Abstract
Scavenger receptor class B type I (SR-BI) is an essential receptor for hepatitis C virus (HCV) and a cell surface high-density-lipoprotein (HDL) receptor. The mechanism of SR-BI-mediated HCV entry, however, is not clearly understood, and the specific protein determinants required for the recognition of the virus envelope are not known. HCV infection is strictly linked to lipoprotein metabolism, and HCV virions may initially interact with SR-BI through associated lipoproteins before subsequent direct interactions of the viral glycoproteins with SR-BI occur. The kinetics of inhibition of cell culture-derived HCV (HCVcc) infection with an anti-SR-BI monoclonal antibody imply that the recognition of SR-BI by HCV is an early event of the infection process. Swapping and single-substitution mutants between mouse and human SR-BI sequences showed reduced binding to the recombinant soluble E2 (sE2) envelope glycoprotein, thus suggesting that the SR-BI interaction with the HCV envelope is likely to involve species-specific protein elements. Most importantly, SR-BI mutants defective for sE2 binding, although retaining wild-type activity for receptor oligomerization and binding to the physiological ligand HDL, were impaired in their ability to fully restore HCVcc infectivity when transduced into an SR-BI-knocked-down Huh-7.5 cell line. These findings suggest a specific and direct role for the identified residues in binding HCV and mediating virus entry. Moreover, the observation that different regions of SR-BI are involved in HCV and HDL binding supports the hypothesis that new therapeutic strategies aimed at interfering with virus/SR-BI recognition are feasible.
Hepatitis C virus (HCV) is a global blood-borne pathogen, with 3% of the world's population chronically infected. Most infections are asymptomatic, yet 60 to 80% become persistent and lead to severe fibrosis and cirrhosis, hepatic failure, or hepatocellular carcinoma (3). Currently available therapies are limited to the administration of pegylated alpha interferon in combination with ribavirin, which are expensive and often unsuccessful, with significant side effects (23, 36). Thus, the development of novel therapeutic approaches against HCV remains a high priority (18, 40, 60). Targeting the early steps of HCV infection may represent one such option, and much effort is being devoted to uncovering the mechanism of viral attachment and entry.
The current view is that HCV entry into target cells occurs after attachment to specific cellular receptors via its surface glycoproteins E1 and E2 (27). The molecules to which HCV initially binds might constitute a diverse collection of cellular proteins, carbohydrates, and lipids that concentrate viruses on the cell surface and determine to a large extent which cell types, tissues, and organisms HCV can infect.
CD81, claudin 1 (CLDN1), occludin (OCLN), and scavenger receptor class B type I (SR-BI) were previously shown to play essential roles in HCV cell entry (15, 22, 26, 35, 42, 43, 50, 63, 64).
Recent reports suggest that CD81 engagement triggers intracellular signaling responses, ultimately leading to actin remodeling and the relocalization of CD81 to tight junctions (TJ) (11). Thus, CD81 may function as a bridge between the initial interaction of the virus with receptors on the basolateral surface of the hepatocyte and the TJ where two of the HCV entry molecules, CLDN1 and OCLN, are located. CD81 acts as a postbinding factor, and the TJ proteins CLDN1 and OCLN seem to be involved in late steps of HCV entry, such as HCV glycoprotein-dependent cell fusion (9, 11, 22). The discovery of TJ proteins as entry factors has added complexity to the model of HCV entry, suggesting parallels with other viruses like coxsackievirus B infection, where an initial interaction of the viral particle with the primary receptor decay-accelerating factor induces the lateral movement of the virus from the luminal surface to TJ, where coxsackievirus B binds coxsackievirus-adenovirus receptor and internalization takes place (17).
Much less is known about the specific role of SR-BI in virus entry: neither the specific step of the entry pathway that SR-BI is involved in nor the protein determinants that mediate such processes are known. SR-BI is a lipoprotein receptor of 509 amino acids (aa) with cytoplasmic C- and N-terminal domains separated by a large extracellular domain (1, 13, 14). It is expressed primarily in liver and steroidogenic tissues, where it mediates selective cholesteryl ester uptake from high-density lipoprotein (HDL) and may act as an endocytic receptor (45, 46, 51, 52). SR-BI was originally identified as being a putative receptor for HCV because it binds soluble E2 (sE2) through interactions with E2 hypervariable region 1 (HVR1) (8, 50). RNA interference studies as well as the ability to block both HCV pseudoparticles (HCVpp) and cell culture-derived HCV (HCVcc) infections with anti SR-BI antibodies have confirmed its involvement in the HCV entry process (7, 8, 15, 26, 33, 63). Intriguingly, lipoproteins were previously shown to modulate HCV infection through SR-BI (12). It was indeed previously demonstrated that two natural ligands of SR-BI, HDL and oxidized low-density lipoprotein, can improve and inhibit HCV entry, respectively (57, 59). Moreover, small-molecule inhibitors of SR-BI-mediated lipid transfer (block of lipid transfer BLT-3 and BLT-4) abrogate the stimulation of HCV infectivity by human serum or HDL, suggesting that the enhancement of viral infection might be dependent on the lipid exchange activity of SR-BI (20, 58).
We previously generated high-affinity monoclonal antibodies (MAbs) specific for human SR-BI and showed that they were capable of inhibiting the binding of SR-BI to sE2 and blocking HCVcc infection of human hepatoma cells (15). The HDL-induced enhancement of infection had no impact on the ability of the anti-SR-BI MAbs to block HCV infection, and the antibodies were effective in counteracting HCV infection even in the absence of lipoproteins. These data demonstrated that SR-BI participates in the HCV infection process as an entry receptor by directly interacting with viral glycoproteins. Here we have used one of the anti-SR-BI MAbs to show that SR-BI participates in an early step of HCV infection. By assays of binding of sE2 to SR-BI molecules from different species and to SR-BI mutants, we identified species-specific SR-BI protein residues that are required for sE2 binding. The functional significance of these observations was confirmed by the finding that SR-BI mutants with reduced binding to sE2 were also impaired in their ability to restore the infectivity of an SR-BI-knocked-down Huh-7.5 cell line. Finally, we demonstrated that SR-BI mutants with impaired sE2 binding can still form oligomeric structures and that they can bind the physiological ligand HDL and mediate cholesterol efflux, suggesting that distinct protein determinants are responsible for the interaction with HDL and the HCV particle.
MATERIALS AND METHODS
Cell lines and antibodies.
Human hepatoma Huh-7.5, HEK-293, HEK-293T, and mouse neuronal N2a (ATCC) cells were grown in Dulbecco's modified essential medium (Invitrogen) supplemented with 10% fetal bovine serum.
Anti-SR-BI MAbs were generated and produced as previously described (15). Anti-SR-BI (ab396) polyclonal antibody was purchased from Abcam. Monoclonal anti-c-Myc or anti-V5 antibodies were obtained from Invitrogen. Anti-CD81 MAb was purchased from Santa Cruz Biotechnology (clone 1.3.3.22). Negative control mouse immunoglobulin G1 (IgG1) was obtained from Serotec, and matched isotype control human IgG was purchased from the Binding Site.
HCVcc production and synchronized infection.
