECs0110-0112 |
| 2 | ROD_03641-03671 | Chaperone-usher γ4 fimbriae | |||
| 3 | ROD_03351, 10951-11021 | crlcsgGFEDBAC | Curli fimbriae | E2348_C_0233, 1129-1136 | ECs0267, 1414-1412 |
| 4 | ROD_11771-11781 | Chaperone-usher β fimbriaee | |||
| 5 | ROD_18141-18181 | lpfEDCBA | Long polar fimbriae (chaperone-usher γ1 fimbriae) | ||
| 6 | ROD_19341-19381 | Chaperone-usher σ fimbriae | |||
| 7 | ROD_22311-22341 | Chaperone-usher γ4 fimbriae | |||
| 8 | ROD_27771-27801 | Chaperone-usher γ4 fimbriae | |||
| 9 | ROD_29101-29191 | fimBEAICDFGHK | Chaperone-usher γ1 fimbriae | E2348_C_4619-4627a | ECs5271-5279a |
| 10 | ROD_29201-29241 | Alternate chaperone-usher (α) fimbriae | E2348_C_0247-0249b | ECs0321-0323b | |
| 11 | ROD_29351-29391 | Chaperone-usher γ4 fimbriaed | |||
| 12 | ROD_34961-35021 | Chaperone-usher π fimbriae | |||
| 13 | ROD_41241-41291 | kfcHGFEDC | Kfc (K99-like factor involved in Citrobacter colonization) chaperone-usher κ fimbriae | ||
| 14 | ROD_41381-41551 | flp1 rcpCA tadZABCD tadVEFG | Tight adherance (tad) locus (type IV fimbriae) | ||
| 15 | ROD_44281-44321 | hofMNOPQ | Type IV fimbriae | E2348_C_3635-3639 | ECs4233-4237 |
| 16 | ROD_46461-46571 | cfcABCDEFGHIJPV | CFC (colonization factor Citrobacter) type IV fimbriae | ||
| 17 | ROD_50611-50651 | Chaperone-usher γ4 fimbriaed | |||
| 18 | ROD_p1161-p1201 | Chaperone-usher κ fimbriae | |||
| 19 | ROD_p1291-p1301 | Chaperone-usher γ1 fimbriaed,e |
fimK is missing in EPEC and EHEC.
No orthologues of ROD_29231 or ROD_29241.
Chaperone-usher fimbrial operons have been grouped into clades and subclades according to the fimbrial usher protein classification scheme (68).
Contains pseudogenes.
Part of operon is missing.
Of the nonfimbrial adhesins encoded by C. rodentium genes, LifA (47) and intimin (83) are essential for colonization. We also found two LifA homologues that are divergent from LifA at the N terminus but almost identical at the C terminus and that had 97% amino acid identity to each other. In addition, two intact genes and one pseudogene encoding intimin-like proteins were identified outside of the LEE (see Table S4 in the supplemental material).
Other potential virulence factors.
Other predicted proteins encoded by C. rodentium genes that are potentially involved in virulence include homologues of the dispersin Aap of enteroaggrative E. coli (EAEC) (58) and virulence factor CexE of ETEC (76), the virulence-related outer membrane protein PagC (66), and the Yersinia enterocolitica porin required for colonization, SpfA (36).
Other factors previously reported to be important for the virulence of C. rodentium include the transcription factors Ler, GrlA, GrlR, RegA, and RpoS (2, 19, 21, 36); the quorum-sensing locus (CroIR); and the Pho regulon (15, 16). In addition, C. rodentium genes encode three putative hemolysin expression regulatory proteins, two on the chromosome and the third on pCROD2, and the putative hemolysin activator HlyBC, which is presumably involved in the regulation of the putative hemolysin/hemagglutinin encoded by the downstream ROD_49941.
Plasmids.
C. rodentium carries four plasmids (Table 2), the largest of which, pCROD1, is nonconjugative; encodes two toxin/antitoxin (TA) addiction systems, PemIK and CcdAB; and has a replication locus similar to those of plasmids belonging to the incompatibility group IncFII, such as the E. coli plasmid R100 (NR1) (94). In addition to functions associated with plasmid replication and maintenance, pCROD1 genes, including a colicin S4-like biosynthesis operon and one intact and one degenerate chaperone-usher fimbrial operon, encode proteins associated with virulence, including three putative autotransporters (Table 5).
