Abstract
We have used deep sequencing to analyze the pattern of viral microRNA (miRNA) expression observed in the B-cell line BC-3, which is latently infected with Kaposi's sarcoma-associated herpesvirus (KSHV). We recovered 14.6 × 106 total miRNA cDNA reads, of which a remarkable 92% were of KSHV origin. We detected 11 KSHV miRNAs as well as all 11 predicted miRNA* or passenger strands from the miRNA duplex intermediate. One previously reported KSHV miRNA, miR-K9, was found to be mutationally inactivated. This analysis revealed that the 5′ ends of 10 of the 11 KSHV miRNAs were essentially invariant, with significantly more variation being observed at the 3′ end, a result which is consistent with the proposal that the 5′-proximal region of miRNAs is critical for target mRNA recognition. However, one KSHV miRNA, miR-K10-3p, was detected in two isoforms differing by 1 nucleotide (nt) at the 5′ end that were present at comparable levels, and these two related KSHV miRNAs are therefore likely to target at least partially distinct mRNA populations. Finally, we also report the first detection of miRNA offset RNAs (moRs) in vertebrate somatic cells. moRs, which derive from primary miRNA (pri-miRNA) sequences that immediately flank the mature miRNA and miRNA* strands, were identified flanking one or both sides of nine of the KSHV miRNAs. These data provide new insights into the pattern of miRNA processing in mammalian cells and indicate that this process is highly conserved during animal evolution.
MicroRNAs (miRNAs) are a recently discovered class of ∼22-nucleotide (nt) noncoding RNAs that play a key role in the posttranscriptional regulation of gene expression in all multicellular eukaryotes (reviewed in reference 3). Most miRNAs are initially transcribed by RNA polymerase II as part of a long capped and polyadenylated precursor transcript called a primary miRNA (pri-miRNA) (6, 20). At this stage, the mature miRNA forms part of one arm of an imperfect ∼80-nt RNA stem-loop structure that is recognized by the nuclear microprocessor complex, consisting of the RNase III enzyme Drosha and its cofactor DGCR8 (8, 11, 15). Drosha cleaves the pri-miRNA to liberate an ∼60-nt-long pre-miRNA hairpin bearing a 2-nt 3′ overhang (19, 38). After export to the cytoplasm, the pre-miRNA is bound by a second RNase III enzyme, called Dicer, which cleaves the pre-miRNA ∼22 bp from the base, leaving a second 2-nt 3′ overhang, to generate the miRNA duplex intermediate (16). One strand of this duplex intermediate—the mature miRNA strand—is then incorporated into the RNA-induced silencing complex (RISC) (14), while the second strand—referred to as the passenger or miRNA* strand—is degraded. Selection of which strand of the miRNA duplex intermediate is incorporated into RISC is largely determined by the stability of base pairing at the 5′ end of each strand, with the less tightly base-paired strand being preferentially selected (17, 29). However, this discrimination is not absolute, and miRNA* strands are also occasionally incorporated into RISC and have been reported to be active as miRNAs (23).
Once incorporated into RISC, miRNAs guide RISC to mRNAs bearing a complementary target sequence (4). The key recognition element is the miRNA seed sequence, extending from position 2 to 8 from the 5′ end of the miRNAs, and most mRNA targets show full complementarity to miRNA seeds, although exceptions with only partial complementarity to the miRNA seed but extensive 3′ complementarity do exist (4).
In addition to large numbers of cellular miRNAs, recent reports have also documented the existence of viral miRNAs. In particular, almost all herpesviruses examined so far have been shown to express viral miRNAs in latently and/or productively infected cells (34). One herpesvirus that has attracted particular interest is Kaposi's sarcoma-associated herpesvirus (KSHV), which has been reported to express at least 12 distinct miRNAs in latently infected B cells (7, 25, 28). Eleven of these, miR-K1 to miR-K11, were initially detected in studies that used conventional sequencing to analyze small RNAs expressed in latently KSHV-infected B cells, while a 12th KSHV miRNA, miR-K12, was predicted computationally and then confirmed by Northern analysis (13).
In this report, we have used deep sequencing to analyze the expression of KSHV miRNAs in detail. Our interest was firstly to determine whether KSHV encodes more than the 12 miRNAs identified so far—each of the previous studies missed at least two viral miRNAs, so this was a possibility—and to unequivocally identify the major sequence variant for each viral miRNA, especially at the 5′ end, for future functional studies. We were also interested in the level of sequence variation seen in viral miRNAs, compared to known cellular miRNAs, and whether this analysis would provide more general insights into miRNA processing in mammalian cells. Our analysis recovered 11 of the 12 known KSHV miRNAs, as well as all 11 predicted miRNA* strands, but did not identify additional KSHV miRNAs. One KSHV miRNA, miR-K9, was found to be mutationally inactivated in the KSHV-infected BC-3 cell line that was analyzed. Strikingly, we report the first detection of miRNA offset RNAs (moRs) (30), encoded by KSHV, in mammalian somatic cells and also report that one KSHV miRNA, miR-K10, exists in two functional isoforms differing by 1 nt at the 5′ end.
