Abstract
The human cytomegalovirus (HCMV) gene UL21a was recently annotated by its conservation in chimpanzee cytomegalovirus. Two large-scale mutagenic analyses showed that mutations in overlapping UL21a/UL21 resulted in a severe defect of virus growth in fibroblasts. Here, we characterized UL21a and demonstrated its role in HCMV infection. We mapped a UL21a-specific transcript of ∼600 bp that was expressed with early kinetics. UL21a encoded pUL21a, a protein of ∼15 kDa, which was unstable and localized predominantly to the cytoplasm during HCMV infection or when expressed alone. Interestingly, pUL21a was drastically stabilized in the presence of proteasome inhibitor MG132, but its instability was independent of a functional ubiquitin-mediated pathway, suggesting that pUL21a underwent proteasome-dependent, ubiquitin-independent degradation. A UL21a deletion virus was attenuated in primary human newborn foreskin fibroblasts (HFFs) and embryonic lung fibroblasts (MRC-5), whereas a marker-rescued virus and mutant viruses lacking the neighboring or overlapping genes UL20, UL21, or UL21.5-UL23 replicated at wild-type levels. The growth defect of UL21a-deficient virus in MRC-5 cells was more pronounced than that in HFFs. At a high multiplicity of infection, the UL21a deletion virus synthesized viral proteins with wild-type kinetics but had a two- to threefold defect in viral DNA replication. More importantly, although pUL21a was not detected in the virion, progeny virions produced by the mutant virus were ∼10 times less infectious than wild-type virus, suggesting that UL21a is required for HCMV to establish efficient productive infection. We conclude that UL21a encodes a short-lived cytoplasmic protein and facilitates HCMV replication in fibroblasts.
Human cytomegalovirus (HCMV), the prototypical betaherpesvirus, is a ubiquitous pathogen that infects the majority of the world's population. HCMV is usually asymptomatic in immunocompetent individuals, except in rare cases where it causes mononucleosis. However, HCMV can cause severe disease and death in immunocompromised individuals such as AIDS patients and transplant recipients. Importantly, HCMV is the most common viral cause of birth defects leading to mental retardation, blindness, and hearing loss (5). In addition, HCMV infection is also a possible risk factor in the development of vascular diseases such as atherosclerosis, transplant vascular sclerosis, and coronary restenosis after angioplasty surgery (17, 21, 23, 26, 34, 35, 46).
HCMV contains a 240-kb double-stranded DNA genome that encodes at least 166 putative open reading frames (ORFs) and several miRNAs (8, 12, 13, 15, 18, 28, 29). With the advent of the infectious bacterial artificial chromosome (BAC) clone-based genetic system for HCMV (3, 44), the functions of many HCMV genes have started to be elucidated. Genome-scale mutagenesis approaches have been used to delineate the functions of genes encoded by HCMV (14, 43). These systematic studies have identified a subset of candidate viral genes that are important for HCMV to establish infection in tissue culture models of primary human cells including fibroblasts. Nonetheless, products of more than half of the annotated viral genes have not been experimentally identified and characterized (25).
Little is known about the gene products produced from the viral genomic region where UL21a resides. UL21.5 is the only gene within this region that has been characterized in detail. UL21.5 encodes a late transcript that is 400 to 500 bp in length, spliced, and incorporated into virions (4, 32) (see Fig. 1A). The protein product of UL21.5, pUL21.5, is a soluble receptor decoy for CC chemokines, selectively binds to RANTES, and prevents binding with its cognate receptors (27, 38). UL23 is a member of the US22 gene family and encodes a tegument protein (1). In addition, HCMV also encodes a miRNA UL22A-1 with unknown targets that is expressed with early gene kinetics from a locus adjacent to UL21.5 (15, 18). However, no gene products emanating from UL21 or UL21a have been identified.
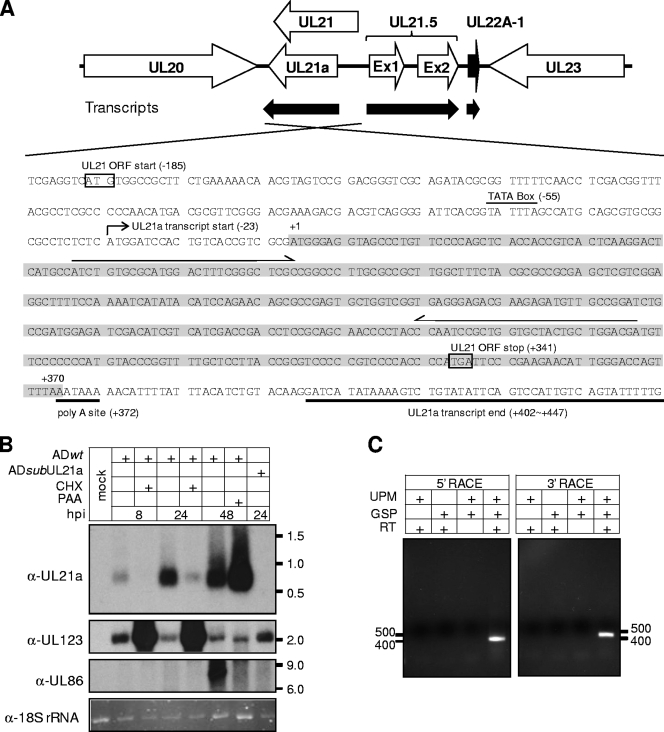
FIG. 1.
UL21a encodes a single unspliced transcript with early gene kinetics. (A) HCMV genomic region spanning UL20 to UL23. The top panel shows the schematic structure of the viral genomic region. Annotated viral ORFs are indicated by open boxed arrows. The HCMV-encoded micro RNA UL22A-1 is indicated by the black boxed arrow. Also shown are the transcripts from this region that have been identified in previous studies or in the present study. The bottom panel shows the genomic sequence of the sense strand where UL21a resides. The mapped start and termination sites of the UL21a transcript are indicated. The UL21a ORF is highlighted in gray. Also shown are the putative TATA box, poly(A) site (both indicated with lines), the start and stop codons of the putative UL21 ORF (both indicated with boxes), and gene specific primers used for 5′ or 3′ RACE (both indicated with arrows). (B) Northern blot analysis of transcripts arising from the UL21a/UL21 gene locus. HFFs were either mock infected or infected with wild-type virus (ADwt) or UL21a deletion virus (ADsubUL21a) at an MOI of 1 in the presence or absence of 100 μg of CHX/ml or 200 μg of PAA/ml. Cells were harvested at indicated times, total RNA was isolated, and the UL21a-specific transcript or UL123 (encoding IE1) and UL86 (encoding MCP) control transcripts were analyzed by Northern blotting with UL21a, UL123, and UL86 strand-specific antisense probes. Also shown is the ethidium bromide staining of 18S rRNA as the loading control. Molecular size markers (in kb) are indicated. (C) RACE analysis of UL21a-specific transcripts. HFFs were infected with ADwt at an MOI of 1, cells were harvested at 40 hpi, total RNA were isolated, cDNA was generated by reverse transcription (RT), and 5′ or 3′ portions of the sequences of the UL21a-specific transcripts were amplified by 5′ or 3′ RACE using universal primer mix (UPM) and UL21a gene-specific primers (GSP) (see Fig. 1A and Materials and Methods), respectively. RACE products were analyzed by agarose gel electrophoresis, individual product was cloned, their sequences were determined, and the assembled UL21a transcript was indicated in Fig. 1A. Molecular size markers (in bp) are indicated.
The original annotation of the HCMV genome recognized the putative gene UL21 that was predicted to encode the largest open reading frame (ORF) between neighboring genes UL20 and UL21.5 (8). Subsequently, an elegant comparative analysis by Davison et al. showed that the UL21 ORF had no homologue in the closely related chimpanzee cytomegalovirus (CCMV) (12). Instead, gene UL21a, which encodes an alternative reading frame that starts 185 bp downstream of the start codon and stops 29 bp downstream of the stop codon of the putative UL21 ORF, was highly conserved in CCMV and therefore added to the annotation of the HCMV genome (12) (Fig. 1A and data not shown). Furthermore, UL21a is entirely conserved among all HCMV strains that have been sequenced, including AD169, Towne, Toledo, PH, Merlin, and TR (12-14, 28, 29). However, no experimental evidence has been documented as to whether HCMV expresses UL21a, UL21, or both.
