Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs which posttranscriptionally regulate gene expression. The current release of the miRNA registry lists 16 viruses which encode a total of 146 miRNA hairpins. Strikingly, 139 of these are encoded by members of the herpesvirus family, suggesting an important role for miRNAs in the herpesvirus life cycle. However, with the exception of 7 miRNA hairpins known to be shared by Epstein-Barr virus (EBV) and the closely related rhesus lymphocryptovirus (rLCV), the known herpesvirus miRNAs show little evidence of evolutionary conservation. We have performed a global analysis of miRNA conservation among gammaherpesviruses which is not limited to family members known to encode miRNAs but includes also those which have not been previously analyzed. For this purpose, we have performed a computational prediction of miRNA candidates of all fully sequenced gammaherpesvirus genomes, followed by sequence/structure alignments. Our results indicate that gammaherpesvirus miRNA conservation is limited to two pairs of viral genomes. One is the already-known case of EBV and rLCV. These viruses, however, share significantly more miRNAs than previously thought, as we identified and experimentally verified 10 novel conserved as well as 7 novel nonconserved rLCV pre-miRNA hairpins. The second case consists of rhesus rhadinovirus (RRV), which is predicted to share at least 9 pre-miRNAs with the closely related Japanese macaque herpesvirus (JMHV). Although several other gammaherpesviruses are predicted to encode large numbers of clustered miRNAs at conserved genomic loci, no further examples of evolutionarily conserved miRNA sequences were found.
MicroRNAs (miRNAs) are small (∼22-nucleotide [nt]-long), noncoding RNAs which are produced from precursor stem-loop structures (pre-miRNAs) via successive cleavage by the RNase III-like enzymes Drosha and Dicer. miRNAs can inhibit the translation of mRNA transcripts with complete or partial sequence complementarity and, in recent years, have emerged as key regulators of cellular gene expression networks (for recent reviews on miRNA biogenesis and function, see references 2, 5, 9, 17, and 34). Besides more than 9,000 miRNAs of animal or plant origin, the current release (v13.0) of the miRNA registry (13, 14) also lists 146 viral miRNAs. Interestingly, 139 of these are encoded by members of the herpesvirus family. As for their cellular counterparts, the targets of the vast majority of these miRNAs and therefore their role in the viral life cycle remain unknown. Among the small contingent of known targets (reviewed in reference 32), however, are several cellular transcripts involved in the regulation of immune responses or apoptosis, suggesting that one way in which herpesviruses employ miRNAs is to combat antiviral host defense pathways. There are also viral miRNAs which share their seed sequence (nt 2 to 7 of the mature miRNA, which are of crucial importance for target recognition) with cellular miRNAs (12, 28, 37). These miRNAs may have evolved to phenocopy cellular miRNAs, thereby allowing the virus to gain access to existing host miRNA/target gene expression networks. Their constitutive expression during latent infection, however, could also be an important factor in the onset and/or progression of virus-associated cancers (12, 28). Finally, a number of miRNAs have been found to regulate the expression of viral lytic transcripts, which may indicate that another function of herpesvirus miRNAs is to maintain a latent state of infection (32).
Although the fact that miRNAs are abundantly expressed by herpesviruses suggests a common and important role in the herpesvirus life cycle, there appears to be very little evolutionary conservation of viral miRNA sequences. So far, the two gammaherpesviruses Epstein-Barr virus (EBV) and rhesus lymphocryptovirus (rLCV) are the only herpesviruses known to share evolutionarily conserved miRNAs: out of the 25 and 16 pre-miRNAs known to be encoded by EBV and rLCV (8, 15, 22, 38), respectively, 7 have been found to show signs of evolutionary conservation (8), suggesting that the latter may regulate the expression of conserved targets. The remaining pre-miRNAs appear to be unique, although they retain their organization in two discrete clusters within the viral genome. Besides EBV and rLCV, three other gammaherpesviruses have been found to encode miRNAs: 12, 7, and 9 pre-miRNAs were identified in the genomes of Kaposi's sarcoma-associated herpesvirus (KSHV), the related rhesus rhadinovirus (RRV), and murine gammaherpesvirus 68 (MHV-68), respectively (7, 15, 21, 24, 25). Similarly to the EBV and rLCV miRNAs, the KSHV and RRV miRNAs are found at homologous genomic locations. However, none of them are conserved in sequence.