MATERIALS AND METHODS
Cells, TPA induction, and RNA.
BC-3 cells were maintained in RPMI medium supplemented with 20% fetal bovine serum (FBS). BC-1 and BJAB cells were maintained in RPMI medium supplemented with 10% FBS. Where appropriate, cells were induced with tetradecanoyl phorbol acetate (TPA) at a final concentration of 20 ng/ml for 16 h. Total RNA was isolated from both induced and uninduced cells by using Trizol (Invitrogen), according to the manufacturer's directions.
cDNA library preparation and data analysis.
cDNA libraries for Solexa/Illumina sequencing were prepared as previously described (34). Briefly, small RNAs were isolated by preparative gel electrophoresis and sequentially ligated to 3′ and then 5′ linkers. Primers complementary to the linker sequences were used for reverse transcription and PCR in order to generate cDNA libraries for deep sequencing. Raw sequencing data were filtered to remove reads lacking identifiable 3′ linker sequences and/or reads falling outside of the predicted miRNA size range. A list of final usable reads was then collapsed down into a list of unique sequences which was analyzed against the KSHV genome (NC_003409) and the mature miRNA database from miRBase (release 9.2) by MegaBLAST using the formatdb, megablast, blastoutparse, and filter_alignment scripts of the miRDeep software package (9).
qRT-PCR for Rta and Mta mRNA.
Rta and Mta mRNA levels were analyzed using the Power SYBR green quantitative reverse transcription-PCR (qRT-PCR) system (Applied Biosystems), according to the manufacturer's directions, using 275 ng of total RNA. Total RNA was DNase treated and reverse transcribed using a random 9-mer primer mix. All values are expressed as a multiple of the background signal detected using RNA derived from the KSHV-negative BJAB B-cell line. Values were normalized to levels of β-actin, and assays were performed in triplicate. The following primers were used for PCR: Rta, forward primer 5′-AGGCATCCCAAGGCATTA-3′ and reverse primer 5′-ATGCGGCTGTCAGAAATG-3′; Mta, forward primer 5′-ACACTGGAAGACGAGCAA-3′ and reverse primer 5′-GCAGTTGAGAACGACCTT-3′; β-actin, forward primer 5′-CACACCTTCTACAATGAGCTGCGTG-3′ and reverse primer 5′-ATGATCTGGGTCATCTTCTCGCGGT-3′.
Stem-loop qRT-PCR for KSHV miRNAs.
Viral miRNA levels were quantified using TaqMan MicroRNA assays (Applied Biosystems) according to the manufacturer's directions using 10 μg of total RNA per RT reaction. Values are expressed as fold expression over an RNA sample derived from uninfected BJAB cells. All values were normalized to levels of β-actin, and assays were performed in triplicate. Custom primers for each miRNA were designed against the most common isoform identified by deep sequencing.
Sequencing of the miR-K9 locus.
The following primers were used to amplify a portion of the KSHV DNA genome encompassing the pri-miR-K9 hairpin from both BC-3 and BC-1 cells: 5′-GGCCAACATAAAGTGTGGATGGCC-3′ and 5′-ATCGAACAACCACGCTTGGCTAAC-3′. The resultant DNA fragments were then sequenced using standard techniques.
RESULTS
Deep sequencing of KSHV miRNA species.
Previous efforts to identify KSHV-encoded miRNAs by conventional sequencing used the primary effusion lymphoma (PEL) B-cell lines BC-1 and BCBL1, which are latently infected with KSHV and Epstein-Barr virus (EBV) or with KSHV alone, respectively (7, 25, 28). For deep sequencing of small RNAs of both viral and cellular origin in a latently KSHV-infected cell context, we selected a third latently KSHV-infected PEL cell line, BC-3, which is not coinfected with EBV and which has been shown to spontaneously produce low levels of KSHV virions in culture (1). We initially harvested total RNA from BC-3 cells and then isolated ∼18- to 24-nt-long RNAs by gel electrophoresis. These RNAs were sequentially ligated to 3′ and 5′ linkers and subjected to reverse transcription followed by PCR amplification. The resultant cDNAs were then directly sequenced using a Solexa/Illumina apparatus.
Deep sequencing resulted in 26,663,462 total sequence reads, of which 19,398,417 were used for subsequent analysis, based on the detection of the intact 3′ linker sequence and an appropriately sized cDNA insert. Of these, 13,469,484 were found to derive from one of the 12 known KSHV pre-miRNAs, while 1,151,608 were found to represent cellular miRNA sequences. The KSHV miRNA therefore represented 92% of the total miRNA pool present in BC-3 cells. The remaining 4,775,545 reads either represented fragments of cellular mRNAs or noncoding RNAs (rRNAs, tRNAs, snRNAs, etc.) or were not present in the databases used. A significant level (1 to 2%) of unassignable reads is expected, as the deep sequencing technology is somewhat error prone and the sequenced cDNA samples had undergone both reverse transcription and PCR amplification, which also generates sequencing errors. Finally, we also detected 1,780 KSHV sequencing reads which did not map to any of the 12 known KSHV pre-miRNAs. Almost all of these represented fragments of the ∼10.5-kb primary miRNA that gives rise to all 12 KSHV miRNAs in latently infected cells (5). Importantly, we did not repeatedly recover any additional unique KSHV sequence distinct from the known miRNAs, as would be expected if KSHV encoded additional miRNAs.