UL21a appears specific to primate CMVs since its homologues are found in CCMV and rhesus CMV (33) but are not present in the more distantly related viruses such as murine CMV (data not shown). UL21a is predicted to encode a protein of 123 amino acids (aa) with the calculated molecular mass of 14.3 kDa. It shares no apparent homology with any known cellular or viral proteins in the database, suggesting that the function of this protein is likely to be unique to CMV biology. Two large-scale mutagenic analyses started to reveal the importance of this previously uncharacterized UL21a/UL21 locus in HCMV infection (14, 43). In the AD169 strain, two independent insertional mutations within UL21a resulted in a growth defect of ∼50-fold in human foreskin fibroblasts (HFFs) (43). In the Towne strain, a substitution mutation targeting UL21 simultaneously deleted the overlapping UL21a, and a more severe growth defect of >20,000-fold was observed with this mutant virus (14). It is possible that several factors, such as the nature of the mutations, the difference of virus strains, and the assays for growth analysis, are responsible for the different degrees of growth attenuation seen in mutant AD169 and Towne viruses. Nonetheless, these studies suggest an important role of the UL21a/UL21 locus for the virus to establish efficient infection.
In the present study, we mapped the transcript arising from the UL21a locus and characterized its protein product, termed pUL21a. Analysis of recombinant HCMV virus lacking UL21a, UL21, or its neighboring genes indicates that UL21a, but not UL21 or other neighboring genes, is required for HCMV growth in primary human fibroblasts. Although we could not detect pUL21a in the virion, the UL21a deletion virus produced progeny that are ∼10 times less infectious than wild-type virus, suggesting that UL21a is required for HCMV to initiate efficient productive infection.
MATERIALS AND METHODS
Plasmids and antibodies.
pYD-C235 is a pLPCX-derived retroviral vector (Clontech) that expresses a DsRed gene driven by an internal ribosome entry site 2 (IRES2) (36). pYD-C423, pYD-C428, and pYD-C429 were created by amplifying the UL21a, UL99 (encoding pp28), and UL44 coding sequences and cloning the PCR products upstream of the IRES2 of pYD-C235, respectively. pYD-C255 contained a GalK/kanamycin dual-expression cassette that was used for the first step of linear recombination (31) (see below).
To generate a polyclonal antibody to pUL21a, the entire UL21a coding sequence was cloned upstream of a His6 tag in the expression vector pET-22b (Novagen). The His-tagged fusion protein was produced in Escherichia coli, purified by using Ni-NTA agarose beads (Qiagen), and used as an immunogen to generate the rabbit antisera (Covance). The rabbit antisera were subsequently affinity-purified by using the UL21a recombinant protein (Covance), resulting in the high-titer, highly specific anti-pUL21a polyclonal antibody that was tested for its specific interaction with the immunogen or the virus-produced pUL21a by immunoblot assay. Additional primary antibodies used in the present study included: anti-β-actin (clone AC15; Abcam); anti-p53 (clone OP-03, Calbiochem); anti-UL44 (clone 10D8; Virusys); anti-MCP (anti-major capsid protein) (generous gift from Wade Gibson, John Hopkins University); and anti-IE1, anti-pp65, anti-pUL69, and anti-pp28 (36) (generous gifts from Thomas Shenk, Princeton University).
Cells and viruses.
Primary human newborn foreskin fibroblasts (HFFs), embryonic lung fibroblasts (MRC-5), HeLa cells, and mouse ts20 cells (10) (a generous gift from Harvey Ozer, UMDNJ-New Jersey Medical School) were propagated in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, nonessential amino acids, and penicillin-streptomycin. For transient expression, HeLa cells were transfected by using Lipofectamine (Invitrogen), and HFFs were electroporated by using Amaxa Technologies (Lonza) with plasmids expressing the proteins of interest according to the manufacturers' instructions.
Various BAC-HCMV clones were constructed to reconstitute recombinant HCMV viruses. The BAC-HCMV clone pAD-GFP was used as the parental clone to produce wild-type virus (ADwt). pAD-GFP carries the full-length genome of HCMV strain AD169 and contains a simian virus 40 (SV40) early promoter-driven green fluorescent protein (GFP) gene in place of the viral US4-US6 region (36, 44). All other recombinant BAC clones were constructed by using a linear recombination protocol in the bacterial strain SW102 (31). A GalK/kanamycin dual marker cassette was amplified by PCR from pYD-C255 with a pair of 70-bp primers that had 5′-terminal 50-bp sequences homologous to the viral sequences of targeted sites (see Table 1 for all primers). The marker cassette was subsequently recombined into pAD-GFP at the locus of interest by linear recombination to generate substitution or insertional mutant BAC-HCMV clones. Resulting transformants were selected on kanamycin-containing LB plates to identify clones carrying the marker cassette. One of the substitution mutant BAC-HCMV clones, pADsubUL21a, carried the marker cassette in place of UL21a. To create a UL21a marker rescue BAC-HCMV clone (pADrevUL21a), the UL21a sequence was amplified by PCR and subsequently recombined into pADsubUL21a to replace the marker cassette by linear recombination. The resulting recombinants were selected on 2-deoxy-galactose (DOG)-containing minimal media plates for the loss of GalK/kanamycin. To create a UL21a clean-deletion BAC-HCMV clone (pADdlUL21a), a pair of overlapping 70-bp primers were generated that together spanned 50 bp both upstream and downstream of the UL21a ORF. The PCR fragment generated from these primers was recombined into pADsubUL21a to replace the marker cassette by linear recombination. To insert the GFP-S tag at the N terminus of the UL21a ORF in the viral genome, the GalK/kanamycin marker was amplified by PCR and inserted immediately downstream of the UL21a start codon by linear recombination in pAD/Cre (44), a BAC clone carrying the full-length AD169 genome without GFP, resulting in the clone ADinGalK/kan-UL21a. The GFP-S tag sequence was amplified from the plasmid pIC113 (9) and recombined into the viral genome of ADinGalK/kan-UL21a, in frame at the N terminus of the UL21a ORF, to replace the marker sequence by linear recombination, resulting in ADinGFP-UL21a. All recombinant BACs were verified by restriction digestion, Southern blotting, PCR, and direct sequencing analysis.
TABLE 1.
Primers used to create substitutions and insertions in the HCMV genome
| Targeted HCMV gene | Type of genetic alteration | Primer pair sequences (5′-3′) used to introduce alterationa | Resulting recombinant HCMV virus |
|---|---|---|---|
| UL21a | Substitution | agccatgcagcgtgcggcgcctctctcatggatccactgtcaccgtcgcgCCTGTTGACAATTAATCATCG | ADsubUL21a |
| atacagacttttatatgatccttgtacagatgtaaataaaatgtttttatCTCAGCAAAAGTTCGATTTA | |||
| UL20 | Substitution | tggaacggtctttatatatacaaacgccgttatgttcagtgtccggcaagCCTGTTGACAATTAATCATCG | ADsubUL20 |
| tatggaaaatatgtagtccgtaccgcttggggctcaaagttcaaagtccgCTCAGCAAAAGTTCGATTTA | |||
| UL21.5-UL23 | Substitution | gcaagaacgtaactctcagtcagggggggtccaccaccgacggagacgaaCCTGTTGACAATTAATCATCG | ADsubUL21.5-UL23 |
| gaccgaccacatctactctgactcgttgacctttgtggccgagagcatcaCTCAGCAAAAGTTCGATTTA | |||
| UL21a | N-terminal insertion | agccatgcagcgtgcggcgcctctctcatggatccactgtcaccgtcgcgCCTGTTGACAATTAATCATCG | ADinGalK/kan-UL21a |
| ggcatgagtccttgagtgacggtggtgagctggggaacagggctacctccCTCAGCAAAAGTTCGATTTA | |||
| UL21a | N-terminal insertion | agccatgcagcgtgcggcgcctctctcatggatccactgtcaccgtcgcgATGGTGAGCAAGGGCGAGGAG | ADinGFP-UL21a |
| ggcatgagtccttgagtgacggtggtgagctggggaacagggctacctccCTCGAGACTAGTACCTCCACC |
Virus-specific sequences are in lowercase letters, GalK-kanamycin marker-specific sequences are in uppercase letters, and GFP-S tag-specific sequences are underlined and in uppercase letters.