The backbone of the 39-kb plasmid, pCROD2, is syntenic with those of plasmids R6K (IncX1) and pOLA52 (IncX2) (67), sharing 22 CDSs (average 47% amino acid identity) with the former and 31 CDSs (average 59% amino acid identity) with the latter out of the 51 pCROD2 CDSs. Included in the shared regions is the plasmid replication locus, suggesting that pCROD2 also belongs to incompatibility group IncX. pCROD2 carries a 16-kb conjugative plasmid transfer locus (ROD_p2371-ROD_p2531); however, transfer of this plasmid might be impaired due to the disruption of triD (ROD_p2461) by IS102. Two addiction systems are likely to be involved in the stability of this plasmid, the TA system stbDE and the putative TA locus hicAB. pCROD2 also encodes a putative H-NS DNA-binding protein (ROD_p2271) that has been linked to mitigating the impact of low-G+C DNA acquired by lateral gene transfer in other systems (22).
Plasmids pCROD3 and pCROD4 are small (3.9 and 3.2 kb, respectively) and nonconjugative. The sequence of the smallest plasmid is identical to that of the previously sequenced 3,172-bp pCRP3 from C. rodentium strain DBS100 (19).
DISCUSSION
The data that we have described here highlight several clear messages. The phylogenetic analysis of C. rodentium showed that although it clusters with members of the Enterobacteriaceae, C. rodentium is more distantly related to E. coli than E. coli is to Salmonella. Moreover, the genomic evidence we have presented also shows that the sequenced species currently within the genus Citrobacter do not cluster, and our analysis indicates that the genus is in fact polyphyletic.
It is apparent from the comparisons of the gene sets of C. rodentium, EPEC, and EHEC that a large percentage (∼32%) of the C. rodentium genome is unique. These distinguishing functions include those known to be important for virulence in other systems, including T6SSs and associated effectors and multiple fimbrial operons, as well as other exported adhesins. Also included in this group are some novel T3SS effector proteins.
The majority of functions shared between C. rodentium, EPEC, and EHEC are also found in the nonpathogenic K-12, which suggests that these genes encode core enterobacterial functions. However, there are a significant number of genes found in C. rodentium, EPEC, and EHEC that are absent from K-12. The majority of these are located on mobile genetic elements and are also recognizable as key virulence determinants, including lifA, LEE, and many of the C. rodentium T3SS effector proteins. It is the acquisition of these factors that is likely to have been responsible for their convergently evolved common virulence strategy.
Specialized transduction by lambdoid phages has been shown to be important for the acquisition of virulence determinants by both EPEC and EHEC (88). It is clear from this analysis that this is also the likely mechanism by which many of the C. rodentium effectors have been acquired. It is important to note that although the cargo genes are found at analogous sites in the genome, comparisons of the phage genes thought to have brought these shared effectors into the genome show that they probably came from distinct phages and so are likely to be part of a large pool of temperate phages disseminating a related set of effector genes to a diverse set of hosts, underscoring their importance in pathogen evolution. It is access to this gene pool that has allowed C. rodentium, at least in part, to convergently evolve with the pathogenic E. coli strains.
It is likely that the acquisition of the LEE and its associated effector proteins had a dramatic effect on the pathogenic potential of C. rodentium in the mouse and perhaps even the niche it occupies within this host. Certainly the presence of such a large number of pseudogenes and IS elements in other bacteria, such as S. Typhi and Yersinia pestis, has been tightly linked to a change in lifestyle associated with the occupation of a new niche (71, 73). Although the numbers of pseudogenes in C. rodentium and EPEC are comparable, 60% of EPEC pseudogenes are in regions that are thought to have been laterally acquired. The inverse is true of C. rodentium, where 57% of pseudogenes fall in the core regions.