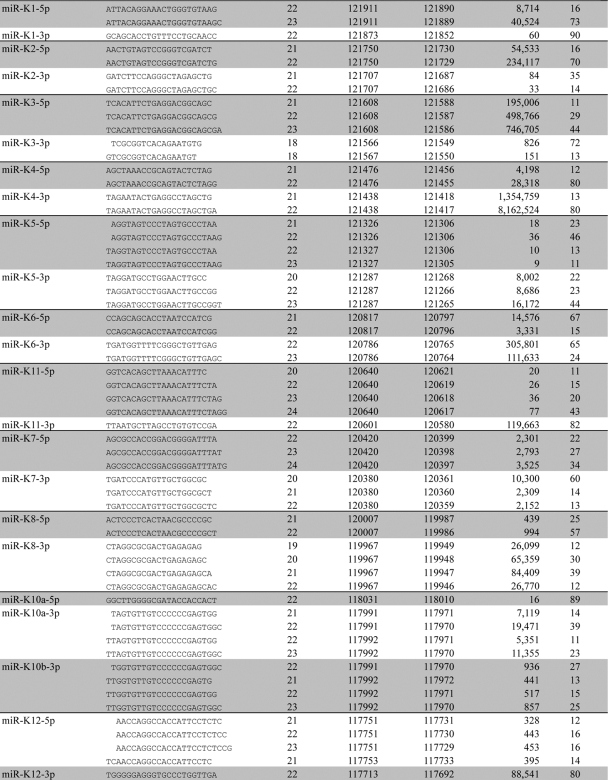
An overview of the major miRNA and miRNA* sequence variants and their origins is provided in Table 1, which shows that we recovered not only 11 of the known KSHV miRNAs but also all 11 matching miRNA* sequences, most of which have not been previously detected. We also recovered the previously reported edited version of miR-K10a-3p, called miR-K10b-3p, which contains a G residue in place of the genomically encoded A in the miRNA seed (25). We did not, however, recover any examples of miR-K9, which has previously been identified in the KSHV-infected cell lines BC-1 and BCBL1, as well as in vivo (7, 22, 25, 28). As described below, this is due to the mutational inactivation of miR-K9 in the KSHV genome present in BC-3 cells.
TABLE 1.
Major KSHV miRNA and miRNA* sequence variants
Major species of each KSHV miRNA comprising at least 10% of the total mature miRNA population are included.
Genomic coordinates provided based on NC_003409 genome.
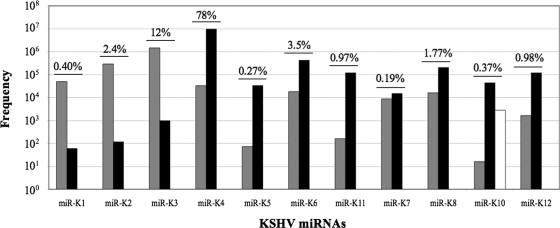
Although all 12 KSHV miRNAs derive from a single pri-miRNA in latently infected cells (5), we observed dramatic differences in their levels of expression, as determined by the number of sequence reads recovered (Fig. 1). Specifically, miR-K4 was found to represent ∼78% of the 13,469,484 KSHV miRNA sequence reads obtained, with miR-K3 representing a further ∼12%. The remaining nine KSHV miRNAs represented from 2.4% to 0.19% of the total number of sequence reads. This remarkable discrepancy could reflect differential stability and/or differential processing of the single KSHV pri-miRNA. However, as the prevalence of the miRNA* strands tended to correlate with the prevalence of the mature miRNA strand (Fig. 1), we feel that the latter mechanism is likely to be more important.
FIG. 1.
Frequency of KSHV miRNA and miRNA* strands recovered from deep sequencing of short RNAs derived from uninduced BC-3 cells. Gray bars represent 5p miRNAs recovered, black bars represent 3p miRNAs recovered, and the white bar indicates edited miRNAs. The percent contribution of each set of miRNAs to the total number of viral miRNAs detected is indicated.
A comparison of the recovery of miRNA versus miRNA* strands for each KSHV miRNA shows that in most cases the former is present at 102- to 103-fold-higher levels than the latter. The major exception to this generalization is miR-K7, where the dominant miR-K7-3p strand was recovered only ∼2-fold more frequently than the less common miR-K7-5p strand (Fig. 1).
RT-PCR analysis of viral miRNA expression.