In vitro-transcribed genomic J6/JFH RNA was delivered to Huh-7.5 cells by electroporation (34). Infectious HCV particles (HCVcc) were recovered 72 h postelectroporation, and the virus-containing supernatant was clarified by centrifugation and stored at −80°C. For synchronized infections, 105 cells were seeded in poly-l-lysine-coated 24-well tissue culture plates 24 h prior to infection. Medium was changed to 4°C with or without antibody 1 h prior to infection, and cultures were placed at 4°C. Medium was then replaced with 250 μl HCVcc-containing supernatant with or without antibody (anti-CD81 was used at 2 μg/ml, and anti-SR-BI was used at 10 μg/ml) for 1.5 h, and cultures were again placed at 4°C. Cultures were washed twice with cold phosphate-buffered saline; fresh 37°C medium, with or without antibodies, was then added, and cultures were placed in 37°C tissue culture incubators. At various time points, antibodies were added to the medium. At 120 min after the shift to 37°C, the medium was changed to fresh medium without antibodies. RNA was extracted from cellular lysates 3 days postinfection, and the number of intracellular HCV genomes was determined by reverse transcription (RT)-quantitative PCR (qPCR). Briefly, we performed a 384-well plate-based assay and used 20 ng total RNA per well. To quantify HCV in RNA samples, the following oligonucleotides and probe were used: HCV sense primer GCGAAAGGCCTTGTGGTACT, HCV antisense primer CACGGTCTACGAGACCTCCC, and probe CCTGATAGGGTGCTTGCGAGTGCC (5′ 6-carboxyfluorescein-3′ 6-carboxytetramethylrhodamine. HCV copies were normalized to rRNA amounts by using a commercially available 18S rRNA mix (Applied Biosystems).
Cloning of SR-BI coding sequences.
Cloning strategies for human and mouse SR-BI were previously described (50). Tupaia SR-BI cDNA was obtained by RT-PCR (Superscript II kit; Invitrogen) of purified primary tupaia hepatocyte RNA (RNeasy; Qiagen) with human SR-BI-specific primers (forward primer 5′-ATG GGG CCC CAG GCG CGCAGA CAT GGG C-3′ and reverse primer 5′-AGC GGG GTG TAG GGG CTG GGGGGC CGG-3′). The PCR products from two independent reactions were cloned (pcRZero Blunt kit; Invitrogen) and sequenced. Full-length tupaia SR-BI cDNA was cloned into expression vector pcDNA3 (Invitrogen) (4). Tamarin SR-BI cDNA was obtained by RT-PCR (Superscript II; Invitrogen) of purified RNA from tamarin liver with SR-BI-specific primers taSenseATG (5′-GAC ATG GGC TGC TCC TCC AGA GCG CGC-3′) and tamAS230 (5′-TAG AAG GGG ATG GGG ATC TCC CG-3′). Full-length tamarin SR-BI cDNA was cloned into the vector pCR-Blunt II (Invitrogen), sequenced, and then cloned into the expression vector pcDNA3 by HindIII-XbaI digestion.
Production of sE2 glycoproteins.
Expression plasmids encoding HCV E2 (aa 384 to 661) protein genotype 1a (strain H77), 1b (strain BK), or 6 (isolate 6.540) and containing a His6 tag were transfected into HEK-293T cells by using a calcium phosphate transfection kit (Clontech). At 48 h posttransfection, cell supernatants were clarified by centrifugation at 4,000 rpm for 30 min at 4°C and then concentrated 100-fold by using Centricon Plus 80 filters (Amicon) supplemented with 5% glycerol, 25 mM HEPES, and 1× protease inhibitors (Boehringer) and stored in aliquots at −80°C.
Mutagenesis of human SR-BI.
Twelve synthetic genes coding for SR-BI mutant receptors were generated by Geneart GmbH from synthetic oligonucleotides. The fragments were cloned into pcDNA3-human SR-BI using HindIII and SfiI restriction sites, and the final constructs were verified by sequencing. Wild-type SR-BI and E210A mutant receptors fused to either c-Myc or V5 tags were also generated by Geneart GmbH from synthetic oligonucleotides. The fragments were cloned into the pcDNA3 vector by using HindIII and XmaI restriction sites, and the final constructs were verified by sequencing.
Measurement of E2 and MAb binding to SR-BI and flow cytometric analysis.
A total of 4 × 105 N2a cells were transfected with wild-type or mutant receptors by using Lipofectamine 2000 (Invitrogen). At 48 h posttransfection, cells were lysed in a solution containing 1% Triton X-100, 50 mM Tris-HCl (pH 8), and 150 mM NaCl in the presence of protease inhibitor cocktail (Roche). Cell lysates were analyzed by immunoblotting with an SR-BI rabbit antiserum (ab396; Abcam), probed with peroxidase-labeled anti-rabbit IgG (Pierce), and visualized by using an enhanced chemiluminescence system to confirm the expression of each mutant receptor. N2a cells were assayed 48 h posttransfection, and sE2 binding (genotype 1a, strain H77; genotype 1b, strain BK; or genotype 6, isolate 6.540) to SR-BI mutant receptors was measured by flow cytometry as previously described (47). A saturating amount of the soluble recombinant E2 glycoproteins was added to N2a cells transfected with the different SR-BI molecules for 1 h at room temperature, and binding was quantified by flow cytometry after incubation with anti-His mouse MAb (Qiagen) and anti-mouse IgG1-phycoerythrin (PE) (Serotec). As a control, an isotype-matched IgG was preincubated on cells before sE2 binding. Background was defined as the E2 binding to N2a cells transfected with empty vector. Mean fluorescence intensity (MFI) values subtracted of background MFI were expressed as a percentage of wild-type receptor binding after normalization for the expression level determined by Western blotting (WB).
Transfected N2a cells were also assayed for binding to MAb C11 and MAb C167 by flow cytometry. Forty-eight hours after transfection, MAbs (0.5 μg/ml) were added to cells for 45 min at room temperature in fluorescence-activated cell sorter (FACS) buffer. C11 and C167 binding was revealed by incubating cells with biotinylated anti-human IgG4 mouse MAb (BD Pharmingen) followed by streptavidin-PE (Serotec). As a control, isotype-matched human IgG4 was used, and the resulting MFI values were subtracted of MFI values obtained with anti-SR-BI MAbs. FACS acquisitions and analysis were performed by use of a FACSCalibur system (Becton Dickinson) and CellQuest software.
Immunoprecipitation of SR-BI oligomers.
HEK-293 cells were transiently transfected with expression plasmids encoding wild-type SR-BI fused to either c-Myc or V5 tags and a mutated (E210A) SR-BI construct fused to the c-Myc tag. Forty-eight hours posttransfection, 5 × 106 cells were lysed in 1 ml of immunoprecipitation buffer (20 mM HEPES [pH 7.5], 1 mM EGTA [pH 8.0], 1 mM EDTA [pH 8.0], 150 mM NaCl, 1% Triton X-100, protease inhibitor cocktail). Lysates were incubated overnight with monoclonal anti-c-Myc or anti-V5 antibodies coupled to protein G-Sepharose 4 Fast Flow columns. After several washings, bound proteins were eluted and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein blots were incubated with antibody to the alternative tag used for immunoprecipitation, probed with peroxidase-labeled anti-mouse IgG (Promega), and visualized by using an enhanced chemiluminescence system.
Generation of an SR-BI-negative Huh-7.5 cell line.