It is not just the relative number of pseudogenes lost by C. rodentium that draws parallels with S. Typhi and Y. pestis; C. rodentium has also lost metabolic, colonization, and virulence-associated functions. If there had been a change of niche, it would be expected that genes important for the previous lifestyle would be lost. Certainly the observation that the majority of pseudogenes lie in the C. rodentium backbone is consistent with this. A specific example that illustrates this point is the loss of function in the fimbrial genes that has occurred exclusively in operons unique to C. rodentium compared to EPEC and EHEC.
C. rodentium is the etiological agent of transmissible colonic hyperplasia in mice. It colonizes the gut mucosa via A/E lesions and may also reach deeper tissues (e.g., the liver and spleen). We have recently shown that espT can mediate invasion of C. rodentium into mammalian cells (10a); it is not yet known if EspT plays a role in in vivo dissemination or in the hyperplastic or immunological responses. C. rodentium is nontoxigenic, using an infection strategy that appear to rely mainly on its T3SS effector repertoire. Although we have shown that C. rodentium is genetically distinct from EPEC and EHEC and does not share the same host, the three bacteria have similar infection strategies and share virulence genes that are found on mobile genetic elements. Accordingly, it is unlikely that C. rodentium acquired the virulence loci directly from EPEC or EHEC, and the data point to the existence of a common ancestral source. Nonetheless, the availability of the complete C. rodentium genome annotation will enhance future studies of virulence mechanisms in A/E pathogens using a natural and unique host-pathogen interaction model.
Supplementary Material
Acknowledgments
We thank Simon Harris for assistance with the phylogenetic analyses. We thank the core sequencing and informatics teams at the Sanger Institute for their assistance and the Wellcome Trust for its support of the Sanger Institute Pathogen Genomics group.
Footnotes
Published ahead of print on 6 November 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Arbeloa, A., M. Blanco, F. Moreira, R. Bulgin, C. Lapez, G. Dahbi, J. Blanco, A. Mora, M. P. Alonso, R. C. Mamani, T. A. Gomes, J. Blanco, and G. Frankel. 2009. Distribution of espM and espT among enteropathogenic and enterohemorrhogic Escherichia coli. J. Med. Microbiol. 58:988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barba, J., V. H. Bustamante, M. A. Flores-Valdez, W. Deng, B. B. Finlay, and J. L. Puente. 2005. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J. Bacteriol. 187:7918-7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthold, S. W., G. L. Coleman, P. N. Bhatt, G. W. Osbaldiston, and A. M. Jonas. 1976. The etiology of transmissible murine colonic hyperplasia. Lab Anim. Sci. 26:889-894. [PubMed] [Google Scholar]
- 4.Barthold, S. W., G. W. Osbaldiston, and A. M. Jonas. 1977. Dietary, bacterial, and host genetic interactions in the pathogenesis of transmissible murine colonic hyperplasia. Lab Anim. Sci. 27:938-945. [PubMed] [Google Scholar]
- 5.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Boisen, N., F. Ruiz-Perez, F. Scheutz, K. A. Krogfelt, and J. P. Nataro. 2009. Short report: high prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am. J. Trop. Med. Hyg. 80:294-301. [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan, M. J., Z. M. Li, J. L. Cowell, M. E. Bisher, A. C. Steven, P. Novotny, and C. R. Manclark. 1988. Identification of a 69-kilodalton nonfimbrial protein as an agglutinogen of Bordetella pertussis. Infect. Immun. 56:3189-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan, P. C., T. E. Fritz, R. J. Flynn, and C. M. Poole. 1965. Citrobacter freundii associated with diarrhea in laboratory mice. Lab Anim. Care 15:266-275. [PubMed] [Google Scholar]