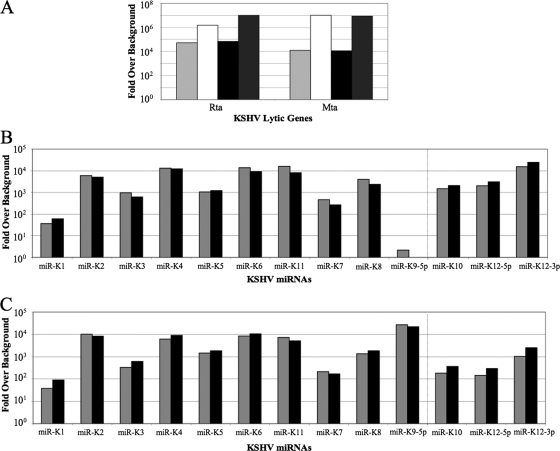
To confirm and extend these deep sequencing data, we performed qRT-PCR to detect the various known KSHV miRNAs in BC-3 cells and also in BC-1 cells. In both cases, we analyzed both uninduced cells and cells treated with TPA, which induces entry of a substantial percentage of cells into the productive KSHV replication cycle (36). As shown in Fig. 2A, TPA treatment indeed strongly induced expression of the KSHV mRNAs encoding the immediate-early Rta and Mta proteins, both of which play a key role in initiating the viral productive replication cycle (21, 24, 32). We note, however, that both mRNAs are also detectable in uninduced cells, consistent with the known low level of spontaneous induction of productive KSHV replication seen with both BC-3 and BC-1 (1, 7).
FIG. 2.
KSHV gene expression in uninduced and induced BC-3 and BC-1 cells. All values are expressed as fold expression over an uninfected control sample and are normalized to cellular β-actin mRNA levels. (A) Rta and Mta mRNA expression in uninduced (light gray) and TPA-induced (white) BC-3 cells and uninduced (black) and TPA-induced (dark gray) BC-1 cells. (B) Expression levels of KSHV miRNAs in uninduced (gray) and induced (black) BC-3 cells. (C) Same as panel B except using RNA derived from uninduced and TPA-induced BC-1 cells.
As shown in Fig. 2B, we were able to detect all mature KSHV miRNA species except miR-K9 in BC-3 cells, consistent with the sequencing data presented in Table 1. TPA treatment had little or no effect on the level of expression of miR-K1 through miR-K8 plus miR-K11, which are expressed under the control of a latent KSHV promoter, but did modestly induce expression of miR-K10 and miR-K12, which both lie 3′ to a promoter induced during productive replication (Fig. 2B) (5, 10). Similar data were also obtained for BC-1 cells (Fig. 2C), with the notable distinction that miR-K9 was, as expected, readily detectable in BC-1 cells.
Perhaps surprisingly, there was only a moderate correlation between the levels of KSHV miRNA expression detected by deep sequencing (Fig. 1) and those detected by qRT-PCR (Fig. 2B). Possible reasons for this include differences in the ligation efficiency of linkers to the various viral small RNAs and differences in RT and/or PCR primer annealing efficiency. Nevertheless, the overall correlation between these two very different techniques was reasonable, given these technical constraints.
KSHV miR-K9 is mutationally inactivated in BC-3 cells.
Based on both deep sequencing data (Table 1) and qRT-PCR (Fig. 2B), it was clear that the KSHV miRNA miR-K9 was absent from BC-3 cells. To illuminate why this might be the case, we used PCR to clone and then sequence the relevant segment of the KSHV genome, using DNA isolated from BC-3 cells. As shown in Fig. 3A, we observed a substantial number of missense mutations, and two single nucleotide deletions, in the predicted pri-miR-K9 sequence in BC-3 compared to the canonical KSHV genomic sequence present in BC-1 cells, which express wild-type miR-K9. In particular, these data predict seven mutations in the mature miR-K9-5p miRNA. Analysis of the “pri-miR-K9” sequence present in BC-3 cells nevertheless predicted that this should fold into a stable RNA stem-loop structure (Fig. 3B). As the predicted miR-K9 sequence in BC-3 cells is substantially different from the sequence present in BC-1 cells and most other KSHV isolates, we were concerned that we might have missed a novel variant of miR-K9 during analysis of our deep sequencing data. However, reexamination of these data using the sequence information shown in Fig. 3 again failed to identify any mature “miR-K9” variants in BC-3 cells. It is unclear why the predicted pri-miR-K9 stem-loop shown in Fig. 3 is not recognized by the microprocessor, but it has been reported that the microprocessor requires a large (≥10-nt) terminal loop for efficient pri-miRNA recognition and cleavage (38).
FIG. 3.
Comparison of the BC-1 and BC-3 miR-K9 genomic loci. (A) Alignment of the KSHV miR-K9 sequences found in BC-1 and BC-3 cells. Conserved nucleotides are indicated with asterisks, and the 5p and 3p strands of miR-K9 are boxed in gray. (B) Predicted secondary structures of the miR-K9 pri-miRNA stem-loops transcribed from the KSHV genomes present in BC-1 and BC-3 cells. In the BC-1 hairpin, the previously reported dominant miRNA is indicated in bold, Drosha cleavage sites are indicated by black arrowheads, and Dicer cleavage sites are indicated by gray arrowheads. In the defective BC-3 stem-loop structure, the mutated “miR-K9” is indicated in bold.