To generate the construct pLVTHM/shSR-BI, a short hairpin RNA (shRNA) duplex sequence (5′-CGCGTCCCCAGAGGCTCGTCAACAAGCATTCAAGAGATGCTTGTTGACGAGCCTCTTTTTTGGAAAT-3′) targeting the 3′ untranslated region of SR-BI mRNA was cloned into vector pLVTHM downstream of the H1 promoter. Lentiviral vectors pLVTHM, pLV-tTRKRAB-Red, pCMV-DR8.74, and pMD2G-VSVG were provided by D. Trono (Addgene) (61). For the production of the lentiviral particles, HEK-293T cells were cotransfected by calcium phosphate precipitation with pLVTHM/shSR-BI or pLV-tTRKRAB-Red, pCMV-DR8.74, and pMD2G-VSVG. Concentrated lentiviral stocks were obtained by ultracentrifugation through a 20% sucrose cushion. Huh-7.5 cells were cotransduced with two different lentiviruses carrying the transgenes pLVTHM/shSR-BI and pLV/tTRKRAB with a multiplicity of infection (MOI) of 50. Dually transduced Huh-7.5 cells were sorted by FACS (green fluorescent protein positive/dsRED positive) after 4 days of induction with 100 ng/ml doxycycline (Sigma) to generate Huh-7.5/shSR-BIkd cells.
Complementation of an SR-BI-negative Huh-7.5 cell line with wild-type and mutant SR-BI receptors and HCVcc infection.
cDNAs coding for human SR-BI, mouse SR-BI, the SW0 or E210A mutant, or empty vector were cloned into pTRIP lentiviral vectors and used to transduce Huh-7.5/shSR-BIkd cells. Cells were maintained in medium containing 100 ng/ml doxycycline for 96 h before transduction with SR-BI-expressing lentivirus. Four days after transduction, HCVcc was added to cells (about 0.1 MOI) for 6 h. Virus-containing medium was then removed and replaced with fresh medium containing doxycycline. The infection was allowed to occur for 32 h, RNA was then extracted, and the number of HCV copies in each well was evaluated by RT-qPCR.
HDL binding and cholesterol efflux assays.
HDL lipoproteins were isolated from plasma of fasting, healthy, normolipidemic individuals by sequential ultracentrifugal flotation. Purified HDL were biotinylated by using the FluoReporter MiniBiotin-XX labeling kit (Invitrogen). CHO-K1 (ATCC) cells were transfected by Nucleofector technology using the Cell Line Nucleofector Kit T (Amaxa) and plasmids encoding either wild-type human or mouse SR-BI receptors, the SW0 or E210A mutant receptors, or the empty pcDNA3 vector. Transfected cells were analyzed for the level of SR-BI expression at the plasma membrane, their capacity to bind HDL particles by flow cytometry, and cholesterol efflux efficiency in the presence of HDL. HDL binding to SR-BI was evaluated 24 h posttransfection by incubating 5 × 105 transfected CHO cells in suspension with biotinylated HDL (20 μg protein/ml) for 30 min at 4°C in phosphate-buffered saline-0.2% bovine serum albumin. The cells were then washed and fixed with 5% formalin neutral buffered solution (Sigma), and streptavidin-PE (eBiosiences) was added before FACS acquisitions on an FC 500 flow cytometer (CXP software; Beckman Coulter). The level of SR-BI expression at the plasma membrane was evaluated by incubating the transfected cells in suspension with MAb C11 (2 μg/ml) for 30 min at room temperature, followed by an incubation with a fluorescein-labeled anti-human IgG antibody (Vector Laboratories). Fluorescein isothiocyanate fluorescence was then measured by flow cytometry. Cellular cholesterol efflux was measured 48 h posttransfection as described previously (15).
Statistical analysis.
Results are presented as means ± standard errors. The statistical significance of the differences between the means of the experimental groups was tested by the Student t test for unpaired data. A difference was considered statistically significant when the P value was <0.05.
RESULTS
SR-BI is involved in an early step of HCV infection.
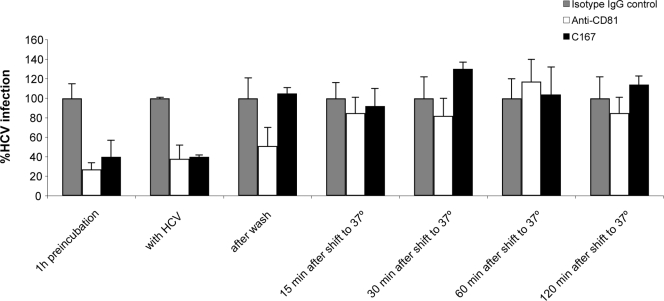
We previously showed that SR-BI is involved in sE2 binding and that antibodies against the extracellular domain of SR-BI can block sE2 interactions and virus infection (15, 50). Similar data were obtained with CD81 (8, 16, 24, 29, 41, 42). To characterize the sequence of events during HCV entry with respect to binding to these two receptors, we investigated the inhibitory capacities of antibodies to CD81 (clone 1.3.3.22; Santa Cruz Biotechnology) and to SR-BI (MAb C167) (15) when administered at different times during the early phase of infection. HCVcc (J6/JFH) was incubated for 90 min at 4°C with Huh-7.5 human hepatoma cells to allow virus attachment and synchronous infection after a shift to 37°C. Anti-receptor MAbs were added at different times to the virus-containing medium before and after the shift to 37°C, as indicated in Fig. 1. Neither antibody could block virus entry after the temperature shift. However, the anti-CD81 MAb inhibited infection with a similar potency irrespective of whether it was added before, during, or immediately after virus binding, while MAb C167 could no longer be effective if it was added after virus binding (Fig. 1). This indicates that SR-BI is involved in an early step of virus/cell recognition, while CD81 acts subsequent to initial virus binding.
FIG. 1.
SR-BI is involved in the early phase of HCV entry. HCV infection was measured 72 h postinfection by RT-qPCR. J6/JFH HCVcc (HCV) was added to Huh-7.5 cells and incubated for 90 min at 4°C. Cells were incubated with an irrelevant isotype IgG control, anti-SR-BI MAb (C167), or anti-CD81 at different times with respect to the addition of the virus, as indicated at the bottom. Values represent the averages of quadruplicates and are expressed as percentages of HCV infection relative to the cells treated with the isotype IgG control at each time point.
Identification of SR-BI regions essential for sE2 recognition.
We compared the efficiencies of binding of sE2 to SR-BI molecules from species with different susceptibilities to HCV infection, human and tupaia, which can be infected by HCV, and tamarin and mouse that are resistant to HCV infection. The transient transfection of expression plasmids encoding SR-BI molecules from these four species into mouse neuronal N2a cells led to high and comparable levels of cell surface receptor expression, which was evaluated with conformational antibodies against the extracellular domain (see Materials and Methods). The binding of sE2 from the H77 strain to the different receptors was measured by cytofluorimetric analysis of transfected cells.
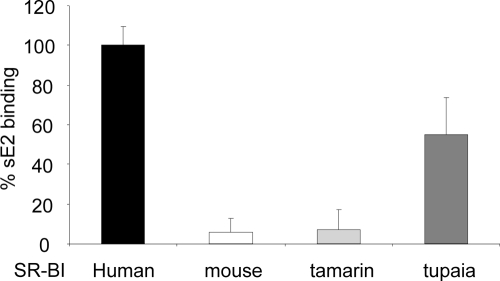
Tupaia SR-BI binds sE2 although with a reduced efficiency (about 50%) compared to that of the human ortholog (Fig. 2). Conversely, mouse SR-BI and tamarin SR-BI were unable to recognize sE2 (Fig. 2). Thus, the profile of sE2 binding to SR-BI molecules from different species correlates with their susceptibility to HCV infection.
FIG. 2.