To reconstitute virus, 2 to 4 μg of the BAC-HCMV DNA and 1 μg of the pp71-expression plasmid were transfected into MRC-5 fibroblasts by electroporation (44). Culture medium was changed 24 h later, and virus was harvested by collecting cell-free culture supernatant when the entire monolayer of infected cells was lysed. Alternatively, virus was also produced by collecting cell-free culture supernatant from HFFs infected at a multiplicity of infection (MOI) of 0.005. Virus-containing culture supernatants were then purified by ultracentrifugation through a 20% d-sorbitol cushion at an average relative centrifugal force of 53,000 × g for 1 h, resuspended in Dulbecco modified Eagle medium with 10% fetal calf serum, and saved as viral stocks. HCMV virus titers were determined in duplicate by plaque assay (31), and their DNA content was determined by real-time quantitative PCR (see below).
Analysis of viral growth kinetics.
HFFs or MRC-5 cells were seeded in 12-well plates overnight to produce a subconfluent monolayer. Cells were then inoculated with recombinant HCMV viruses for 1 h at an MOI of 0.001 for multistep growth analysis or an MOI of 1 for single-step growth analysis. The inoculum was removed, infected monolayers were rinsed with phosphate-buffered saline, and fresh medium was replenished. At various times postinfection, cell-free virus was collected by harvesting medium from infected cultures. In addition, infected cells were pelleted and then lysed by a freeze-thaw cycle followed by sonication, cell debris was cleared by low-speed centrifugation, and supernatants were saved as cell-associated virus samples. Titers of each virus sample were determined by plaque assay. Moreover, plaque size was measured by using ImageJ software (National Institutes of Health), and the P value associated with the Student paired t test with a two-tailed distribution was calculated by scoring at least 50 plaques per infection.
Analysis of DNA, RNA, and proteins.
To prepare virion DNA for quantification by real-time quantitative PCR, 100 μl of cell-free virus (or 10 μl of purified virus) was treated with DNase I (30 U; Roche) at 37°C for 30 min to remove contaminating DNA, followed by incubation at 75°C for 20 min to stop the reaction. The samples were then incubated overnight at 55°C in lysis buffer (400 mM NaCl, 10 mM Tris [pH 8.0], 10 mM EDTA, 0.1 mg of proteinase K/ml, 0.2% sodium dodecyl sulfate [SDS]), and DNA was extracted with phenol-chloroform and treated with 20 μg of RNase A/ml for 1 h at 37°C. DNA was extracted again with phenol-chloroform, precipitated with ethanol, and resuspended in nuclease-free water (Ambion). To prepare intracellular DNA from infected cells for quantification, HCMV-infected HFFs were collected at various times postinfection, resuspended in lysis buffer, and incubated at 55°C overnight. DNA was extracted with phenol-chloroform, treated with 100 μg of RNase A/ml for 1 h at 37°C, extracted again with phenol-chloroform, precipitated with ethanol, and resuspended in nuclease-free water.
Viral DNA was quantified by real-time quantitative PCR as previously described (31) by using a TaqMan probe (Applied Biosystems) and primers specific for the HCMV UL54 gene (30). Cellular DNA was quantified with SYBR green PCR Master Mix (Clonetech) and a primer pair specific for the human β-actin gene (5′-CTC CAT CCT GGC CTC GCT GT-3′ and 5′-GCT GTC ACC TTC ACC GTT CC-3′). The accumulation of viral DNA was normalized by dividing UL54 gene equivalents by β-actin equivalents. The accumulation of wild-type viral DNA at 2 h postinfection (hpi) was arbitrarily set as 1.
mRNA transcripts expressed during HCMV infection were analyzed by Northern blotting and rapid amplification of cDNA ends (RACE) as previously described (31, 44). For Northern blot analysis, single-stranded DNA probes were prepared by using PCR-generated templates and a Strip-EZ PCR kit (Ambion) according to the manufacturer's instructions. Primer pairs used to generate the templates were as follows: 5′-ATG GGA GGT AGC CCT GTT CC-3′ and 5′-TTA AAA CTG GTC CCA ATG TTC TT-3′ (for the UL21a probe), 5′-GTA GCC TAC ACT TTG GCC ACC-3′ and 5′-TTA CTG GTC AGC CTT GCT TCT A-3′ (for the UL123 probe), and 5′-GCG CGC CAG TAC TTT AAC ACA G-3′ and 5′-TCA CGA GTT AAA TAA CAT GGA TTG-3′ (for the UL86 probe). Primers used to generate the 32P-labeled strand-specific antisense probes were as follows: 5′-TCGTCCAGCAGTAGCACCAGCGGATTGG-3′ (for the UL21a probe), 5′-TTA CTG GTC AGC CTT GCT TCT A-3′ (for the UL123 probe), and 5′-TCA CGA GTT AAA TAA CAT GGA TTG-3′ (for the UL86 probe). RACE analysis was performed by using SMART PCR cDNA synthesis kit (Clontech). Gene-specific primers 5′-TCG TCC AGC AGT AGC ACC AGC GGA TTG G-3′ and 5′-ATC TGT GCG CAT GGA CTT TCG GGC TCG C-3′ were used for 5′ and 3′ RACE, respectively (Fig. 1A).
To analyze protein accumulation by immunoblotting, total cell extracts were prepared by lysing phosphate-buffered saline-washed infected cells in SDS-containing sample buffer. Virion proteins were prepared by purifying virions by ultracentrifugation through sorbitol cushion and resuspending in SDS-containing sample buffer. Proteins were resolved by electrophoresis on a SDS-containing polyacrylamide gel, transferred to polyvinylidene difluoride membranes, hybridized with primary antibodies, reacted with horseradish peroxidase-conjugated secondary antibodies, and visualized by using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) (42).
To analyze intracellular protein localization by immunofluorescence, cells grown on glass coverslips were fixed in 2% paraformaldehyde, permeabilized with 0.1% Triton X-100 for 5 min, incubated with the primary antibody, and subsequently labeled with the secondary antibody. Labeled cells were counterstained with TO-PRO-3 (Molecular Probes) to visualize the nuclei and then mounted on slides with Prolong Gold antifade reagent (Molecular Probes). Images were captured by using Zeiss LSM Image software with a Zeiss LSM 510 META confocal laser scanning microscope (31).
Proteasome inhibition assays.
The protease inhibitors Z-Leu-Leu-Leu-CHO (MG132), N-acetyl-Leu-Leu-Met (ALLM), and N-acetyl-Leu-Leu-Nle-CHO (ALLN; Calbiochem) were resuspended in dimethyl sulfoxide (DMSO; Sigma) at a concentration of 10 mM as stock solutions. In overexpression experiments, protease inhibitors were added to culture medium 48 h after HeLa cells were transfected with expression plasmids, and cell lysates were collected 24 h later for immunoblot analysis. In HCMV infection experiments, MG132 was added to culture medium of infected HFFs 10 h before cell lysates were collected.
RESULTS
UL21a encodes a single transcript with early gene expression kinetics.
No products have been previously identified from the UL21/UL21a gene locus. Since UL21a is highly conserved at the amino acid level among primate CMVs, we hypothesized that this gene encoded a protein product.