Sequence variation at the ends of KSHV miRNAs.
As noted above, target mRNA recognition by miRNAs is usually highly dependent on full sequence complementarity between the mRNA and the miRNA seed sequence, located between nucleotides 2 and 8 from the 5′ end of the miRNA (4). As a result, sequence differences at the 5′ end can greatly affect the ability of an miRNA to target a given mRNA. Conversely, while there have been reports documenting a role for the 3′ end of miRNAs in target selection, these sequences are generally less important for target discrimination. Accordingly, deep sequencing of miRNAs in Drosophila and in Caenorhabditis elegans has documented the presence of very little sequence variation at the 5′ ends of most cellular miRNAs, while variation at the 3′ ends was fairly common (26, 27).
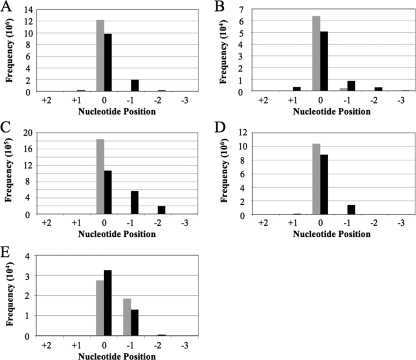
As shown in Fig. 4A, a combined analysis of the mature KSHV miRNAs listed in Table 1 also revealed almost no sequence variation at their 5′ ends and yet significant variation at the 3′ end. Analysis of the sequence variation of the miRNA* strands reported in Table 1 revealed a somewhat higher level of sequence variation (Fig. 4B), which is curious, given that the mature miRNA and miRNA* strands are generated by the same processing events. We did not see any difference in the level of 5′ sequence heterogeneity dependent on whether the mature miRNA derived from the 5p arm of the pre-miRNA, in which case the 5′ end would have been generated by Drosha cleavage (Fig. 4C), or from the 3p arm, in which case the 5′ end would have been generated by Dicer cleavage (Fig. 4D). However, when the 3′ end was generated by Dicer (Fig. 4C), somewhat more sequence variability was observed than when the miRNA 3′ end was generated by Drosha (Fig. 4D).
FIG. 4.
Sequence variation in KSHV miRNAs recovered from deep sequencing of BC-3 cells with 5′ ends indicated in gray and 3′ ends indicated in black. In each case, the end of the most abundant form of each miRNA is designated nucleotide position 0 and all other, less abundant isoforms are numbered accordingly. (A) Collective sequence variation of the mature miRNA strands recovered. (B) Same as panel A except with analysis of the less abundant miRNA* strands. (C) Collective sequence variation of all 5p miRNA strands recovered from deep sequencing. (D) Same as panel C except with analysis of the 3p miRNA strands. (E) Sequence variation detected at the 5′ and 3′ ends of mature miR-K10-3p.
Although 10 of the 11 KSHV miRNAs showed a tightly constrained 5′-terminal sequence, one exception was noted. Specifically, we observed a high level of sequence variation at the 5′ end of the mature miR-K10-3p miRNA, with ∼40% of all the cDNAs sequenced being 1 nt shorter at the 5′ end. This difference was the same for both the edited and unedited forms of miR-K10-3p (Table 1) and has the potential to strongly affect miR-K10-3p function.
Comparison of the dominant sequence variants of the 11 miRNAs recovered from BC-3 cells with the sequences recorded on the miRBase website (12) revealed two important discrepancies. The first is that miRBase gives the sequence of miR-K8-3p—which is the dominant form of miR-K8—as 5′-UAGGCGCGACUGAGAGAGCACG-3′, while our data recovered 5′-CUAGGCGCGACUGAGAGAGCA-3′ as the most prevalent form. This form is 1 nt longer at the 5′ end and 2 nt shorter at the 3′ end than the version in miRBase. Of note, an additional 1 nt at the 5′ end of miR-K8-3p was also reported by two of the three groups that used conventional sequencing to analyze the KSHV miRNAs present in BC-1 and BCBL1 cells (7, 28), and so this is unlikely to represent a BC-3-specific phenomenon.
A more dramatic sequence difference with miRBase is seen with miR-K12. In miRBase, the dominant form of miR-K12 is predicted to be miR-K12-5p. However, our cloning data reveal that the dominant form of miR-K12 is actually miR-K12-3p, which was recovered ∼100-fold more often than miR-K12-5p (Table 1 and Fig. 1, 2, and 5). We note that miR-K12 has not been sequenced previously and was instead detected only by Northern analysis (13), so that the previously reported mature miRNA sequence was actually hypothetical.
FIG. 5.