E2 binding to SR-BI molecules from different species. N2A cells were transfected with SR-BI cDNAs cloned from livers of different species that were either permissive (human and tupaia SR-BI molecules) or nonpermissive (mouse and tamarin SR-BI molecules) to HCV infection. sE2 binding (genotype 1a, strain H77) was measured 48 h after transfection by flow cytometry. Background was defined as E2 binding to N2a cells transfected with the empty vector. MFI values, subtracted of background MFI values, were expressed as percentages of sE2 binding relative to sE2 binding measured in cells transfected with human SR-BI.
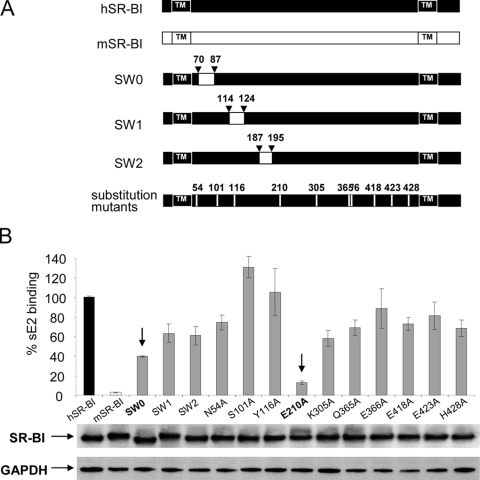
On the basis of the amino acid sequence alignment of the mouse and human SR-BI molecules, we designed a number of interspecies chimeras (Fig. 3). Swapping mutants SW0 (aa 70 to aa 87), SW1 (aa 114 to aa 124), and SW2 (aa 187 to aa 195) contained highly divergent regions of the mouse sequence in the context of the human receptor. The other mutant proteins contained a single alanine substitution of residues different in the mouse sequence but conserved in tupaia SR-BI (Fig. 3A).
FIG. 3.
E2 binding to SR-BI mutant receptors. (A) The human SR-BI (hSR-BI) and mouse SR-BI (mSR-BI) proteins are schematically depicted at the top, and the transmembrane (TM) domains are indicated. Swap mutants SW0, SW1, and SW2 were generated by replacing the human SR-BI sequence (black) with the murine counterpart (white), and the numbers corresponding to the positions of the amino acids exchanged are shown. Ten single-point mutants were also designed by alanine scanning and are indicated by lanes and numbers. (B) N2A cells were transfected with SR-BI mutant receptors, and sE2 binding (genotype 1a, strain H77) was measured by flow cytometry. Background was defined as the E2 binding to N2a cells transfected with an empty vector. MFI values, with background MFI subtracted, were expressed as percentages of wild-type human receptor binding after normalization for the protein expression level determined by WB (middle). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
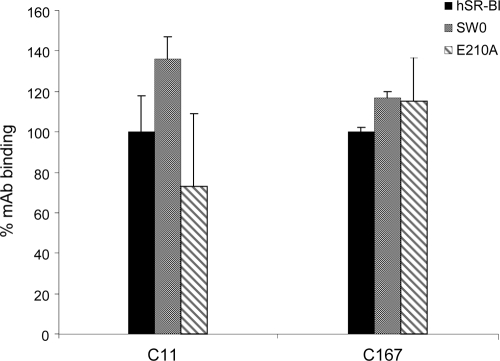
The expression plasmids encoding the SR-BI mutant receptors were transfected into the recipient mouse N2a cell line. All mutants and wild-type proteins showed comparable levels of expression (Fig. 3B, middle). N2a cells transfected with expression plasmids encoding wild-type and mutant receptors were also equally recognized by conformational MAbs C11 and C167 in a cell surface FACS assay, indicating that mutagenesis had not resulted in drastic changes in the overall display and conformation of the SR-BI molecule (Fig. 4 and data not shown). E2 binding of these mutants, expressed as a percentage of binding with respect to the wild-type human receptor, is shown in Fig. 3B (top). Swapping mutant SW0 showed a 60% reduction in E2 binding, while the SW1 and SW2 mutants were less impaired (40% reduction of ligand binding). Mutant E210A showed a 90% reduction of E2 binding, while the remaining point mutants did not significantly affect E2 recognition (Fig. 4).
FIG. 4.
Binding of MAbs C11 and C167 to mutant SR-BI receptors. C11 and C167 binding to N2a cells transfected with human wild-type SR-BI (hSR-BI), SW0, or the E210A mutant was measured 48 h after transfection by flow cytometry. Values are expressed as percentages of MAb binding relative to the binding of the MAb in cells transfected with the wild-type human receptor.
We also observed that the impaired-binding phenotype of the E210A mutant receptor was not limited to E2 from HCV genotype 1a but was also found for other genotypes (genotype 1b, strain BK, and genotype 6, isolate 6.540), thus confirming the crucial role of E210 in the recognition of the viral ligand (data not shown).
SR-BI mutants with reduced sE2 binding are impaired for HCVcc infection.
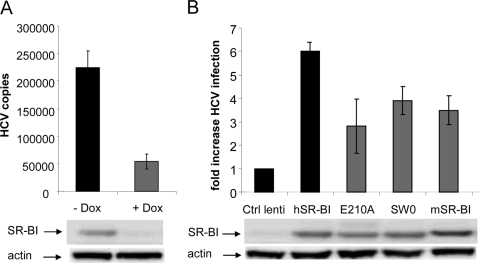
Human hepatoma Huh-7.5 cells support efficient infection and replication by HCV (10, 34). Therefore, to assess whether SR-BI regions that mediate sE2 interactions are indeed important for HCV infection, we generated an Huh-7.5 derivative cell line where endogenous SR-BI expression could be reversibly downregulated. A doxycycline-inducible SR-BI-specific shRNA sequence was cloned into a lentiviral vector and stably transduced in Huh-7.5 cells (see Materials and Methods). The resulting cell line (Huh-7.5/SR-BIkd) showed a significant reduction in the level of SR-BI expression upon the induction of the transduced shRNA with doxycycline (Fig. 5A, middle). The silencing of SR-BI resulted in a significantly lower susceptibility to HCVcc infection, to about 20% of the uninduced Huh-7.5/SR-BIkd cells (Fig. 5A, top), confirming that SR-BI is important for HCV infection of human hepatic cells.
FIG. 5.
HCVcc infection of an SR-BI-negative Huh-7.5 cell line complemented with wild-type and mutant SR-BI receptors. (A) J6/JFH HCVcc (HCV) was added to Huh-7.5/shSR-BIkd cells pretreated or not treated with doxycycline (Dox), and HCV infection was measured 32 h postinfection by RT-qPCR. Data are expressed as the numbers of HCV copies normalized to the number of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) copies per 50 ng of total RNA. The SR-BI protein expression level in Huh-7.5/shSR-BIkd cells with or without doxycycline was determined by WB and is shown (middle). (B) HCVcc infection of Huh-7.5/shSR-BIkd cells treated with doxycycline and transduced with either a control lentivirus (Ctrl lenti), human SR-BI (hSR-BI), mouse SR-BI (mSR-BI), or the E210A or SW0 mutant was measured. Results are expressed as increases in HCV infection compared to cells transduced with the control lentivirus. The relative levels of SR-BI protein expression determined by WB are shown at the bottom.