We carried out Northern blot analysis to assess potential transcripts arising from the UL21a gene locus during HCMV infection (Fig. 1B). Using a UL21a-specific antisense probe, we detected a single specific transcript of ∼600 bp during wild-type virus infection. The accumulation of this transcript was evident as early as 8 hpi and became more abundant by 24 hpi. The transcript was absent in infection of ADsubUL21a, the recombinant virus in which the entire UL21a putative coding sequence was replaced by a marker sequence (see Fig. 5A), indicating its UL21a-specific origin. The expression kinetics of the transcript was determined by inhibition assays for protein translation and DNA replication. Cycloheximide (CHX), a protein synthesis inhibitor, increased the accumulation of the immediate-early transcript of UL123, whereas it significantly reduced the accumulation of the UL21a transcript at 8 and 24 hpi, ruling out UL21a as an immediate-early gene. Phosphonoacetic acid (PAA), a DNA synthesis inhibitor, blocked the accumulation of the 8-kb transcript of the late gene UL86 (7) but did not decrease production of the UL21a transcript at 48 hpi, ruling out UL21a as a late or leaky late gene. Together with its temporal expression pattern, these results indicate that UL21a is transcribed with early gene kinetics.
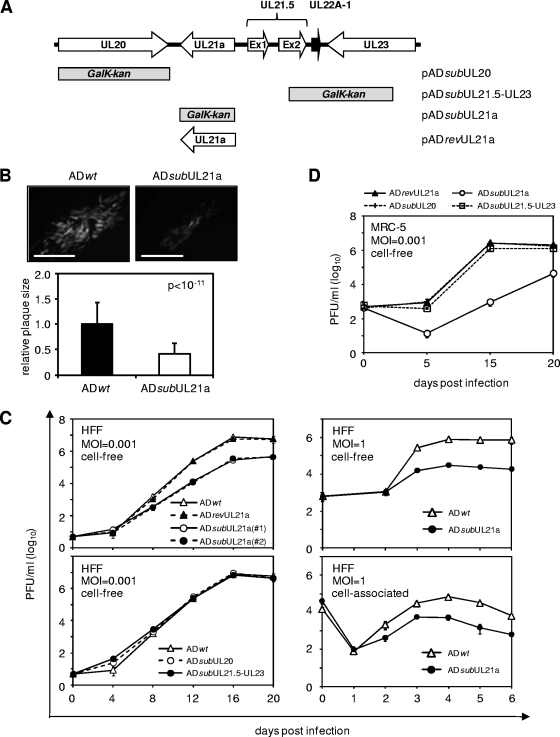
FIG. 5.
Deletion of UL21a results in attenuated growth of HCMV in fibroblasts. (A) Schematic structure of the viral genomic region in recombinant BAC-HCMV clones where genetic alterations were introduced. Viral ORFs and known miRNAs are indicated by open and black boxed arrows, respectively. The boxes below the first line represent the locations of viral sequences that were replaced by the GalK/kanamycin marker sequence in the substitution mutants, as indicated. Note that the UL21a sequence was reintroduced into pADsubUL21a to replace the marker sequence and generate the marker-rescued clone pADrevUL21a. (B) Representative images of plaques (as indicated by virus-driven GFP expression) from HFFs that were infected with wild-type or UL21a deletion virus, overlaid with agarose, and examined under a fluorescence microscope at 12 dpi. Scale bars (0.1 mm) are shown. Average sizes of 50 plaques from wild-type or mutant virus infection, and the P value by using the Student paired t test are also shown. (C and D) Growth kinetic analysis of recombinant HCMV viruses. HFFs (C) or MRC-5 primary human embryonic lung fibroblasts (D) were infected with indicated viruses at an MOI of 0.001 or 1, cell-free and cell-associated viruses were collected at different days postinfection, and their yields were determined by plaque assay.
To map the ends of the UL21a transcript, we performed both 5′- and 3′-RACE analysis of HCMV-infected HFFs using two UL21a-specific primers that would produce overlapping RACE products (Fig. 1A). Both RACE reactions produced a single band, providing additional evidence that UL21a encodes a single transcript (Fig. 1C). Sequence analysis of RACE products indicated that the UL21a transcript was unspliced and initiated 23 bp upstream of the start codon. All of the three 3′-RACE products terminated at different locations within 30 to 75 bp downstream of the stop codon, suggesting that the transcript can be polyadenylated at multiple sites within this region (Fig. 1A). Notably, 5′-RACE analysis did not detect a product that could encode the N-terminal portion of the putative UL21 ORF (Fig. 1A), even after extending the reaction for an additional 10 cycles (data not shown). Therefore, we mapped the UL21a transcript but found no evidence for the presence of a UL21-specific transcript.
pUL21a is expressed but targeted for degradation in a proteasome-dependent manner during HCMV infection.
To directly detect the protein products encoded by UL21a, we created a rabbit polyclonal antibody against a recombinant protein derived from the UL21a coding sequence. The antisera were affinity purified with the UL21a recombinant protein to generate a specific polyclonal antibody. The antibody was sensitive since it could detect the recombinant protein at the level of 10−8 nmol when used in immunoblot analysis (data not shown). Importantly, this antibody detected a single protein, termed pUL21a, with an apparent molecular mass of ∼15 kDa in the lysates of cells infected with wild-type virus but not in mock-infected cells or in cells infected with UL21a deletion virus (Fig. 2A). Therefore, pUL21a was encoded by UL21a, was expressed at the predicted size, and was lost in the deletion mutant virus. The accumulation of pUL21a became visible 10 hpi, peaked at 24 to 48 hpi, and then gradually decreased. Despite the strong sensitivity of the antibody, the signal detected in infected cell lysates was weak, even at 24 hpi (Fig. 2A and data not shown), suggesting low levels of pUL21a accumulated during HCMV infection.
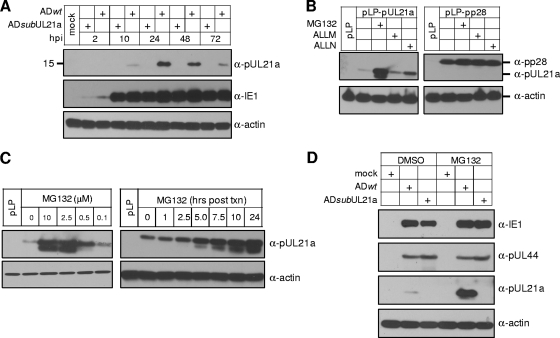
FIG. 2.
pUL21a is expressed but undergoes proteasome-dependent degradation during HCMV infection. (A) Kinetics of pUL21a accumulation in HCMV infection. HFFs were infected with ADwt or ADsubUL21a at an MOI of 1, cells were collected at indicated times, and total lysates were analyzed by immunoblotting with an affinity-purified rabbit polyclonal antibody raised against a UL21a recombinant protein. (Blotting was performed under a long exposure to reveal the presence of pUL21a at 10 hpi). The antibody to viral protein IE1 or cellular protein actin was used as infection or loading control, respectively. (B) Proteasome inhibitor stabilizes the accumulation of overexpressed pUL21a. HeLa cells were transfected with vector expressing pUL21a (pLP-pUL21a) or another HCMV protein pp28 (pLP-pp28), and protease inhibitor as indicated or DMSO control was added at 48 h to the final concentration of 10 μM. Total cell lysates were collected 24 h later and analyzed by immunoblotting with the antibody to pUL21a or pp28 (blotting was performed under a short exposure to reveal differentiated levels of pUL21a in the presence or absence of MG132). (C) Dose- and time-dependent effects of proteasome inhibitor on the accumulation of overexpressed pUL21a. HeLa cells transfected with pLP-pUL21a were incubated with MG132 at different concentration as indicated (μM) for 24 h (left panel) or with 2.5 μM MG132 for different times as indicated (right panel), and cell lysates were prepared for immunoblot analysis with the α-pUL21a antibody (short exposure). (D) HFFs were infected with the indicated HCMV virus at an MOI of 1, MG132, or DMSO control was added at 38 hpi to the final concentration of 10 μM, cells were collected at 48 hpi, and total lysates were analyzed by immunoblotting with the antibody to viral proteins IE1, pUL44, or pUL21a (short exposure).