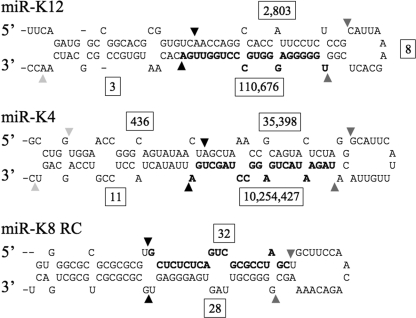
Predicted structures of the KSHV miR-K12, miR-K4, and miR-K8 RC pri-miRNA stem-loops. The most common miRNA recovered is indicated in bold. Drosha cleavage sites are indicated by black triangles, and Dicer cleavage sites are indicated by dark gray triangles. The predominant termini observed for the moRs are indicated by light gray triangles. Boxed numbers indicate the frequency with which each processing fragment was recovered by deep sequencing.
MicroRNA offset RNAs, loops, and reverse complements.
Additional analysis of the small RNA sequence reads revealed several short RNAs that are likely to represent by-products of KSHV pri- or pre-miRNA processing or that lie antisense to the known KSHV miRNAs (Table 2). The most common KSHV RNA sequences obtained, other than the miRNA and miRNA* strands themselves, were ∼22-nt RNAs that lie immediately 5′ or immediately 3′ to one of the KSHV miRNAs. These so-called moRs (30) were initially observed upon deep sequencing of small RNAs expressed in the simple chordate Ciona intestinalis, where a substantial number of small RNAs that were found to immediately flank mature miRNA sequences were recovered. moRs have also been detected at very low levels in Drosophila (27) but have not previously been reported in mammalian somatic cells. Because moRs derive from RNA sequences that lie directly adjacent to either the mature miRNA or the miRNA* strand, it is apparent that one end of each moR must arise from Drosha cleavage (Fig. 5). However, how the other end arises is currently unclear. In our hands, we obtained moRs derived from nine of the KSHV pre-miRNAs, with both 5p and 3p moRs obtained in the case of miR-K4 and miR-K10 (Table 2). As shown in Fig. 5, these are far less prevalent than the adjacent miRNA or miRNA* strands and their level of RISC association is currently unclear.
TABLE 2.
Frequency of non-miRNA KSHV fragments
| miRNA | Frequency of fragmenta: |
||||
|---|---|---|---|---|---|
| moR-5p | Loop | moR-3p | 5p RC | 3p RC | |
| miR-K1 | 546 | ||||
| miR-K2 | 3 | ||||
| miR-K3 | 11 | 11 | |||
| miR-K4 | 436 | 11 | 7 | ||
| miR-K5 | 5 | ||||
| miR-K6 | 23 | 1 | |||
| miR-K11 | 2 | ||||
| miR-K7 | |||||
| miR-K8 | 4 | 4 | 28 | 32 | |
| miR-K10 | 97 | 49 | |||
| miR-K12 | 8 | 3 | 20 | ||
moR-5p, moR flanking an miRNA 5p strand; loop, terminal loop of the pre-miRNA; moR-3p, moR flanking an miRNA 3p strand; 5p RC, 5p strand of an miRNA reverse complement transcript; 3p RC, 3p strand of an miRNA reverse complement transcript.
In addition to a fairly large number of KSHV moRs, we also obtained a number of short RNAs that appear to represent the terminal loops of KSHV pre-miRNAs and that therefore presumably represent unstable by-products of Dicer cleavage of viral pre-miRNAs (Table 2). Finally, we also detected a small number of “reverse complement” (RC) RNAs, in particular in the case of miR-K8 (Fig. 5). RC miRNAs have previously been reported in both Drosophila and herpes simplex virus type 1 (HSV-1) (31, 33, 35) and arise when both strands of a genomic region are transcribed. Bidirectional transcription of a single miRNA locus can result in the generation of “antisense” pri-miRNA hairpins that are then recognized by the microprocessor, although not all antisense pri-miRNA sequences are able to fold appropriately. As shown in Fig. 5, the miR-K8 RC stem-loop looks like a normal pri-miRNA stem-loop and the miR-K8-5p RC and miR-K8-3p RC RNAs that we identified are also located where one would expect. Although the sequences that encode the KSHV miRNAs do not lie antisense to any known viral transcript, these must presumably exist at a low level in latently and/or productively KSHV-infected cells.
DISCUSSION
We have performed deep sequencing of small RNAs expressed in the latently KSHV-infected PEL B-cell line BC-3 in order to more fully define the exact miRNA coding potential of KSHV and also to ask whether an in-depth analysis of viral miRNA expression, which is expected to be more homogeneous than the cellular miRNA expression pattern, might reveal insights into miRNA processing in vertebrate somatic cells.
Our analysis resulted in the sequencing of 14,621,092 miRNA cDNAs, of which a remarkable 92% were found to be of KSHV origin. This is a significantly higher percentage than observed previously in the KSHV-infected PEL cell lines BC-1 and BCBL1, where viral miRNAs were found to represent ∼30 to 40% of the total miRNA pool (7, 25). This high level of viral miRNA expression, which is similar to but even more extreme than what has previously been reported for viral miRNAs encoded by the avian herpesvirus Marek's disease virus (37), suggests that cellular miRNA function is likely to have been seriously compromised in these cells. As cellular miRNAs are known to globally act as tumor suppressors (18), it is possible that competitive inhibition of cellular miRNA function by highly expressed viral miRNAs may contribute to viral oncogenesis.