We then constructed lentiviral vectors encoding wild-type human and mouse SR-BI molecules and the two mutants SW0 and E210A. None of the SR-BI sequences cloned in the lentiviral vectors included the target of the SR-BI shRNA used to silence the endogenous gene. Huh-7.5/SR-BIkd cells were kept in culture in the presence of doxycycline 4 days prior to infecting them with the lentiviral vectors encoding the different SR-BI constructs at an MOI of 0.5. An empty lentiviral vector was used as a negative control. Four days posttransduction, cells were infected with HCVcc at an MOI of 0.1. All SR-BI lentiviral vectors expressed comparable levels of wild-type and mutant receptors, as measured by WB with whole-cell extracts of transduced cells (Fig. 5B, middle). Thirty-two hours after HCVcc infection, Huh-7.5/SR-BIkd cells transduced with the different vectors were lysed, and total RNA was extracted for HCV RNA quantification by RT-qPCR. Lentivirus-mediated transduction with wild-type human SR-BI resulted in a sixfold increase in the relative permissiveness to HCVcc infection with respect to SR-BI-knocked-down cells transduced with the control lentiviral vector (Fig. 5B, top). In contrast, murine SR-BI and derivatives of human SR-BI only partially rescued the infectivity of silenced Huh-7.5/SR-BIkd cells, with the E210A mutant showing almost a 60% reduction in HCVcc infection compared to cells complemented with wild-type human SR-BI. At present, we cannot explain why mouse SR-BI can partially rescue HCV infection upon transduction in Huh-7.5/SR-BIkd cells while showing a strong reduction in the binding of the sE2 glycoprotein. Such a discrepancy could be due to a partially different conformation between sE2 and the full-length protein on the viral particles or the presence of other structural elements of the HCV envelope like the E1 protein.
SR-BI determinants necessary for sE2 recognition are not involved in receptor oligomerization.
Since human SR-BI forms oligomers on the cell surface (48, 49), we wanted to investigate whether the impaired-sE2-binding phenotype of the E210A mutant could be due to a defect in receptor oligomerization. Therefore, we generated C terminus tagged versions of wild-type and mutant receptors, adding two different tag sequences: V5 or c-Myc.
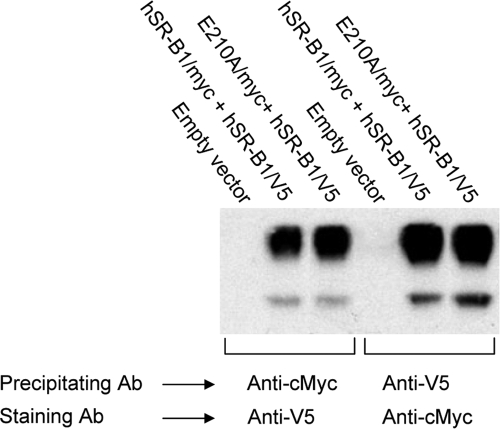
We first cotransfected HEK-293 cells with expression plasmids encoding the two tagged forms of the wild-type receptor. Cell lysates were then immunoprecipitated with the anti-c-Myc antibody and revealed by WB analysis with anti-V5 antibody or vice versa. Coimmunoprecipitation of the two tagged proteins was observed in both experiments (Fig. 6, lanes 2 and 4). We then cotransfected HEK-293 cells with expression plasmids encoding V5-tagged wild-type SR-BI and c-Myc-tagged E210A mutant SR-BI. Also, in this case, cell lysates were immunoprecipitated with the anti-c-Myc antibody and revealed by WB analysis with anti-V5 antibody or vice versa (Fig. 6, lanes 3 and 6). After immunoprecipitation with the anti-V5 antibody recognizing the wild-type protein, the coexpressed E210A mutant was quantitatively recovered. Similarly, efficient coimmunoprecipitation of the wild-type protein was observed with an antibody recognizing the c-Myc-tagged E210A mutant. These data suggested that the capacity to form oligomers is not impaired in the E210A mutant.
FIG. 6.
Heterooligomerization of wild-type SR-BI with mutant receptors. HEK-293 cells were cotransfected with constructs encoding c-Myc- and V5-tagged versions of wild-type human SR-BI (hSR-BI/myc + hSR-BI/V5) (lanes 2 and 5), c-Myc-tagged E210A and V5-tagged wild-type human SR-BI (E210A/myc + hSR-BI/V5) (lanes 3 and 6), or empty vector pcDNA3 (lanes 1 and 4). Tagged SR-BI proteins precipitated with either anti-V5 (lanes 1 to 3) or anti-c-Myc (lanes 4 to 6) antibodies were stained with anti-c-Myc antibody (Ab). Molecular mass markers are indicated on the left. Bands migrating with an apparent molecular mass of 85 kDa correspond to tagged SR-BI. The faster-migrating bands correspond to the heavy chain of the immunoprecipitating antibody revealed by peroxidase-labeled anti-mouse IgG.
Different regions of SR-BI mediate binding to sE2 and HDL.
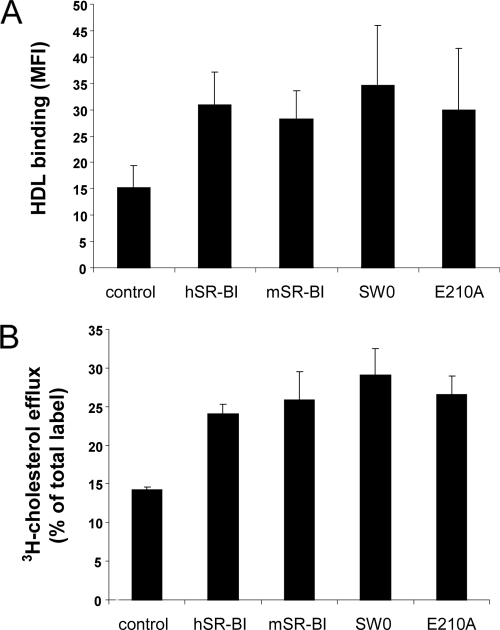
We previously showed that expressing human SR-BI in CHO cells correlated with a marked increase in HDL binding to the cell membrane compared to untransfected cells (15). To verify whether the SR-BI protein elements involved in HCV recognition are different from those responsible for binding to the physiological ligand HDL, we performed HDL binding assays with CHO cells transiently transfected with expression plasmids encoding mutant SR-BI or the empty control plasmid. Cell surface expression levels of the different SR-BI molecules were measured by FACS analysis with conformational antibody C11 (data not shown). Both SW0 and E210A mutants showed HDL binding levels similar to those of wild-type human and mouse SR-BI proteins (Fig. 7A). A number of studies have shown that when cells express SR-BI, the bidirectional flux of free cholesterol (FC) between cells and HDL particles is accelerated (30, 38, 53-56). Thus, we further assessed whether the SW0 and E210A mutants were functional in facilitating cellular FC efflux to HDL. The transient overexpression of either human or mouse SR-BI in [3H]cholesterol-labeled CHO cells led to a major increase in FC efflux to HDL. Similar results were obtained with SW0 and E210A mutants (Fig. 7B).
FIG. 7.