We were intrigued by the apparent low levels of pUL21a because the accumulation of the UL21a transcript appeared abundant during infection (Fig. 1B), and thus hypothesized that pUL21a was targeted for degradation. We tested this hypothesis by expressing pUL21a in the presence or absence of protease inhibitors. MG132 is a specific inhibitor of the proteasome, ALLN is a calpain inhibitor that also acts as a weak inhibitor of the proteasome, and ALLM is a calpain inhibitor that does not inhibit the proteasome at the concentrations used here (22). When proteins were expressed from the HCMV immediate-early promoter in HeLa cells in a transient-expression experiment, the proteasome inhibitor MG132 drastically enhanced the accumulation of pUL21a, whereas the calpain inhibitor ALLM had a minimal effect (Fig. 2B). ALLN increased the accumulation of pUL21a to some extent, a finding consistent with its weak inhibitory activity to the proteasome. As a control, all three inhibitors had a minimal effect on the accumulation of another HCMV protein, pp28. The degradation of pUL21a could be markedly inhibited by MG132 at a concentration as low as 0.5 μM (Fig. 2C). At a concentration of 2.5 μM, treatment with MG132 for as little as 5 h had a clear effect in the rescue of pUL21a accumulation (Fig. 2C). Interestingly, when pUL21a accumulation was stabilized, a second, faster-migrating protein band was also detected by the α-pUL21a antibody, suggesting that an additional form of pUL21a might be produced. These results indicate that the proteasome-mediated degradation of pUL21a is independent of other viral factors. Furthermore, such degradation appears to be pUL21a-specific since the viral protein pp28 was stable independent of proteasome inhibition.
We then tested whether pUL21a was also subject to rapid degradation in the context of viral infection. HFFs were infected with ADwt or ADsubUL21a, treated with MG132 or DMSO control at 38 hpi, and analyzed for the accumulation of viral proteins at 48 hpi by immunoblot analysis (Fig. 2D). Again, whereas MG132 had a marginal effect on the accumulation of HCMV proteins IE1 and pUL44, it drastically increased the accumulation of pUL21a during HCMV infection. Therefore, pUL21a is targeted for proteasome-dependent degradation during HCMV infection.
Degradation of pUL21a is unaltered in cells lacking a functional ubiquitin-conjugation system.
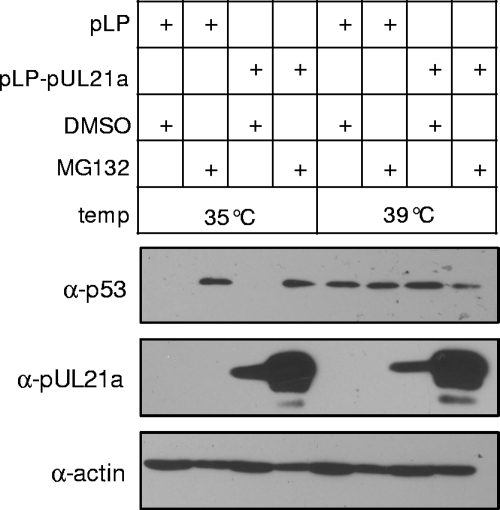
The most common mechanism by which proteins are targeted to the 26S proteasome is via polyubiquitination at internal lysine residues and, in some cases, ubiquitination can occur at cysteine, serine, and threonine residues (6, 40) or at the N terminus of the protein (2, 11). The UL21a ORF contains no lysines or cysteines and, furthermore, we could not detect pUL21a-ubiquitin conjugates in cells treated with MG132 (data not shown). We hypothesized that pUL21a degradation was ubiquitin independent, a mechanism that has been recently reported for a limited number of proteins (24, 45). To test this hypothesis, we transiently transfected pUL21a-expressing vector into mouse ts20 cells that carried a temperature-sensitive E1 ubiquitin-activating enzyme (10) and examined the accumulation of pUL21a at both permissive (35°C) and restrictive (39°C) temperatures. In these cells, the ubiquitin pathway was disabled at the restrictive temperature, thus stabilizing proteins, such as p53, that were targeted for the ubiquitin-mediated degradation (10). In this experiment, the ubiquitin pathway was active at 35°C, reducing p53 to the undetectable level and, as anticipated, MG132 treatment stabilized p53 (Fig. 3). At 39°C, p53 was stabilized, and MG132 treatment had no effect on its accumulation, indicating that the ubiquitin conjugation pathway was indeed disabled at this restrictive temperature. In contrast, the pUL21a level was equally low at both temperatures, but the protein level was greatly enhanced by MG132, suggesting that the degradation of pUL21a is proteasome dependent and ubiquitin independent.
FIG. 3.
Degradation of pUL21a is unaltered in cells lacking a functional ubiquitin-conjugation system. Mouse ts20 cells were transfected with pLP-pUL21a or the empty vector pLP, and cultured at 35°C. At 24 h later, DMSO control or MG132 was added to indicated cell culture at the final concentration of 10 μM, one set of cells were kept at 35°C, and the second set were shifted to 39°C. Total cell lysates were prepared after 20 h of drug treatment and analyzed by immunoblotting for accumulation of p53 and pUL21a. The antibody to actin was used as a loading control.
pUL21a specifically localizes to the cytoplasm.
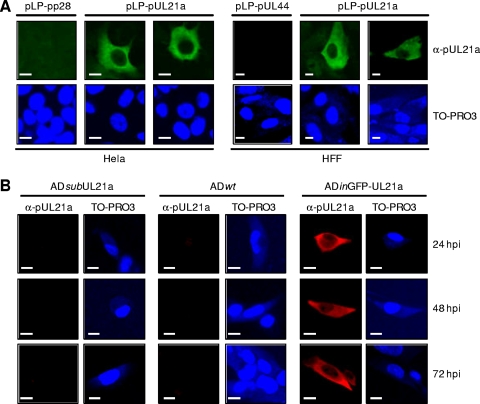
Next, we sought to determine the intracellular localization of pUL21a by indirect immunofluorescence. To determine the localization pattern of pUL21a in the absence of any other viral proteins, HeLa or HFF cells were transfected with the plasmid expressing both a CMV immediate-early promoter-driven viral protein (i.e., pUL21a, pp28, or pUL44) and IRES-driven DsRed. The overexpressed pUL21a was detected throughout the cytoplasm by the α-pUL21a antibody but was almost completely excluded from the nucleus of DsRed-positive cells (Fig. 4A and data not shown). The antibody was specific to pUL21a since it did not cross-react with other overexpressed viral proteins (i.e., pp28 and pUL44). We then analyzed the localization of pUL21a during virus infection. Unfortunately, the low level of pUL21a accumulation during infection relative to that in overexpression (data not shown) and perhaps also the low sensitivity of α-pUL21a antibody to the nondenatured protein prevented us from detecting the native pUL21a during infection by immunofluorescence analysis, even in the presence of MG132 (Fig. 4B and data not shown). To circumvent this issue, we created a recombinant HCMV virus, ADinGFP-UL21a, which accumulated a functional, GFP-S tagged variant of pUL21a at much higher levels than the native protein during infection by an unknown mechanism (see Materials and Methods and data not shown). Similar to what was observed in overexpression experiments, the tagged UL21a protein was detected predominantly in the cytoplasm by the α-pUL21a antibody during the entire course of ADinGFP-UL21a infection (Fig. 4B). In a few infected and transfected cells, some pUL21a-specific staining was also observed in the nucleus (data not shown). pUL21a is only 123 aa long with an apparent molecular mass of ∼15 kDa, a size that would allow it to pass through nuclear pores freely. The low level of pUL21a staining in the nucleus of these cells might represent the background level of passive diffusion of the protein through the nuclear pores. Nonetheless, even in these cells, the distribution of pUL21a in the cytoplasm was much more abundant than that in the nucleus. In both overexpression and infection experiments, pUL21a distribution appeared to be diffuse with no apparent colocalization with cytoplasmic organelles.
FIG. 4.
pUL21a predominantly localizes to the cytoplasm of both overexpressing and infected cells. (A) HeLa cells (left panel) or HFFs (right panel) were transfected with plasmid expressing pUL21a or control viral proteins (pp28 or pUL44), seeded onto coverslips at 48 h posttransfection, and analyzed for the subcellular localization of pUL21a 24 h later by immunofluorescence with the α-pUL21a antibody. Cells were also counterstained with TO-PRO3 to visualize the nuclei of the cells. Scale bars (10 μm) are shown. (B) HFFs were infected with ADsubUL21a, ADwt, or ADinGFP-UL21a at an MOI of 0.1 and then analyzed by immunofluorescence with the α-pUL21a antibody at indicated times postinfection.