- 10.Bulgin, R. R., A. Arbeloa, J. C. Chung, and G. Frankel. 2009. EspT triggers formation of lamellipodia and membrane ruffles through activation of Rac-1 and Cdc42. Cell Microbiol. 11:217-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Bulgin, R., A. Arbeloa, D. Goulding, G. Dougan, V. Crepin, B. Raymond, and G. Frankel. The T3SS effector EspT defines a new category of invasive enteropathogenic E. coli (EPEC) which form intracellular actin pedestals. PLoS Pathogens, in press. [DOI] [PMC free article] [PubMed]
- 11.Carter, P. B. 1975. Animal model of human disease. Yersinia enteritis. Animal model: oral Yersinia enterocolitica infection of mice. Am. J. Pathol. 81:703-706. [PMC free article] [PubMed] [Google Scholar]
- 12.Carver, T., N. Thomson, A. Bleasby, M. Berriman, and J. Parkhill. 2009. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 14.Charpentier, X., and E. Oswald. 2004. Identification of the secretion and translocation domain of the enteropathogenic and enterohemorrhagic Escherichia coli effector Cif, using TEM-1 beta-lactamase as a new fluorescence-based reporter. J. Bacteriol. 186:5486-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng, C., S. M. Tennant, K. I. Azzopardi, V. Bennett-Wood, E. L. Hartland, R. M. Robins-Browne, and M. Tauschek. 2009. Contribution of the pst-phoU operon to cell adherence by atypical enteropathogenic Escherichia coli and virulence of Citrobacter rodentium. Infect. Immun. 77:1936-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coulthurst, S. J., S. Clare, T. J. Evans, I. J. Foulds, K. J. Roberts, M. Welch, G. Dougan, and G. P. Salmond. 2007. Quorum sensing has an unexpected role in virulence in the model pathogen Citrobacter rodentium. EMBO Rep. 8:698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das, S., and K. Chaudhuri. 2003. Identification of a unique IAHP (IcmF associated homologous proteins) cluster in Vibrio cholerae and other proteobacteria through in silico analysis. In Silico Biol. 3:287-300. [PubMed] [Google Scholar]
- 18.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dodd, C. E., and D. Jones. 1982. A numerical taxonomic study of the genus Shigella. J. Gen. Microbiol. 128:1933-1957. [DOI] [PubMed] [Google Scholar]
- 21.Dong, T., B. K. Coombes, and H. E. Schellhorn. 2009. Role of RpoS in the virulence of Citrobacter rodentium. Infect. Immun. 77:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyle, M., M. Fookes, A. Ivens, M. W. Mangan, J. Wain, and C. J. Dorman. 2007. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science 315:251-252. [DOI] [PubMed] [Google Scholar]
- 23.Dutta, P. R., R. Cappello, F. Navarro-Garcia, and J. P. Nataro. 2002. Functional comparison of serine protease autotransporters of enterobacteriaceae. Infect. Immun. 70:7105-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ediger, R. D., R. M. Kovatch, and M. M. Rabstein. 1974. Colitis in mice with a high incidence of rectal prolapse. Lab Anim. Sci. 24:488-494. [PubMed] [Google Scholar]
- 26.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 27.Elsinghorst, E. A., and J. A. Weitz. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filloux, A., A. Hachani, and S. Bleves. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570-1583. [DOI] [PubMed] [Google Scholar]
- 29.Folkesson, A., S. Lofdahl, and S. Normark. 2002. The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res. Microbiol. 153:537-545. [DOI] [PubMed] [Google Scholar]
- 30.Frankel, G., and A. D. Phillips. 2008. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cell Microbiol. 10:549-556. [DOI] [PubMed] [Google Scholar]
- 31.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73:2573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girardeau, J. P., Y. Bertin, and C. Martin. 2009. Genomic analysis of the PAI ICL3 locus in pathogenic LEE-negative Shiga toxin-producing Escherichia coli and Citrobacter rodentium. Microbiology 155:1016-1027. [DOI] [PubMed] [Google Scholar]
- 33.Gruenheid, S., I. Sekirov, N. A. Thomas, W. Deng, P. O'Donnell, D. Goode, Y. Li, E. A. Frey, N. F. Brown, P. Metalnikov, T. Pawson, K. Ashman, and B. B. Finlay. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 51:1233-1249. [DOI] [PubMed] [Google Scholar]