Evaluation of HDL binding to SR-BI mutant receptors and their capacity to mediate cellular FC efflux to HDL. CHO cells were transfected to transiently express either wild-type human SR-BI (hSR-BI) or mouse SR-BI (mSR-BI) receptors or the SW0 or E210A mutant molecules. Control cells were transfected with the empty vector pcDNA3. (A) Twenty-four hours posttransfection, the cells were incubated with biotinylated HDL (20 μg protein/ml for 30 min at 4°C). HDL binding was revealed by the addition of streptavidin-PE and analyzed by flow cytometry. The MFI measured is reported. Values ± standard deviations represent the means of data from five to six independent experiments. MFI values for CHO cells expressing either wild-type or mutant receptors were statistically significantly different from that measured with control cells (P < 0.02). (B) HDL-mediated cellular FC efflux in transfected CHO cells labeled with [3H]cholesterol was determined. Forty-eight hours posttransfection, the cells were incubated in serum-free medium containing HDL (20 μg protein/ml) for 2 h. Cholesterol efflux is expressed as the percentage of total [3H]cholesterol radioactivity released into the medium. Data represent the means (± standard deviations) of data from four independent experiments. FC efflux values determined for CHO cells expressing either wild-type or mutant receptors were statistically significantly different from that measured with control cells (P < 0.0001).
Thus, our data support the notion that the human SR-BI residues mutated in the SW0 and E210A variants are necessary for HCV interactions but are not involved in SR-BI-mediated HDL metabolism.
DISCUSSION
A variety of receptor candidates have been proposed for HCV, including CD81, heparan sulfate proteoglycans (HSPGs), the TJ-associated proteins CLDN1 and OCLN, and SR-BI (5, 6, 22, 42, 43, 50). However, it is not yet understood whether these molecules act in combination or sequentially to mediate attachment, entry into target cells, and release of the viral genome into the cytoplasm. In particular, little is known about how and when these molecules interact with the virus during the infection process and which of them contributes to the tropism and species specificity of HCV infection. Some pieces of information on the kinetics of HCV entry have been indirectly gathered by the usage of tools such as truncated forms of the receptors (CD81 large extracellular loop), molecular mimics (heparin), and neutralizing antibodies. The finding that the heparan sulfate homolog heparin inhibits HCV infection only when administered before or during virus binding implies that HSPGs play a role in the initial attachment of HCV to target cells (5, 32). Several groups demonstrated the ability of anti-CD81 antibodies to inhibit HCV entry at a postbinding step, suggesting that CD81 functions as an HCV entry coreceptor after the docking of the virus to attachment molecules (16, 22, 31). Intriguingly, CD81 clustering and engagement, although induced by surrogate systems such as anti-CD81 antibodies or sE2 and not through the binding of viral particles, have been shown to trigger intracellular events ultimately leading to the relocalization of the CD81-bound E2 to the TJ, where two other receptors, CLDN1 and OCLN, are located (11). So far, there is no clear evidence that CLDN1 and OCLN bind the virus directly. Even though anti-CLDN1 blocking antibodies are not available, kinetic studies with antibodies directed against a foreign epitope inserted into the first extracellular loop (EL1) of CLDN1 indicate that CLDN1 acts late in the entry process, likely after virus binding to the cell surface and the interaction of the virus with CD81 (22). The downregulation of both CLDN1 and OCLN resulted in a reduction of HCV glycoprotein-dependent cell-to-cell fusion, indicating their possible role in the fusion process (9, 22).
Our time course experiments of the inhibition of HCVcc infection with MAbs against CD81 and SR-BI confirm that interactions of HCV with both cell surface proteins are crucial for the efficient infection of Huh-7.5 cells. The anti-SR-BI MAb used in this study (C167) was originally identified for its ability to block the binding of sE2 to SR-BI displayed on the cell surface, suggesting that this antibody binds a region of SR-BI that is important for virus recognition (15). Consistent with this finding, we show that MAb C167 inhibits HCVcc infection when added before or together with the virus but not after virus attachment to the cells. In contrast, the anti-CD81 MAb was still capable of reducing HCVcc infection when it was added after the binding of the virus to Huh-7.5 cells (Fig. 1). Thus, our data support the hypothesis that SR-BI is responsible for the initial recognition of the virus envelope, possibly in concert with HSPGs (5). Since we used a MAb, we cannot rule out the possibility that SR-BI is also involved in a postbinding step. In fact, by using anti-SR-BI rat polyclonal antibodies possibly recognizing different receptor determinants, Zeisel et al. previously showed an inhibition of HCVcc infection up to 1 h after virus binding to the cell (63).
CD81 appears to be necessary for viral entry into hepatic cells via its large extracellular loop, and specific residues involved in virus-receptor interactions have been identified, although the profile of binding of HCV E2 to CD81 derived from different species does not show a clear correlation with susceptibility to infection (2, 21, 24, 28, 37, 41). Unlike CD81, the binding of sE2 to SR-BI seems to be predictive of an infection-producing interaction between HCV and host cells. We cloned SR-BI cDNAs from species with different susceptibilities to HCV infection in mammalian expression vectors and tested their abilities to bind sE2 by transient transfection into the mouse N2 neuronal cell line. The tree shrew Tupaia belageri has been shown to be susceptible to HCV infection (62). Tupaia SR-BI can bind sE2 but with a reduced efficiency (about 50%) compared to that of the human receptor. Conversely, mouse SR-BI was totally deficient in E2 binding, which correlates with the lack of permissiveness of this species to HCV infection. Similarly, attempts to experimentally infect tamarins have led to the conclusion that these monkeys are resistant to HCV infection (25). Consistently, tamarin SR-BI was completely unable to bind the viral ligand. Interestingly, sequence analysis of the SR-BI gene from chimpanzees, which is the only other species that is susceptible to HCV infection, revealed a complete identity of amino acid residues with the human protein (our unpublished observations).
The tridimensional structure of the SR-BI ectodomain is unknown; therefore, in order to identify regions and residues that are important for viral ligand binding, we used a mutagenesis approach guided by a comparative analysis of the amino acid sequence of SR-BI from the four different species. We generated mutants of the human protein containing highly divergent regions of mouse SR-BI. In addition, we constructed single alanine substitutions of residues that are different in the mouse and tamarin sequences but conserved in human and tupaia SR-BI proteins. A detailed analysis of the capacity of the binding of the mutant receptors to the viral glycoprotein E2 was performed, leading to the identification of a domain (from aa 70 to aa 87) and a single residue (E210) that are required for sE2 recognition. We also confirmed that all the mutant proteins were correctly folded and expressed on the surface of the transfected cells. Moreover, we demonstrated that the reduced sE2 binding of the SR-BI E210A mutant is not due to an impaired ability to form oligomers.
To confirm that SR-BI derivatives with a reduced binding of sE2 have a functional defect in mediating HCVcc infection, we performed complementation experiments. Other groups previously tested the role of the different HCV receptor candidates in nonhuman cell lines of nonhepatic origin or in human liver endothelial cells but only upon the ectopic expression of some of the other entry molecules (19, 43). However, such modifications might affect the overall localization of the HCV receptors, the formation of entry-competent complexes, and intracellular signaling, thereby making it difficult to evaluate a specific and direct contribution of a given molecule/mutant in the infection process. We reasoned that a phenotypic analysis of SR-BI variants would have more functional relevance if done in a more physiological environment. Hence, we generated a novel Huh-7.5 cell line where the silencing of the endogenous SR-BI gene was achieved by lentivirus-mediated inducible RNA interference. We used an shRNA that targeted a sequence in the 3′ untranslated region of the SR-BI mRNA, therein allowing trans-complementation studies with SR-BI mutants. Thus, for the first time, the determinants of SR-BI involved in HCV infection could be addressed through gain-of-function studies in the context of a fully HCV-permissive human hepatoma cell line without the need for manipulating the expression of other entry receptors and overcoming the barriers of low levels of RNA replication, assembly, and release that would have been encountered by use of other cell lines. We confirmed that the defect in sE2 binding of mouse SR-BI and the human SW0 and E210A derivatives correlates with a reduced ability to mediate HCVcc infectivity in the SR-BI knockdown cell line.