UL21a facilitates HCMV replication in fibroblasts.
Previously, two genome-wide mutagenic analyses tentatively classified overlapping UL21a/UL21 as augmenting genes because mutations in this locus resulted in attenuated growth of HCMV in fibroblasts (14, 43). Since the transcript and the protein product arising from UL21a have now been identified, we created mutant viruses where UL21a, UL21, and its neighboring genes were individually deleted, and determined whether the UL21a gene was directly required for growth of the virus.
We constructed three deletion mutant BAC-HCMV clones (pADsubUL20, pADsubUL21a, and pADsubUL21.5-UL23) by using a linear recombination-based BAC mutagenesis approach (Fig. 5A) (see Materials and Methods). Each of these clones was derived from the parental BAC clone (pAD-GFP) that carries the genome of the HCMV AD169 strain, expresses an SV40 early promoter-driven GFP gene, and reconstitutes a wild-type control virus ADwt (31, 44). The mutant BAC clones were used to generate mutant viruses, namely, ADsubUL20, ADsubUL21a, and ADsubUL21.5-UL23, respectively, upon transfection into human fibroblasts. In pADsubUL21a or pADsubUL20, the entire coding sequence of UL21a or UL20 was replaced by a GalK/kanamycin marker cassette. In pADsubUL21.5-UL23, the viral genomic sequence encoding the C-terminal half of the UL21.5 ORF (i.e., aa 64 to 103), the miRNA UL22A-1, and the C-terminal half of the UL23 ORF (i.e., aa 121 to 284) was replaced by the marker cassette. In addition, the UL21a gene in ADsubUL21a was subsequently repaired, producing the marker-rescued BAC clone, pADrevUL21a, that was used to generate the marker-rescued virus, ADrevUL21a (Fig. 5A). All recombinant BAC clones were examined for integrity and desired alterations by using EcoRI endonuclease digestion, Southern blotting, PCR, and direct sequencing analysis (data not shown). All other recombinant BAC clones used in the present study were examined with the same level of analysis and were shown to carry the intact viral genome and contain the precise modification at the correct locus.
We then tested the roles of neighboring genes UL20, UL21a/UL21, and UL21.5-UL23, in HCMV growth in HFFs. The first indication of the importance of UL21a in HCMV growth was the observation that the UL21a deletion virus ADsubUL21a generated much smaller plaques and produced progeny virus with titers ∼30-fold lower than those with the wild-type virus (data not shown). On average, the size of plaque produced by ADsubUL21a was no more than 40% of those produced by ADwt at 12 days postinfection (dpi) (Fig. 5B). A multistep growth analysis demonstrated that two independent isolates of UL21a deletion virus were attenuated in growth, producing ∼30-fold less infectious virus than wild-type virus by 16 dpi (Fig. 5C). The marker-rescued virus ADrevUL21a replicated indistinguishably from wild-type virus. Furthermore, ADdlUL21a, a mutant virus in which UL21a was deleted without the insertion of GalK/kanamycin marker sequence, replicated indistinguishably from ADsubUL21a virus (data not shown). In contrast, mutant viruses lacking neighboring genes UL20 (ADsubUL20) or UL21.5-UL23 (ADsubUL21.5-UL23) replicated at wild-type levels, a finding consistent with previous reports (Fig. 5C) (14, 38, 43). These results indicated that UL21a deletion virus is markedly attenuated in growth. This defect is the direct result of loss of UL21a rather than aberrant expression of neighboring genes resulting from the deletion of UL21a or inadvertent second-site mutations generated during mutagenesis.
Under single-step growth condition, the UL21a deletion virus was also attenuated, producing 25-fold less cell-free infectious virus than the wild-type virus at 4 dpi (Fig. 5C). This attenuation was not the result of a defect in the release of infectious virus from host cells because the accumulation of cell-associated infectious virus was similarly reduced in UL21a deletion virus (Fig. 5C).
Intriguingly, we found that the growth defect of UL21a deletion virus was even more pronounced in MRC-5 human embryonic lung fibroblasts (Fig. 5D). As anticipated, ADsubUL20 and ADsubUL21.5-UL23 replicated indistinguishably from ADrevUL21a in MRC-5 fibroblasts. In contrast, at an MOI of 0.001, ADsubUL21a grew at a much reduced rate with delayed kinetics. It produced >2,500-fold less infectious virus than marker-rescued virus ADrevUL21a at 15 dpi, the time when ADrevUL21a reached its peak yield. The growth of ADsubUL21a did not reach its peak until 20 dpi, producing a titer that was still >60-fold lower than the peak yield of ADrevUL21a (Fig. 5D). Thus, the growth of the UL21a deletion virus appeared to be differentially attenuated in HFFs and MRC-5 fibroblasts, which to our knowledge has not previously been reported for any other HCMV mutant viruses.
Since UL21a overlaps with the originally annotated putative gene UL21 (8), it was formally possible that the growth defect of pUL21a deletion virus was the result of the inadvertent deletion of UL21. However, UL21 might not represent a real viral gene because the putative ORF that it encoded was not conserved among primate CMVs and because our analysis found no evidence for the presence of UL21-specific transcripts (Fig. 1). In order to exclude any possible involvement of UL21 in the phenotype of UL21a deletion virus, we took advantage of the recombinant HCMV virus, ADinGFP-UL21a, in which the insertion of GFP at the N terminus of the UL21a ORF fortuitously disrupted the putative UL21 ORF in a manner similar to ADsubUL21a (data not shown) (Fig. 1A). Importantly, ADinGFP-UL21a replicated at wild-type levels despite the expected disruption of UL21 in this virus (data not shown). Collectively, our data provide experimental evidence that the loss of UL21a, not UL21, is directly responsible for the growth defect of ADsubUL21a.
UL21 deletion virus produces progeny with reduced infectivity.
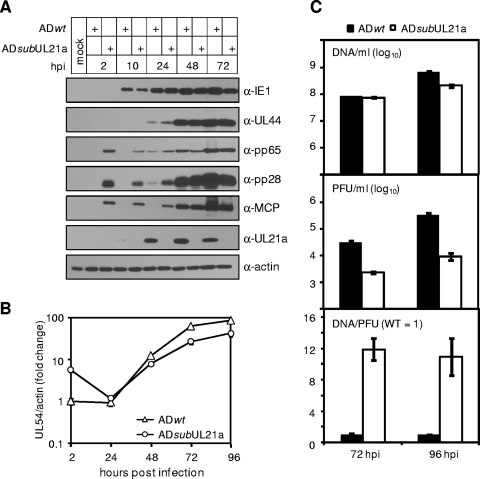
To determine the step of the viral replication cycle that is compromised in the absence of UL21a, we first analyzed viral gene expression and DNA replication in the absence of UL21a. At an MOI of 1, UL21a deletion virus produced representative viral proteins at near wild-type levels (Fig. 6A). The mutant virus had a small decrease (two- to threefold) in viral DNA replication at late times postinfection (Fig. 6B), correlating to a similar decrease in the release of DNase-resistant virion DNA into the supernatants (Fig. 6C). Interestingly, cells infected with the UL21a deletion virus contained significantly more viral DNA (Fig. 6B) and virion proteins (pp65, pp28, and MCP) (Fig. 6A) at 2 hpi, a time prior to viral gene expression. Consistently, ∼10 times more DNA-containing virions were needed for the mutant virus to achieve the same infectivity as the wild-type virus (Fig. 6C). Together, these results suggest that UL21a may have multiple roles; one is to enhance viral DNA replication, and another is to promote the ability of the virus to establish productive infection.
FIG. 6.