We hypothesized that there may be additional cases of evolutionarily conserved miRNAs, either in the form of yet-unknown miRNAs encoded by viruses which have already been found to express such molecules or in viral genomes which have not been previously investigated. Therefore, we have conducted a global analysis of miRNA conservation across the gammaherpesvirus family which is not limited to known miRNAs but also includes predicted miRNA candidates. For this purpose, we computationally predicted miRNA precursor stem-loops in all fully sequenced gammaherpesvirus genomes using VMir, a computer program which we have previously developed (15, 26, 27, 29-31). We then performed sequence/structure alignments to detect conserved miRNAs. Besides the already-known rLCV miRNAs, we identified and experimentally verified 22 novel rLCV miRNAs derived from 17 discrete pre-miRNA hairpins, 9 of which represent homologues of EBV pre-miRNAs. In addition, we identified 9 candidates which are likely to be conserved between RRV and the related Japanese macaque herpesvirus (JMHV). Both viruses are furthermore predicted to encode several novel nonconserved miRNA candidates located in close proximity to the conserved ones. In contrast, while several other gammaherpesviruses are predicted to encode large numbers of miRNAs at conserved genomic locations, the miRNAs themselves are unrelated in sequence.
MATERIALS AND METHODS
Cell lines.
The rLCV latently infected rhesus macaque cell lines 260-98 and 211-98 (23) were maintained in RPMI medium containing 20% fetal bovine serum, 2 mM glutamine, and antibiotics. The EBV-positive Burkitt's lymphoma-derived B-cell lines Jijoye and Raji as well as the EBV-negative B-cell line BJAB were maintained in RPMI medium containing 10% fetal bovine serum, 2 mM glutamine, and antibiotics. The EBV-positive nasopharyngeal carcinoma (NPC)-derived cell line C666-1 (10) was maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum, 2 mM glutamine, and antibiotics.
Northern blot analysis.
Total RNA was harvested using RNA-Bee (AMS Biotechnology) according to the manufacturer's instructions and analyzed by Northern blotting as described previously (30). Briefly, 14 μg of total RNA was electrophoresed through a 15% acrylamide urea denaturing gel and electroblot transferred to Zeta-Probe GT membranes (Bio-Rad). Blots were hybridized to radiolabeled antisense oligonucleotide probes in ExpressHyb (BD Biosciences Clontech) hybridization buffer and subjected to autoradiography. For each hairpin, two different oligonucleotide probes complementary to the proximal (relative to the hairpin loop) 35 nucleotides of each hairpin arm were used.
Cloning of small RNAs.
miRNAs were cloned using a modified version of the protocol previously described by Pfeffer et al. (21). Briefly, total RNA was run on a denaturing 15% polyacrylamide gel, and the region of the gel containing small RNAs (∼10 to 40 nt) was excised. Gel pieces were eluted overnight with 2 to 3 volumes of 0.3 M NaCl, and RNA was ethanol precipitated and dephosphorylated. Small RNAs were subsequently ligated to a 3′ linker (5′-PrUrUrUCTGTAGGCACCATCAATGTCAAGTCGGAAddC-3′ [P, phosphate; r, ribose; dd, dideoxyribose]) using T4 RNA ligase (Fermentas). Ligation products were gel purified as described above, phosphorylated, and then ligated to a 5′ linker (5′-TGTTACGGCACCTCAGTTGATCAGAGCCCArGrGrG-3′). Subsequently, cDNAs were synthesized using SuperScript III reverse transcriptase (Invitrogen) and PCR amplified and the resulting products were subjected to TA cloning (Invitrogen). To map 5′ termini of predicted miRNAs, the cloned library was subjected to conventional or seminested PCRs using 3′ primers specific for the 3′ region of the predicted miRNAs and linker-specific 5′ primers. The miRNA-specific primers were designed such that at least 4 nucleotides of the 5′ ends of mature miRNA were not present in the primer but were derived from the amplified miRNA sequence. The resulting PCR products were subjected to TA cloning and sequenced.
Computational prediction of conserved pre-miRNA candidates.