Recently, it was shown that only human OCLN and CD81, but not their murine counterparts, allow HCVpp entry into NIH 3T3 and CHO cells, suggesting that these two factors play a role in the species specificity of HCV infection (43). In that same study, mouse CLDN1 and mouse SR-BI were capable of complementing the lack of human protein and rendered NIH 3T3 and CHO cells permissive to HCVpp entry.
In the present work, we show that mouse SR-BI is not able to fully restore an efficient infection when expressed in human hepatic cells where the endogenous receptor has been knocked down. Similarly, human-to-mouse chimeras showed a lower ability to mediate HCVcc entry into Huh-7.5 cells than human SR-B1, suggesting that SR-BI may contribute to the species specificity of HCV infection. On the basis of these results, it is advisable to also include the human version of SR-BI to make humanized mouse models of HCV infection, which would provide an excellent platform for developing effective therapies and vaccines.
Previous observations indicated that SR-BI was important for infection by different HCV subtypes despite the high level of amino acid variability observed across HVR1, which was previously shown to be the principal determinant for SR-BI binding (8, 50). In fact, the chemicophysical properties of HVR1 are highly conserved, suggesting that HCV HVR1, rather than being completely variable, might actually adopt only one or a restricted spectrum of closely related conformations (39, 44). In agreement with this hypothesis, we found that the E210A mutant showed an impaired binding to sE2 glycoproteins of different strains or genotypes. This finding provides a rationale for developing new therapeutics targeting SR-BI instead of the virus to overcome the problem of the development of an extreme level of viral variability. Importantly, we showed that residues involved in HCV recognition are not required for HDL binding or SR-BI-mediated cholesterol efflux, suggesting that the development of new and safe agents selectively inhibiting HCV infection, but with no or low impact on HDL metabolism, is a feasible task.
Acknowledgments
This work was supported by funding from the European Union (grants QLK2-CT-2001-01120 and MRTN-CT-2006-035599) and PHS grant R01 AI072613. M.T.C. was supported by a Women & Science fellowship.
We thank D. Trono (Addgene) for providing reagents, including lentiviral plasmids pLVTHM, pLV-tTRKRAB-Red, pCMV-DR8.74, and pMD2G-VSVG.
Footnotes
Published ahead of print on 14 October 2009.
REFERENCES
- 1.Acton, S., A. Rigotti, K. T. Landschulz, S. Xu, H. H. Hobbs, and M. Krieger. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271:518-520. [DOI] [PubMed] [Google Scholar]
- 2.Allander, T., X. Forns, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology 277:358-367. [DOI] [PubMed] [Google Scholar]
- 3.Alter, M. J. 2007. Epidemiology of hepatitis C virus infection. World J. Gastroenterol. 13:2436-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth, H., R. Cerino, M. Arcuri, M. Hoffmann, P. Schurmann, M. I. Adah, B. Gissler, X. Zhao, V. Ghisetti, B. Lavezzo, H. E. Blum, F. von Weizsacker, A. Vitelli, E. Scarselli, and T. F. Baumert. 2005. Scavenger receptor class B type I and hepatitis C virus infection of primary tupaia hepatocytes. J. Virol. 79:5774-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. [DOI] [PubMed] [Google Scholar]
- 6.Barth, H., E. K. Schnober, F. Zhang, R. J. Linhardt, E. Depla, B. Boson, F. L. Cosset, A. H. Patel, H. E. Blum, and T. F. Baumert. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 80:10579-10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 9.Benedicto, I., F. Molina-Jimenez, B. Bartosch, F. L. Cosset, D. Lavillette, J. Prieto, R. Moreno-Otero, A. Valenzuela-Fernandez, R. Aldabe, M. Lopez-Cabrera, and P. L. Majano. 2009. The tight junction-associated protein occludin is required for a postbinding step in hepatitis C virus entry and infection. J. Virol. 83:8012-8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brazzoli, M., A. Bianchi, S. Filippini, A. Weiner, Q. Zhu, M. Pizza, and S. Crotta. 2008. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 82:8316-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burlone, M. E., and A. Budkowska. 2009. Hepatitis C virus cell entry: role of lipoproteins and cellular receptors. J. Gen. Virol. 90:1055-1070. [DOI] [PubMed] [Google Scholar]
- 13.Calvo, D., D. Gomez-Coronado, M. A. Lasuncion, and M. A. Vega. 1997. CLA-1 is an 85-kD plasma membrane glycoprotein that acts as a high-affinity receptor for both native (HDL, LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arterioscler. Thromb. Vasc. Biol. 17:2341-2349. [DOI] [PubMed] [Google Scholar]
- 14.Calvo, D., and M. A. Vega. 1993. Identification, primary structure, and distribution of CLA-1, a novel member of the CD36/LIMPII gene family. J. Biol. Chem. 268:18929-18935. [PubMed] [Google Scholar]
- 15.Catanese, M. T., R. Graziani, T. von Hahn, M. Moreau, T. Huby, G. Paonessa, C. Santini, A. Luzzago, C. M. Rice, R. Cortese, A. Vitelli, and A. Nicosia. 2007. High-avidity monoclonal antibodies against the human scavenger class B type I receptor efficiently block hepatitis C virus infection in the presence of high-density lipoprotein. J. Virol. 81:8063-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 101:7270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coyne, C. B., and J. M. Bergelson. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119-131. [DOI] [PubMed] [Google Scholar]
- 18.De Francesco, R., and G. Migliaccio. 2005. Challenges and successes in developing new therapies for hepatitis C. Nature 436:953-960. [DOI] [PubMed] [Google Scholar]
- 19.Dreux, M., V. L. D. Thi, J. Fresquet, M. Guerin, Z. Julia, G. Verney, D. Durantel, F. Zoulim, D. Lavillette, F. L. Cosset, and B. Bartosch. 2009. Receptor complementation and mutagenesis reveal SR-BI as an essential HCV entry factor and functionally imply its intra- and extra-cellular domains. PLoS Pathog. 5:e1000310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreux, M., T. Pietschmann, C. Granier, C. Voisset, S. Ricard-Blum, P. E. Mangeot, Z. Keck, S. Foung, N. Vu-Dac, J. Dubuisson, R. Bartenschlager, D. Lavillette, and F. L. Cosset. 2006. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 281:18285-18295. [DOI] [PubMed] [Google Scholar]
- 21.Drummer, H. E., K. A. Wilson, and P. Poumbourios. 2002. Identification of the hepatitis C virus E2 glycoprotein binding site on the large extracellular loop of CD81. J. Virol. 76:11143-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801-805. [DOI] [PubMed] [Google Scholar]
- 23.Feld, J. J., and J. H. Hoofnagle. 2005. Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967-972. [DOI] [PubMed] [Google Scholar]
- 24.Flint, M., T. von Hahn, J. Zhang, M. Farquhar, C. T. Jones, P. Balfe, C. M. Rice, and J. A. McKeating. 2006. Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 80:11331-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garson, J. A., K. Whitby, P. Watkins, and A. J. Morgan. 1997. Lack of susceptibility of the cottontop tamarin to hepatitis C infection. J. Med. Virol. 52:286-288. [PubMed] [Google Scholar]