- 34.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison, J. A., B. Villarreal-Ramos, P. Mastroeni, R. Demarco de Hormaeche, and C. E. Hormaeche. 1997. Correlates of protection induced by live Aro− Salmonella typhimurium vaccines in the murine typhoid model. Immunology 90:618-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hart, E., J. Yang, M. Tauschek, M. Kelly, M. J. Wakefield, G. Frankel, E. L. Hartland, and R. M. Robins-Browne. 2008. RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect. Immun. 76:5247-5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 38.Heimer, S. R., R. A. Welch, N. T. Perna, G. Posfai, P. S. Evans, J. B. Kaper, F. R. Blattner, and H. L. Mobley. 2002. Urease of enterohemorrhagic Escherichia coli: evidence for regulation by fur and a trans-acting factor. Infect. Immun. 70:1027-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemrajani, C., O. Marches, S. Wiles, F. Girard, A. Dennis, F. Dziva, A. Best, A. D. Phillips, C. N. Berger, A. Mousnier, V. F. Crepin, L. Kruidenier, M. J. Woodward, M. P. Stevens, R. M. La Ragione, T. T. MacDonald, and G. Frankel. 2008. Role of NleH, a type III secreted effector from attaching and effacing pathogens, in colonization of the bovine, ovine, and murine gut. Infect. Immun. 76:4804-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho, T. D., B. M. Davis, J. M. Ritchie, and M. K. Waldor. 2008. Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect. Immun. 76:1858-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hormaeche, C. E. 1979. Natural resistance to Salmonella typhimurium in different inbred mouse strains. Immunology 37:311-318. [PMC free article] [PubMed] [Google Scholar]
- 42.Iguchi, A., N. R. Thomson, Y. Ogura, D. Saunders, T. Ooka, I. R. Henderson, D. Harris, M. Asadulghani, K. Kurokawa, P. Dean, B. Kenny, M. A. Quail, S. Thurston, G. Dougan, T. Hayashi, J. Parkhill, and G. Frankel. 2009. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191:347-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itoh, K., K. Maejima, K. Ueda, and K. Fujiwara. 1978. Effect of intestinal flora on megaenteron of mice. Microbiol. Immunol. 22:661-672. [DOI] [PubMed] [Google Scholar]
- 44.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 46.Kelly, M., E. Hart, R. Mundy, O. Marches, S. Wiles, L. Badea, S. Luck, M. Tauschek, G. Frankel, R. M. Robins-Browne, and E. L. Hartland. 2006. Essential role of the type III secretion system effector NleB in colonization of mice by Citrobacter rodentium. Infect. Immun. 74:2328-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klapproth, J. M., M. Sasaki, M. Sherman, B. Babbin, M. S. Donnenberg, P. J. Fernandes, I. C. Scaletsky, D. Kalman, A. Nusrat, and I. R. Williams. 2005. Citrobacter rodentium lifA/efa1 is essential for colonic colonization and crypt cell hyperplasia in vivo. Infect. Immun. 73:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Latasa, C., A. Roux, A. Toledo-Arana, J. M. Ghigo, C. Gamazo, J. R. Penades, and I. Lasa. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322-1339. [DOI] [PubMed] [Google Scholar]
- 49.Leiman, P. G., M. Basler, U. A. Ramagopal, J. B. Bonanno, J. M. Sauder, S. Pukatzki, S. K. Burley, S. C. Almo, and J. J. Mekalanos. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. USA 106:4154-4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levine, M. M., J. P. Nataro, H. Karch, M. M. Baldini, J. B. Kaper, R. E. Black, M. L. Clements, and A. D. O'Brien. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J. Infect. Dis. 152:550-559. [DOI] [PubMed] [Google Scholar]
- 51.Lin, R. J., M. Capage, and C. W. Hill. 1984. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J. Mol. Biol. 177:1-18. [DOI] [PubMed] [Google Scholar]
- 52.Liu, C., L. S. Heath, and C. L. Turnbough, Jr. 1994. Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 8:2904-2912. [DOI] [PubMed] [Google Scholar]
- 53.Liu, J., X. Zhang, S. Zhou, and P. Tao. 2007. Purification and characterization of a 4-hydroxybenzoate decarboxylase from Chlamydophila pneumoniae AR39. Curr. Microbiol. 54:102-107. [DOI] [PubMed] [Google Scholar]