The ab initio prediction software VMir, used to identify miRNA candidates in viral genomes, has been described previously (15, 29). Briefly, VMir slides a sequence window of adjustable size across the viral genomes and then employs the RNAfold algorithm (16) to perform a structure prediction by minimal free energy folding. Pre-miRNA candidates are identified and scored based on comparison to structural features of known pre-miRNA hairpins. The program can be downloaded at http://www.hpi-hamburg.de/research/departments-and-research-groups/anti-viral-defense-mechanism/software-download.html. All user-adjustable prediction parameters employed in this study were set to their default values (see reference 15 for details). Hairpin filtering parameters were as follows: minimum (min.) score, 135; min. window count, 25; min. stem-loop length, 50; maximum (max.) stem-loop length, 220. These settings are of moderate stringency, and 94% of all known gammaherpesvirus miRNAs were retained for further analysis. To identify conserved miRNAs, all gammaherpesvirus genomes were subjected to pairwise alignments using the BLAST (basic local alignment tool) algorithm (1). Overall sequence identity of aligned genomes was determined by calculating the percentage of nucleotides which registered as being conserved in any of the aligned segments from the BLAST output. We used the bl2seq executable from the v2.2.15 BLAST package (available at ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/). All BLAST parameters except for word size (-w) and gap open penalty values were set to their default values. The word size parameter was decreased from its default value of 11 to 7 in order to also detect relatively short segments of consecutively conserved nucleotides (such as a conserved seed region), and the gap open penalty was decreased from 3 to 2 since pre-miRNA hairpins should be able to accommodate small deletions or insertions as long as the hairpin structure is conserved. Within the genomic alignments, we then identified all pairwise aligned sequences of predicted pre-miRNA candidates which met the following criteria. First, the core region of either hairpin region, consisting of the terminal loop and the proximal 35 nt of the 5-prime and 3-prime arms, did not overlap with any annotated CDS GenBank feature. Second, at least one of the hairpin arms had to harbor a conserved region of at least 6 consecutive nucleotides located within 35 nt of the terminal loop (i.e., the region expected to harbor the mature miRNA). Lastly, the predicted apical positions of the aligned hairpin structures had to be located within a distance of maximally 10 nucleotides (i.e., the overall structure arrangement had to be conserved). Candidates which had passed this filtering procedure were then subjected to a second pairwise BLAST alignment of the isolated hairpin sequences. To ensure significant homology, instead of the default value of 10 we used a stringent expect value cutoff (-e parameter) value of 0.01 in this second alignment (this corresponds to a P value of approximately 0.01; while larger expect values cannot be readily compared to P values, small expect and P values of 0.01 or less are nearly identical). Experimentally confirmed pre-miRNAs which had not registered in the above screen but which were located at homologous regions of the rLCV and EBV genomes were also subjected to pairwise alignments using reduced-stringency parameters (expect value cutoff, 0.05; gap open penalty, −1) in order to detect more-distant relationships.
RESULTS
Computational analysis of conserved gammaherpesvirus pre-miRNAs and pre-miRNA candidates.
We have previously developed VMir, a computational algorithm for the ab initio prediction of putative pre-miRNA stem-loop structures in viral genomes, which has been successfully applied to identify miRNAs in the genomes of several viruses of the herpesvirus and polyomavirus families (15, 26, 27, 29-31). The program uses the RNAFold algorithm (16) for the prediction of secondary structures by minimal free energy folding in sequence windows tiled across the viral genome and then scores stem-loop structures by comparing their structural features to those of known pre-miRNA hairpins.
To investigate whether yet-unknown conserved gammaherpesvirus miRNAs exist, we performed a global analysis of miRNA conservation which includes predicted as well as known miRNAs. For this purpose, we first carried out a VMir prediction for all 14 reference sequence gammaherpesvirus genomes currently deposited in GenBank. Four of the analyzed genomes (callitrichine herpesvirus 3 [CaHV-3], rLCV, and EBV types I and II) belong to the lymphocryptovirus genus, while the remainder are rhadinoviruses. VMir predictions were carried out using default conditions and subsequently filtered using modest-stringency parameters (see Materials and Methods for details). All of the 68 known gammaherpesvirus-encoded pre-miRNAs currently listed in the miRNA registry (13, 14) were detected by VMir, and 64 (94%) of these passed the initial filtering step. To prevent the identification of hairpins as being conserved due merely to the presence of overlapping coding regions, we next eliminated hairpins which overlapped with any annotated GenBank CDS feature. This step removed another 3 known miRNA precursors, retaining 90% of all known gammaherpesvirus pre-miRNAs for further analysis. Considering all stem-loop structures, a total of 42,356 predicted hairpins were analyzed, and 810 of these passed our filtering procedure for potential pre-miRNA candidates. Due to the partially inverted nature of RNA sequences able to form fold-back structures, stem-loops are often predicted also for the reverse complement of a hairpin sequence. As a result, frequently both strands of a genomic pre-miRNA score prominently during the computational prediction of pre-miRNA hairpins, although in vivo usually only one of the strands is transcribed and produces a bona fide miRNA (the exceptions to this rule are a few miRNAs produced from bidirectionally transcribed loci in mouse cytomegalovirus and herpes simplex virus type 1 [6, 11, 33]). In our contingent of filtered gammaherpesvirus hairpins, 40 stem-loops represented such complementary-strand equivalents of known pre-miRNAs. As both sense and antisense hairpin sequences will register as being conserved in a sequence alignment, for our further conservation-based analysis we thus assigned our predictions to genomic hairpin loci, grouping sense and antisense hairpins whenever their apical positions (center of the terminal loop) fell within 10 nucleotides of one another. According to this criterion, the 810 gammaherpesvirus stem-loop structures mapped to a total of 607 hairpin loci, 60 of which represented known loci and 547 of which represented candidates for novel gammaherpesvirus miRNA loci. All hairpin loci as well as their individual predicted hairpin structures and sequences are given in data file S1 in the supplemental material.