Analysis of the infection cycle of the UL21a deletion virus. (A) Analysis of viral gene expression. HFFs were infected with ADwt or ADsubUL21a at an MOI of 1, total cell lysates were collected at indicated times and were analyzed by immunoblotting with antibodies to IE1, pUL44, pp65, pp28, MCP, and pUL21a. (Note the difference in tegument and capsid proteins at 2 hpi.) (B) Analysis of viral DNA replication. HFFs were infected as described above, the total cell-associated DNA was isolated at indicated times, and the accumulation of viral genomes was determined by real-time quantitative PCR. (Note the difference in viral DNA at 2 hpi.) (C) Analysis of the infectivity of progeny virions. HFFs were infected as described as described above, cell-free virus was collected at 72 and 96 hpi, and the same volumes of viral samples were measured for their viral genome copies by real-time quantitative PCR (top panel) and for their infectivity by plaque assay (middle panel). The viral genome copy normalized by PFU was presented in the bottom panel. The relative genome copy per PFU of wild-type virus was set to 1.
pUL21a is not detected in HCMV virions.
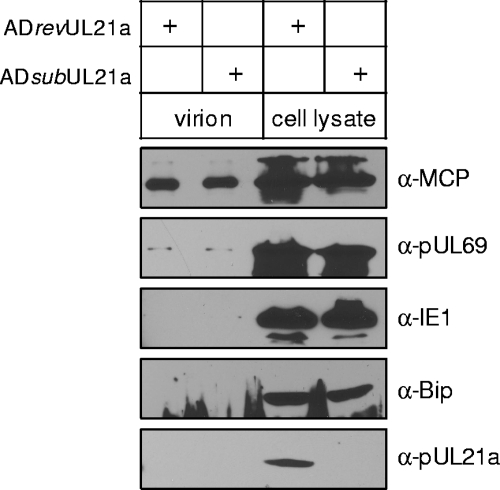
Since UL21a enhanced the infectivity of HCMV virions, it was possible that pUL21a is a tegument or structural protein. However, a proteomics study failed to detect pUL21a in the HCMV virions (37). To confirm this result, supernatants were harvested from ADrevUL21a- or ADsubUL21a-infected cells at 96 hpi, and the virions were partially purified by ultracentrifugation through a sorbitol cushion. These virion samples appeared to be largely free of contamination of cellular debris since they did not contain detectable amounts of the viral protein IE1 or the cellular protein Bip (Fig. 7). On the other hand, the MCP was readily detected and, importantly, pUL69, a protein that is present in the tegument and dense bodies in very low amounts (37, 41), could also be detected in the virion samples of ADrevUL21a (Fig. 7). Under these conditions, however, we could not detect pUL21a in the virion samples. Thus, pUL12a is unlikely to be a component of the HCMV virion.
FIG. 7.
pUL21a is not detected in HCMV virions. 5X106 HFFs were infected with ADrevUL21a or ADsubUL21a at an MOI of 0.2, and total cell lysates and cell-free virions were collected at 96 hpi. Cell-free virions were then partially purified by ultracentrifugation through a sorbitol cushion and quantified for their genome copies by real-time quantitative PCR. Cell lysates from 3.75 × 105 cells and virions from 6 × 109 genome copies were analyzed by immunoblotting for MCP, pUL69, IE1, and Bip. For pUL21a, twice as many virions (1.2 × 1010 genome copies) were used in the experiment.
DISCUSSION
In this study we report the identification and characterization of the products of HCMV gene UL21a. A single transcript of ∼600 bp was transcribed from this gene locus with early gene expression kinetics (Fig. 1B). The transcript initiated exactly 23 bp upstream and terminated within 30 to 75 bp downstream of the predicted UL21a ORF (Fig. 1A). Presumably, a poly(A) tail of 100 to 150 bp was added to the transcript and allowed its final size to reach ∼600 bp. Supporting our mapping analysis, a putative TATA box was found 55 bp upstream and a consensus poly(A) site immediately followed the stop codon of the UL21a ORF. The UL21a-specific polyclonal antibody allowed the detection of a protein, pUL21a, with an apparent molecular mass of ∼15 kDa, which was only present in infection with wild-type virus but lost in infection with UL21a deletion virus (Fig. 2A). pUL21a was detected as early as 10 hpi and reached its peak accumulation by 24 to 48 hpi (Fig. 2A), and therefore its expression was consistent with early gene kinetics of UL21a (Fig. 1B). Thus, our study provides the first experimental evidence for the presence of viral gene UL21a and its product pUL21a. pUL21a was likely the sole product encoded by the UL21a locus because our sequence analysis found no evidence for the presence of miRNAs, other noncoding RNAs, or small proteins predicted to be encoded by the UL21a transcript (data not shown). In addition, extensive third-base wobble was found in the conserved regions of the UL21a protein homologues in CCMV and rhesus CMV (data not shown), suggesting that it was the UL21a protein sequence that was maintained by evolutionary selection pressure.
UL21a was recently added to the annotation of the HCMV genome only based on the presence of its homologue but not the originally annotated UL21 in chimpanzee CMV (12). UL21 overlaps with UL21a, and encodes the largest putative ORF (i.e., 175 aa) in this region, an essential criterion used in the original annotation (8). However, both UL21a and UL21 are conserved among all sequenced isolates of HCMV (29) and have equal likelihoods to encode proteins, as predicted by a Bio-Dictionary-based Gene Finder algorithm (28). In the present study, we mapped the UL21a-specific transcript and identified the protein pUL21a but found no evidence for the presence of a UL21 transcript (Fig. 1). Furthermore, a GFP insertion that disrupted the putative UL21 ORF but preserved the UL21a ORF replicated as efficiently as wild-type virus (data not shown), ruling out any potential role of UL21, if it exists, in HCMV growth in fibroblasts. These results lead us to favor the view that UL21a, and not UL21, is expressed and required for efficient HCMV growth in fibroblasts.
UL21a was important for HCMV to establish efficient infection because the deletion virus was attenuated in growth, particularly in MRC-5 fibroblasts compared to that in HFFs (Fig. 5). The basis for such an elevated defect in UL21a deletion virus growth in MRC-5 cells is unclear, but it is likely due to physiological differences between these two cell types. For instance, MRC-5 are embryonic lung fibroblasts that proliferate more rapidly than HFFs and are able to grow up to >30 passages, whereas HFFs are newborn foreskin fibroblasts that generally undergo senescence much sooner. In addition, an unknown difference between the two cell types, such as differential expression of a specific gene or a set of genes, may also be targeted by UL21a and contribute to, at least in part, the difference that we observed. Nonetheless, analysis of the life cycle of the UL21a deletion virus in HFFs under a high MOI condition reveals a minor defect in viral DNA replication at later times (two- to threefold) and a more pronounced reduction in infectivity of progeny produced (∼10-fold) (Fig. 6). Intriguingly, pUL21 does not appeared to be a tegument or structural protein (Fig. 7) (37), raising the question of how pUL21a enhances the infectivity of the virion. It is conceivable that pUL21a may help proper tegumentation of virion particles. Alternatively, pUL21a may promote efficient viral gene expression or DNA replication, a function that can be compensated for by the addition of excess virions. We are currently searching for viral or cellular factors that pUL21a may interact with and investigating the mechanistic basis of its function.
pUL21a underwent proteasome-dependent degradation during infection (Fig. 2) despite its important role in HCMV growth (Fig. 5). pUL21a degradation was blocked by the treatment of the proteasome inhibitor MG132 and was independent of any other viral factors. The absence of any lysines or cysteines in the pUL21a sequence and the failure to detect pUL21a-ubiquitin conjugates (data not shown) led us to hypothesize that the degradation of pUL21a might be ubiquitin independent, as shown for the parvovirus minute virus NS2 protein and the HCV F protein (24, 45). Our hypothesis was supported by the fact that pUL21a was efficiently degraded in cells that lacked a functional ubiquitin-conjugation system (Fig. 3). Interestingly, HCMV contains a virus-encoded deubiquitinating protease within its virion, which may favor ubiquitin-independent protein degradation in infected cells (39). Notably, the HCMV protein pp71 directs cellular proteins Rb and hDaxx for ubiquitin-independent, proteasome-dependent degradation shortly after infection (19, 20). How then would pUL21a be targeted to the proteasome? pUL21a is rich in prolines (21/123 [17%]) that are dispersed but are more predominant at the C terminus (data not shown), which is reminiscent of a relatively unstructured protein. We speculate that pUL21a may be recognized as a misfolded protein in the cytoplasm and targeted to the proteasome for degradation. In accordance with this hypothesis, an N-terminal GFP tag, which would presumably add significant structure to the protein, dramatically stabilized levels of pUL21a (data not shown).