Analysis of the KSHV miRNA sequences obtained revealed 11 distinct mature miRNAs as well as all 11 matched miRNA* strands. Although all 11 KSHV miRNAs have been previously reported, we noted discrepancies between the primary viral sequence that we obtained and the sequence given in miRBase in two cases. Specifically, as noted above, the dominant 21-nt form of miR-K8 was found to be 1 nt longer at the 5′ end and 2 nt shorter at the 3′ end than the sequence given in miRBase. As the seed sequence—nucleotides 2 to 8 measured from the 5′ end—is the major determinant of mRNA target selection (4), this difference is of major importance for efforts to define mRNAs that are downregulated by miR-K8. An even more significant discrepancy was noted for miR-K12, where our data indicate that miR-K12-3p is the more relevant miRNA strand (Table 1 and Fig. 1), while miRBase proposes miR-K12-5p as the mature miRNA. This discrepancy likely reflects the fact that miR-K12 has not previously been sequenced and was, instead, predicted computationally (13).
Somewhat disappointingly, our data did not reveal any new KSHV miRNA species. Although it might be argued that this analysis used RNA isolated from latently KSHV-infected BC-3 cells, and we might therefore have missed viral miRNAs induced during lytic replication, BC-3 actually shows a low level (1 to 2%) of spontaneous activation of productive viral replication, which should have been sufficient to detect any lytic phase-specific KSHV miRNAs, given that over 1.4 × 107 cDNA sequences were analyzed. Moreover, we have also performed deep sequencing of small RNAs derived from BC-3 cells that had been induced to enter the KSHV productive replication cycle by treatment with TPA, as shown in Fig. 2A, and this analysis also failed to detect any novel KSHV miRNAs, although we did recover far more KSHV mRNA breakdown products than seen with uninduced cells (data not shown).
Unexpectedly, our analysis failed to detect one of the 12 known KSHV miRNAs in BC-3 cells, i.e., miR-K9. Sequence analysis subsequently revealed that the miR-K9 miRNA and pri-miRNA stem-loop are extensively mutated in the KSHV genome present in BC-3 cells (Fig. 3), and this is undoubtedly responsible for the loss of miR-K9 expression. Interestingly, the same constellation of miR-K9 mutations reported in Fig. 3 has been previously reported by Marshall et al. (22), who sequenced the KSHV genomic region encoding the viral miRNAs using viral DNA recovered from five PEL cell lines and 17 KSHV+ patient samples. Of the 22 sequences analyzed, one PEL cell line, called BCP-1, was found to contain the same mutations in miR-K9 seen here in BC-3 cells. However, this earlier report did not address whether these mutations affected miR-K9 expression.
Because of the very large number of viral miRNA cDNA sequences obtained, we were able to analyze the precise characteristics of these miRNAs in a way that has previously been reported only using miRNA cDNA sequences obtained from Drosophila and embryonic stem cells (2, 27). We were therefore able to document that the KSHV miRNAs, like Drosophila miRNAs, show a highly homogeneous 5′ end but a significantly more heterogeneous 3′ end (Fig. 4), as would be predicted given the known importance of the 5′ seed sequence in target mRNA recognition (4). However, one KSHV miRNA proved to be different, in that the 5′ end of miR-K10 was found to exist in two isoforms, differing by a single nucleotide (Fig. 4E). A similar situation has been previously reported in Drosophila, in the case of the cellular miR-210 miRNA, which exists in two essentially equally prevalent isoforms that differ by a single nucleotide at the 5′ end and that have been annotated as miR-210.1 and miR-210.2. This single nucleotide change should not perturb the recognition of mRNA targets that have extensive seed homology to either miR-210 or KSHV miR-K10 but would be likely to affect recognition of mRNAs with more limited seed homology. Therefore, it is possible that miR-K10.1 and miR-K10.2 regulate a distinct but overlapping set of mRNA targets in vivo. This situation is rendered even more complex by the fact that miR-K10 is edited, within the miRNA seed, to generate the isoforms miR-K10a and miR-K10b (Table 1) (25), so that four different isoforms of miR-K10 are actually predicted to be expressed in KSHV-infected cells. No other examples of editing of KSHV miRNAs were noted in this analysis.
A final interesting aspect of the sequencing data presented in this report is that we were able, for the first time, to identify moRs in mammalian somatic cells (Table 2) (30). For example, in the case of miR-K4, we identified 436 reads corresponding to moR-K4-5p and 11 reads corresponding to moR-K4-3p (Fig. 5). While one end of these moRs undoubtedly arises from Drosha cleavage, the protein responsible for cleavage at the other end is unclear. One possibility is that moRs are simply breakdown products derived from the exonucleolytic digestion of flanking pri-miRNA sequences (2), although their relative sequence homogeneity would seem to argue against this. One alternative possibility is that structured pri-miRNA fragments left after Drosha cleavage can occasionally be exported to the cytoplasm and are then cleaved by Dicer, a scenario which predicts that moRs are likely to be RISC associated. However, whether this is indeed the case is currently unclear.