- 54.Luperchio, S. A., J. V. Newman, C. A. Dangler, M. D. Schrenzel, D. J. Brenner, A. G. Steigerwalt, and D. B. Schauer. 2000. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. J. Clin. Microbiol. 38:4343-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma, W., F. F. Dong, J. Stavrinides, and D. S. Guttman. 2006. Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLoS Genet. 2:e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsui, T., T. Yoshida, T. Hayashi, and T. Nagasawa. 2006. Purification, characterization, and gene cloning of 4-hydroxybenzoate decarboxylase of Enterobacter cloacae P240. Arch. Microbiol. 186:21-29. [DOI] [PubMed] [Google Scholar]
- 57.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monteiro, B. T., L. C. Campos, M. P. Sircili, M. R. Franzolin, L. F. Bevilacqua, J. P. Nataro, and W. P. Elias. 2009. The dispersin-encoding gene (aap) is not restricted to enteroaggregative Escherichia coli. Diagn. Microbiol. Infect. Dis. 65:81-84. [DOI] [PubMed] [Google Scholar]
- 59.Mougous, J. D., M. E. Cuff, S. Raunser, A. Shen, M. Zhou, C. A. Gifford, A. L. Goodman, G. Joachimiak, C. L. Ordonez, S. Lory, T. Walz, A. Joachimiak, and J. J. Mekalanos. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mundy, R., F. Girard, A. J. FitzGerald, and G. Frankel. 2006. Comparison of colonization dynamics and pathology of mice infected with enteropathogenic Escherichia coli, enterohaemorrhagic E. coli and Citrobacter rodentium. FEMS Microbiol. Lett. 265:126-132. [DOI] [PubMed] [Google Scholar]
- 61.Mundy, R., T. T. Macdonald, G. Dougan, G. Frankel, and S. Wiles. 2005. Citrobacter rodentium of mice and man. Cell. Microbiol. 7:1697-1706. [DOI] [PubMed] [Google Scholar]
- 62.Mundy, R., L. Petrovska, K. Smollett, N. Simpson, R. K. Wilson, J. Yu, X. Tu, I. Rosenshine, S. Clare, G. Dougan, and G. Frankel. 2004. Identification of a novel Citrobacter rodentium type III secreted protein, EspI, and roles of this and other secreted proteins in infection. Infect. Immun. 72:2288-2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mundy, R., D. Pickard, R. K. Wilson, C. P. Simmons, G. Dougan, and G. Frankel. 2003. Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol. Microbiol. 48:795-809. [DOI] [PubMed] [Google Scholar]
- 64.Muto, T., M. Nakagawa, Y. Isobe, M. Saito, and T. Nakano. 1969. Infectious megaenteron of mice. I. Manifestation and pathological observation. Jpn. J. Med. Sci. Biol. 22:363-374. [DOI] [PubMed] [Google Scholar]
- 65.Nakano, M., T. Iida, and T. Honda. 2004. Urease activity of enterohaemorrhagic Escherichia coli depends on a specific one-base substitution in ureD. Microbiology 150:3483-3489. [DOI] [PubMed] [Google Scholar]
- 66.Nishio, M., N. Okada, T. Miki, T. Haneda, and H. Danbara. 2005. Identification of the outer-membrane protein PagC required for the serum resistance phenotype in Salmonella enterica serovar Choleraesuis. Microbiology 151:863-873. [DOI] [PubMed] [Google Scholar]
- 67.Norman, A., L. H. Hansen, Q. She, and S. J. Sorensen. 2008. Nucleotide sequence of pOLA52: a conjugative IncX1 plasmid from Escherichia coli which enables biofilm formation and multidrug efflux. Plasmid 60:59-74. [DOI] [PubMed] [Google Scholar]
- 68.Nuccio, S. P., and A. J. Baumler. 2007. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71:551-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Osoegawa, K., P. Y. Woon, B. Zhao, E. Frengen, M. Tateno, J. J. Catanese, and P. J. de Jong. 1998. An improved approach for construction of bacterial artificial chromosome libraries. Genomics 52:1-8. [DOI] [PubMed] [Google Scholar]
- 70.Parham, N. J., S. J. Pollard, M. Desvaux, A. Scott-Tucker, C. Liu, A. Fivian, and I. R. Henderson. 2005. Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 43:4076-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 72.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 73.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 74.Penno, C., P. Sansonetti, and C. Parsot. 2005. Frameshifting by transcriptional slippage is involved in production of MxiE, the transcription activator regulated by the activity of the type III secretion apparatus in Shigella flexneri. Mol. Microbiol. 56:204-214. [DOI] [PubMed] [Google Scholar]