We detected a second, faster-migrating form of the UL21a-specific protein during pUL21a overexpression in the presence of MG132 or during HCMV infection where pUL21a was stabilized by the N-terminal GFP tag (Fig. 2 and data not shown). It is possible that one of the forms of pUL21a results from posttranslational modification, such as phosphorylation, since the protein contains multiple serine/threonine residues at its N terminus. In addition, the faster-migrating form may be derived from alternative translation as the UL21a ORF contains multiple methionines. Finally, the faster-migrating form may represent an intermediate product of proteasome degradation that is stabilized by MG132.
What would be the benefit for the virus to target pUL21a to the proteasome? Perhaps the binding of pUL21a to the proteasome is important to regulate general protein degradation during HCMV infection as proposed for the HCV F protein (45), or perhaps pUL21a interacts with certain antiviral factors and directs them for degradation. This would be reminiscent of the human immunodeficiency virus protein Vif that binds to APOBEC3G, leading to polyubiquitination and degradation of both proteins (16). Studies are currently under way to test these hypotheses.
Acknowledgments
We thank Herbert Virgin and the members of his laboratory for helpful discussions and invaluable advice, Harvey Ozer for ts20 cells, Wade Gibson and Thomas Shenk for the antibodies, and members of the Yu lab for critical reading of the manuscript.
This study was supported by a Public Health Service grant (CA-120768) from the National Cancer Institute and in part by a grant from the American Heart Association (0755804Z). D.Y. holds an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund. A.R.F. was a Morse/Berg Fellow of the Department of Molecular Microbiology, Washington University School of Medicine, and is supported by Institutional Training Grant T32-AI007172.
Footnotes
Published ahead of print on 21 October 2009.
REFERENCES
- 1.Adair, R., E. R. Douglas, J. B. Maclean, S. Y. Graham, J. D. Aitken, F. E. Jamieson, and D. J. Dargan. 2002. The products of human cytomegalovirus genes UL23, UL24, UL43, and US22 are tegument components. J. Gen. Virol. 83:1315-1324. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Saadon, R., I. Fajerman, T. Ziv, U. Hellman, A. L. Schwartz, and A. Ciechanover. 2004. The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system: direct evidence for ubiquitination at the N-terminal residue. J. Biol. Chem. 279:41414-41421. [DOI] [PubMed] [Google Scholar]
- 3.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 5.Britt, W. J., and C. A. Alford (ed.). 1996. Cytomegalovirus, 3rd ed. Lippincott-Raven, Philadelphia, PA.
- 6.Cadwell, K., and L. Coscoy. 2005. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science 309:127-130. [DOI] [PubMed] [Google Scholar]
- 7.Chee, M., S. A. Rudolph, B. Plachter, B. Barrell, and G. Jahn. 1989. Identification of the major capsid protein gene of human cytomegalovirus. J. Virol. 63:1345-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 9.Cheeseman, I. M., and A. Desai. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE 2005:pl1. [DOI] [PubMed] [Google Scholar]
- 10.Chowdary, D. R., J. J. Dermody, K. K. Jha, and H. L. Ozer. 1994. Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14:1997-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciechanover, A., and R. Ben-Saadon. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14:103-106. [DOI] [PubMed] [Google Scholar]
- 12.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 13.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn, W., P. Trang, Q. Zhong, E. Yang, C. van Belle, and F. Liu. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 7:1684-1695. [DOI] [PubMed] [Google Scholar]
- 16.Goila-Gaur, R., and K. Strebel. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grattan, M. T., C. E. Moreno-Cabral, V. A. Starnes, P. E. Oyer, E. B. Stinson, and N. E. Shumway. 1989. Cytomegalovirus infection is associated with cardiac allograft rejection and atherosclerosis. JAMA 261:3561-3566. [PubMed] [Google Scholar]
- 18.Grey, F., A. Antoniewicz, E. Allen, J. Saugstad, A. McShea, J. C. Carrington, and J. Nelson. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79:12095-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang, J., and R. F. Kalejta. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334-338. [DOI] [PubMed] [Google Scholar]
- 20.Kalejta, R. F., and T. Shenk. 2003. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proc. Natl. Acad. Sci. USA 100:3263-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuvin, J. T., and C. D. Kimmelstiel. 1999. Infectious causes of atherosclerosis. Am. Heart J. 137:216-226. [DOI] [PubMed] [Google Scholar]
- 22.McCormick, A. L., C. D. Meiering, G. B. Smith, and E. S. Mocarski. 2005. Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J. Virol. 79:12205-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melnick, J. L., E. Adam, and M. E. Debakey. 1993. Cytomegalovirus and atherosclerosis. Eur. Heart J. 14(Suppl. K):30-38. [PubMed] [Google Scholar]
- 24.Miller, C. L., and D. J. Pintel. 2001. The NS2 protein generated by the parvovirus minute virus of mice is degraded by the proteasome in a manner independent of ubiquitin chain elongation or activation. Virology 285:346-355. [DOI] [PubMed] [Google Scholar]
- 25.Mocarski, E. S., and C. T. Courcelle (ed.). 2001. Cytomegaloviruses and their replication, 4th ed., vol. 2. Lippincott-Raven, Philadelphia, PA.
- 26.Muhlestein, J. B., B. D. Horne, J. F. Carlquist, T. E. Madsen, T. L. Bair, R. R. Pearson, and J. L. Anderson. 2000. Cytomegalovirus seropositivity and C-reactive protein have independent and combined predictive value for mortality in patients with angiographically demonstrated coronary artery disease. Circulation 102:1917-1923. [DOI] [PubMed] [Google Scholar]
- 27.Mullberg, J., M. L. Hsu, C. T. Rauch, M. J. Gerhart, A. Kaykas, and D. Cosman. 1999. The R27080 glycoprotein is abundantly secreted from human cytomegalovirus-infected fibroblasts. J. Gen. Virol. 80(Pt. 2):437-440. [DOI] [PubMed] [Google Scholar]
- 28.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. USA 100:13585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2006. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 80:3872-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian, Z., B. Xuan, T. T. Hong, and D. Yu. 2008. The full-length protein encoded by human cytomegalovirus gene UL117 is required for the proper maturation of viral replication compartments. J. Virol. 82:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawlinson, W. D., and B. G. Barrell. 1993. Spliced transcripts of human cytomegalovirus. J. Virol. 67:5502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivailler, P., A. Kaur, R. P. Johnson, and F. Wang. 2006. Genomic sequence of rhesus cytomegalovirus 180.92: insights into the coding potential of rhesus cytomegalovirus. J. Virol. 80:4179-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 35.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:2798S-2804S. [DOI] [PubMed] [Google Scholar]
- 36.Terhune, S., E. Torigoi, N. Moorman, M. Silva, Z. Qian, T. Shenk, and D. Yu. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81:3109-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, D., W. Bresnahan, and T. Shenk. 2004. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc. Natl. Acad. Sci. USA 101:16642-16647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, X., R. A. Herr, W. J. Chua, L. Lybarger, E. J. Wiertz, and T. H. Hansen. 2007. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J. Cell Biol. 177:613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winkler, M., and T. Stamminger. 1996. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J. Virol. 70:8984-8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xuan, B., Z. Qian, E. Torigoi, and D. Yu. 2009. Human cytomegalovirus protein pUL38 induces ATF4 expression, inhibits persistent JNK phosphorylation, and suppresses endoplasmic reticulum stress-induced cell death. J. Virol. 83:3463-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuksek, K., W. L. Chen, D. Chien, and J. H. Ou. 2009. Ubiquitin-independent degradation of hepatitis C virus F protein. J. Virol. 83:612-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, Y. F., M. B. Leon, M. A. Waclawiw, J. J. Popma, Z. X. Yu, T. Finkel, and S. E. Epstein. 1996. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N. Engl. J. Med. 335:624-630. [DOI] [PubMed] [Google Scholar]