- 26.Grove, J., T. Huby, Z. Stamataki, T. Vanwolleghem, P. Meuleman, M. Farquhar, A. Schwarz, M. Moreau, J. S. Owen, G. Leroux-Roels, P. Balfe, and J. A. McKeating. 2007. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J. Virol. 81:3162-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helle, F., and J. Dubuisson. 2008. Hepatitis C virus entry into host cells. Cell. Mol. Life Sci. 65:100-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. U. S. A. 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji, Y., B. Jian, N. Wang, Y. Sun, M. L. Moya, M. C. Phillips, G. H. Rothblat, J. B. Swaney, and A. R. Tall. 1997. Scavenger receptor BI promotes high density lipoprotein-mediated cellular cholesterol efflux. J. Biol. Chem. 272:20982-20985. [DOI] [PubMed] [Google Scholar]
- 31.Koutsoudakis, G., E. Herrmann, S. Kallis, R. Bartenschlager, and T. Pietschmann. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 34.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 35.Liu, S., W. Yang, L. Shen, J. R. Turner, C. B. Coyne, and T. Wang. 2009. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 83:2011-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manns, M. P., H. Wedemeyer, and M. Cornberg. 2006. Treating viral hepatitis C: efficacy, side effects, and complications. Gut 55:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meola, A., A. Sbardellati, B. B. Ercole, M. Cerretani, M. Pezzanera, A. Ceccacci, A. Vitelli, S. Levy, A. Nicosia, C. Traboni, J. McKeating, and E. Scarselli. 2000. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol. 74:5933-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pagler, T. A., S. Rhode, A. Neuhofer, H. Laggner, W. Strobl, C. Hinterndorfer, I. Volf, M. Pavelka, E. R. Eckhardt, D. R. van der Westhuyzen, G. J. Schutz, and H. Stangl. 2006. SR-BI-mediated high density lipoprotein (HDL) endocytosis leads to HDL resecretion facilitating cholesterol efflux. J. Biol. Chem. 281:11193-11204. [DOI] [PubMed] [Google Scholar]
- 39.Penin, F., C. Combet, G. Germanidis, P. O. Frainais, G. Deleage, and J. M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereira, A. A., and I. M. Jacobson. 2009. New and experimental therapies for HCV. Nat. Rev. Gastroenterol. Hepatol. 6:403-411. [DOI] [PubMed] [Google Scholar]
- 41.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 43.Ploss, A., M. J. Evans, V. A. Gaysinskaya, M. Panis, H. You, Y. P. de Jong, and C. M. Rice. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puntoriero, G., A. Meola, A. Lahm, S. Zucchelli, B. B. Ercole, R. Tafi, M. Pezzanera, M. U. Mondelli, R. Cortese, A. Tramontano, G. Galfre, and A. Nicosia. 1998. Towards a solution for hepatitis C virus hypervariability: mimotopes of the hypervariable region 1 can induce antibodies cross-reacting with a large number of viral variants. EMBO J. 17:3521-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhainds, D., and L. Brissette. 2004. The role of scavenger receptor class B type I (SR-BI) in lipid trafficking. Defining the rules for lipid traders. Int. J. Biochem. Cell Biol. 36:39-77. [DOI] [PubMed] [Google Scholar]
- 46.Rigotti, A., H. E. Miettinen, and M. Krieger. 2003. The role of the high-density lipoprotein receptor SR-BI in the lipid metabolism of endocrine and other tissues. Endocr. Rev. 24:357-387. [DOI] [PubMed] [Google Scholar]
- 47.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahoo, D., Y. F. Darlington, D. Pop, D. L. Williams, and M. A. Connelly. 2007. Scavenger receptor class B type I (SR-BI) assembles into detergent-sensitive dimers and tetramers. Biochim. Biophys. Acta 1771:807-817. [DOI] [PubMed] [Google Scholar]
- 49.Sahoo, D., Y. Peng, J. R. Smith, Y. F. Darlington, and M. A. Connelly. 2007. Scavenger receptor class B, type I (SR-BI) homo-dimerizes via its C-terminal region: fluorescence resonance energy transfer analysis. Biochim. Biophys. Acta 1771:818-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Silver, D. L. 2004. SR-BI and protein-protein interactions in hepatic high density lipoprotein metabolism. Rev. Endocr. Metab. Disord. 5:327-333. [DOI] [PubMed] [Google Scholar]
- 52.Silver, D. L., N. Wang, X. Xiao, and A. R. Tall. 2001. High density lipoprotein (HDL) particle uptake mediated by scavenger receptor class B type 1 results in selective sorting of HDL cholesterol from protein and polarized cholesterol secretion. J. Biol. Chem. 276:25287-25293. [DOI] [PubMed] [Google Scholar]
- 53.Treguier, M., M. Moreau, A. Sposito, M. J. Chapman, and T. Huby. 2007. LDL particle subspecies are distinct in their capacity to mediate free cholesterol efflux via the SR-BI/Cla-1 receptor. Biochim. Biophys. Acta 1771:129-138. [DOI] [PubMed] [Google Scholar]
- 54.Trigatti, B., A. Rigotti, and M. Krieger. 2000. The role of the high-density lipoprotein receptor SR-BI in cholesterol metabolism. Curr. Opin. Lipidol. 11:123-131. [DOI] [PubMed] [Google Scholar]
- 55.Trigatti, B. L., M. Krieger, and A. Rigotti. 2003. Influence of the HDL receptor SR-BI on lipoprotein metabolism and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 23:1732-1738. [DOI] [PubMed] [Google Scholar]
- 56.Vinals, M., S. Xu, E. Vasile, and M. Krieger. 2003. Identification of the N-linked glycosylation sites on the high density lipoprotein (HDL) receptor SR-BI and assessment of their effects on HDL binding and selective lipid uptake. J. Biol. Chem. 278:5325-5332. [DOI] [PubMed] [Google Scholar]
- 57.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 58.Voisset, C., A. Op de Beeck, P. Horellou, M. Dreux, T. Gustot, G. Duverlie, F. L. Cosset, N. Vu-Dac, and J. Dubuisson. 2006. High-density lipoproteins reduce the neutralizing effect of hepatitis C virus (HCV)-infected patient antibodies by promoting HCV entry. J. Gen. Virol. 87:2577-2581. [DOI] [PubMed] [Google Scholar]
- 59.von Hahn, T., B. D. Lindenbach, A. Boullier, O. Quehenberger, M. Paulson, C. M. Rice, and J. A. McKeating. 2006. Oxidized low-density lipoprotein inhibits hepatitis C virus cell entry in human hepatoma cells. Hepatology 43:932-942. [DOI] [PubMed] [Google Scholar]
- 60.Webster, D. P., P. Klenerman, J. Collier, and K. J. Jeffery. 2009. Development of novel treatments for hepatitis C. Lancet Infect. Dis. 9:108-117. [DOI] [PubMed] [Google Scholar]
- 61.Wiznerowicz, M., and D. Trono. 2003. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie, Z. C., J. I. Riezu-Boj, J. J. Lasarte, J. Guillen, J. H. Su, M. P. Civeira, and J. Prieto. 1998. Transmission of hepatitis C virus infection to tree shrews. Virology 244:513-520. [DOI] [PubMed] [Google Scholar]
- 63.Zeisel, M. B., G. Koutsoudakis, E. K. Schnober, A. Haberstroh, H. E. Blum, F. L. Cosset, T. Wakita, D. Jaeck, M. Doffoel, C. Royer, E. Soulier, E. Schvoerer, C. Schuster, F. Stoll-Keller, R. Bartenschlager, T. Pietschmann, H. Barth, and T. F. Baumert. 2007. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology 46:1722-1731. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]