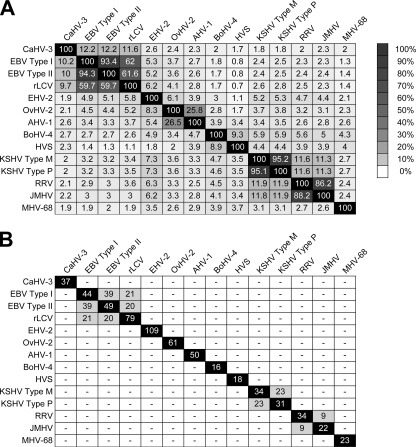
Next, we performed BLAST alignments of all possible pairwise combinations of the gammaherpesvirus genomes for which pre-miRNA candidates had been predicted. Figure 1A shows the overall sequence identity of the various genome combinations as detected by the pairwise alignments. To identify putative conserved pre-miRNAs among the individual genome pairs, we then filtered our candidate miRNA loci for those in which at least one pre-miRNA candidate (i) aligned with at least one putative orthologue such that (ii) at least 6 consecutive nucleotides in one of the hairpin arms were conserved and for which furthermore (iii) a second BLAST alignment of the isolated hairpin sequences yielded an expect value equal to or below 0.01 (see Materials and Methods for details). Figure 1B summarizes the results of the analysis and shows the total number of putative conserved pre-miRNAs for each of the pairwise alignments (cells with white lettering on black background represent self-aligned genomes; the numbers in these cases thus represent the total numbers of loci initially predicted by VMir). A detailed output of all hairpin alignments and their BLAST scores is given in data file S2 in the supplemental material. In general, we did observe a close correlation between overall sequence conservation and conservation of predicted pre-miRNA candidates. Not surprisingly, a high degree of hairpin conservation was observed for the closely related strain variants EBV types I and II as well as KSHV types P and M. As has been noted before, the known KSHV-encoded miRNAs are well conserved in the P and M variants (19). Nine out of a total of 12 known KSHV pre-miRNAs also registered in our analysis, while the remaining 3 had failed the initial filtering process (two folded only in limited sequence context and therefore had been eliminated by our VMir quality filters, and one is located within an open reading frame [ORF]). An additional 14 hairpin loci are conserved between KSHV types P and M, 4 of which map to the region which also harbors the known miRNA cluster. Since none of these hairpins have emerged as pre-miRNAs in 4 independent studies which used cloning or computational prediction and/or microarray approaches to identify small RNAs in KSHV-infected cells (7, 15, 21, 24), we assume that these candidates represent false-positive predictions. Alternatively, it is possible that some of the candidates produce mature miRNAs at levels which are below the detection limits of the techniques used in the above-described studies. All of the 23 EBV-encoded miRNAs which were known at the time of this study also registered in the comparison between EBV types I and II. An additional 15 loci harbor conserved structure predictions which, as discussed above for KSHV, may represent false positives.
FIG. 1.
Global analysis of conserved gammaherpesvirus pre-miRNA candidates. (A) Sequence identity of gammaherpesvirus genomes. The indicated pairs were subjected to pairwise BLAST alignments, and maximum sequence identity was determined as described in Materials and Methods. Sequence identity is shown as a percentage and additionally indicated by the degree of shading. (B) Prediction of conserved pre-miRNA candidates. Potential pre-miRNA hairpins were predicted with VMir, and conserved hairpin loci were identified using sequence/structure alignments (see text for details). Cells shown in black represent self-aligned genomes; in these cases the given numbers represent the total numbers of candidate loci predicted by VMir. All GenBank accession numbers are given in data file S1 in the supplemental material.