- 75.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pilonieta, M. C., M. D. Bodero, and G. P. Munson. 2007. CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli. J. Bacteriol. 189:5060-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 78.Pukatzki, S., S. B. McAuley, and S. T. Miyata. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11-17. [DOI] [PubMed] [Google Scholar]
- 79.Rasko, D. A., M. J. Rosovitz, G. S. Myers, E. F. Mongodin, W. F. Fricke, P. Gajer, J. Crabtree, M. Sebaihia, N. R. Thomson, R. Chaudhuri, I. R. Henderson, V. Sperandio, and J. Ravel. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robson, H. G., and S. I. Vas. 1972. Resistance of inbred mice to Salmonella typhimurium. J. Infect. Dis. 126:378-386. [DOI] [PubMed] [Google Scholar]
- 81.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 82.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schauer, D. B., and S. Falkow. 1993. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect. Immun. 61:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schauer, D. B., B. A. Zabel, I. F. Pedraza, C. M. O'Hara, A. G. Steigerwalt, and D. J. Brenner. 1995. Genetic and biochemical characterization of Citrobacter rodentium sp. nov. J. Clin. Microbiol. 33:2064-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shrivastava, S., and S. S. Mande. 2008. Identification and functional characterization of gene components of Type VI secretion system in bacterial genomes. PLoS One 3:e2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Simpson, N., R. Shaw, V. F. Crepin, R. Mundy, A. J. FitzGerald, N. Cummings, A. Straatman-Iwanowska, I. Connerton, S. Knutton, and G. Frankel. 2006. The enteropathogenic Escherichia coli type III secretion system effector Map binds EBP50/NHERF1: implication for cell signalling and diarrhoea. Mol. Microbiol. 60:349-363. [DOI] [PubMed] [Google Scholar]
- 87.Tatsuno, I., R. Mundy, G. Frankel, Y. Chong, A. D. Phillips, A. G. Torres, and J. B. Kaper. 2006. The lpf gene cluster for long polar fimbriae is not involved in adherence of enteropathogenic Escherichia coli or virulence of Citrobacter rodentium. Infect. Immun. 74:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tobe, T., S. A. Beatson, H. Taniguchi, H. Abe, C. M. Bailey, A. Fivian, R. Younis, S. Matthews, O. Marches, G. Frankel, T. Hayashi, and M. J. Pallen. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA 103:14941-14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vallance, B. A., W. Deng, K. Jacobson, and B. B. Finlay. 2003. Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect. Immun. 71:3443-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang, Y. D., S. Zhao, and C. W. Hill. 1998. Rhs elements comprise three subfamilies which diverged prior to acquisition by Escherichia coli. J. Bacteriol. 180:4102-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wiles, S., S. Clare, J. Harker, A. Huett, D. Young, G. Dougan, and G. Frankel. 2004. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell Microbiol. 6:963-972. [DOI] [PubMed] [Google Scholar]
- 92.Wiles, S., S. Clare, J. Harker, A. Huett, D. Young, G. Dougan, and G. Frankel. 2005. Organ-specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell Microbiol. 7:459. [DOI] [PubMed] [Google Scholar]
- 93.Wiles, S., G. Dougan, and G. Frankel. 2005. Emergence of a ‘hyperinfectious’ bacterial state after passage of Citrobacter rodentium through the host gastrointestinal tract. Cell Microbiol. 7:1163-1172. [DOI] [PubMed] [Google Scholar]
- 94.Williams, L. E., C. Detter, K. Barry, A. Lapidus, and A. O. Summers. 2006. Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Appl. Environ. Microbiol. 72:4899-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zheng, J., and K. Y. Leung. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66:1192-1206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.