Acknowledgments
This work was supported in part by Public Health Service grant AI067968 from the National Institute of Allergy and Infectious Diseases (B.R.C.). J.L.U. was supported by NIH training grant T32-CA009111 from the National Cancer Institute.
Footnotes
Published ahead of print on 4 November 2009.
REFERENCES
- 1.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 2.Babiarz, J. E., J. G. Ruby, Y. Wang, D. P. Bartel, and R. Blelloch. 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22:2773-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 4.Bartel, D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, X., and B. R. Cullen. 2006. Transcriptional origin of Kaposi's sarcoma-associated herpesvirus microRNAs. J. Virol. 80:2234-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, X., C. H. Hagedorn, and B. R. Cullen. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10:1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denli, A. M., B. B. Tops, R. H. Plasterk, R. F. Ketting, and G. J. Hannon. 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432:231-235. [DOI] [PubMed] [Google Scholar]
- 9.Friedlander, M. R., W. Chen, C. Adamidi, J. Maaskola, R. Einspanier, S. Knespel, and N. Rajewsky. 2008. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 26:407-415. [DOI] [PubMed] [Google Scholar]
- 10.Gottwein, E., X. Cai, and B. R. Cullen. 2006. Expression and function of microRNAs encoded by Kaposi's sarcoma-associated herpesvirus. Cold Spring Harbor Symp. Quant. Biol. 71:357-364. [DOI] [PubMed] [Google Scholar]
- 11.Gregory, R. I., K. P. Yan, G. Amuthan, T. Chendrimada, B. Doratotaj, N. Cooch, and R. Shiekhattar. 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432:235-240. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34:D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-296. [DOI] [PubMed] [Google Scholar]
- 15.Han, J., Y. Lee, K. H. Yeom, Y. K. Kim, H. Jin, and V. N. Kim. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18:3016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 17.Khvorova, A., A. Reynolds, and S. D. Jayasena. 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209-216. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, M. S., J. Lu, K. L. Mercer, T. R. Golub, and T. Jacks. 2007. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39:673-677. [DOI] [PubMed] [Google Scholar]
- 19.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 20.Lee, Y., M. Kim, J. Han, K. H. Yeom, S. Lee, S. H. Baek, and V. N. Kim. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23:4051-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall, V., T. Parks, R. Bagni, C. D. Wang, M. A. Samols, J. Hu, K. M. Wyvil, K. Aleman, R. F. Little, R. Yarchoan, R. Renne, and D. Whitby. 2007. Conservation of virally encoded microRNAs in Kaposi sarcoma-associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J. Infect. Dis. 195:645-659. [DOI] [PubMed] [Google Scholar]
- 23.Okamura, K., M. D. Phillips, D. M. Tyler, H. Duan, Y. T. Chou, and E. C. Lai. 2008. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol. 15:354-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmeri, D., S. Spadavecchia, K. D. Carroll, and D. M. Lukac. 2007. Promoter- and cell-specific transcriptional transactivation by the Kaposi's sarcoma-associated herpesvirus ORF57/Mta protein. J. Virol. 81:13299-13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 26.Ruby, J. G., C. Jan, C. Player, M. J. Axtell, W. Lee, C. Nusbaum, H. Ge, and D. P. Bartel. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127:1193-1207. [DOI] [PubMed] [Google Scholar]
- 27.Ruby, J. G., A. Stark, W. K. Johnston, M. Kellis, D. P. Bartel, and E. C. Lai. 2007. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17:1850-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwarz, D. S., G. Hutvagner, T. Du, Z. Xu, N. Aronin, and P. D. Zamore. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199-208. [DOI] [PubMed] [Google Scholar]
- 30.Shi, W., D. Hendrix, M. Levine, and B. Haley. 2009. A distinct class of small RNAs arises from pre-miRNA-proximal regions in a simple chordate. Nat. Struct. Mol. Biol. 16:183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stark, A., N. Bushati, C. H. Jan, P. Kheradpour, E. Hodges, J. Brennecke, D. P. Bartel, S. M. Cohen, and M. Kellis. 2008. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 22:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyler, D. M., K. Okamura, W. J. Chung, J. W. Hagen, E. Berezikov, G. J. Hannon, and E. C. Lai. 2008. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 22:26-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umbach, J. L., and B. R. Cullen. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umbach, J. L., M. F. Kramer, I. Jurak, H. W. Karnowski, D. M. Coen, and B. R. Cullen. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, S. E., F. Y. Wu, H. Chen, M. Shamay, Q. Zheng, and G. S. Hayward. 2004. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 78:4248-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao, Y., Y. Zhao, H. Xu, L. P. Smith, C. H. Lawrie, M. Watson, and V. Nair. 2008. MicroRNA profile of Marek's disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J. Virol. 82:4007-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng, Y., R. Yi, and B. R. Cullen. 2005. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 24:138-148. [DOI] [PMC free article] [PubMed] [Google Scholar]