Among the more distantly related genomes, most miRNAs were predicted to be shared by EBV and rLCV, two viruses which have been noted before to share 7 pre-miRNAs (8). Our analysis, however, suggested a significantly higher number of conserved loci (21 and 20 for EBV types I and II, respectively [Fig. 1 and 2A]), several of which corresponded to novel pre-miRNA predictions. As discussed further below, we were able to experimentally confirm all but one of the novel predictions as bona fide pre-miRNAs. Therefore, the higher degree of sequence diversity (∼40%) between EBV and rLCV was sufficient to eliminate nearly all false-positive pre-miRNA predictions, indicating that such hairpins are significantly less conserved than are authentic pre-miRNAs.
FIG. 2.
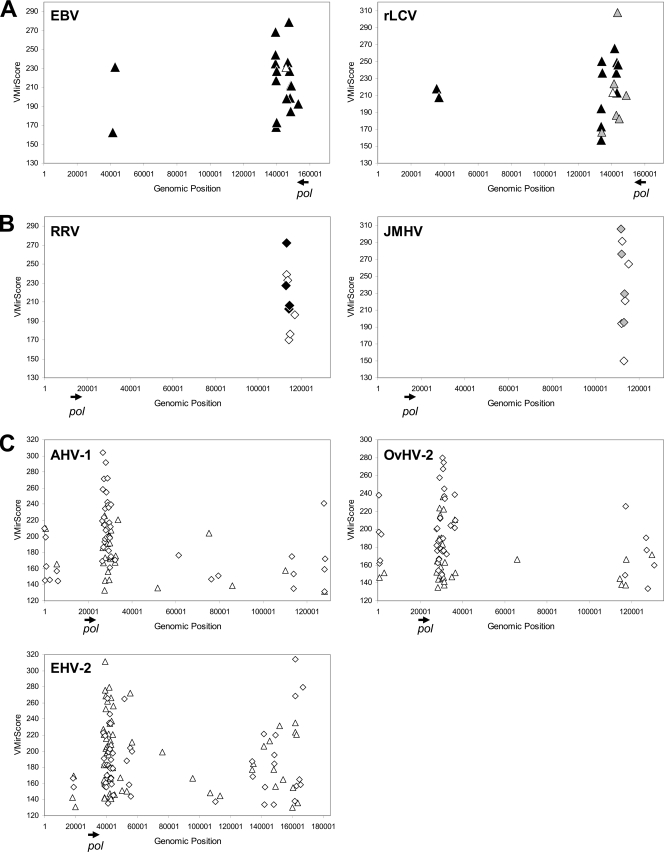
VMir prediction of conserved and nonconserved gammaherpesvirus pre-miRNAs. VMir prediction of conserved (A and B) and nonconserved (C) pre-miRNA hairpins in select gammaherpesvirus genomes. Pre-miRNAs are shown according to their genomic location (horizontal axis) and hairpin score assigned by VMir (vertical axis). Triangles and diamonds indicate hairpins in direct and reverse orientations, respectively. The genomic position and orientation of the open reading frame encoding the DNA polymerase are indicated by arrows beneath each of the diagrams. (A and B) The diagrams depict known or predicted pre-miRNAs in direct orientation conserved between rLCV and EBV (A) or those in reverse orientation conserved between RRV and JMHV (B). Known pre-miRNAs are shown as black symbols, novel predicted orthologues of known miRNAs are shown as gray symbols, and homologous hairpins which represent novel predictions in each of the paired genomes are indicated by open symbols. (C) Genomic positions of nonconserved pre-miRNA candidates which map to noncoding and nonrepetitive regions of the AHV-1, OvHV-2, and EHV-2 genomes.
In addition to the EBV/rLCV miRNAs, our analysis also suggested the existence of 9 pre-miRNA loci conserved between RRV and JMHV. RRV, a rhadinovirus isolated from rhesus macaques (Macaca mulatta), is distantly related to KSHV and has been previously found to encode 8 miRNAs (25). Although the RRV miRNAs are found at the same genomic location as that of the KSHV-encoded miRNAs, they do not exhibit any sequence homology (25), a finding which is confirmed by our analysis (Fig. 1B). While RRV and KSHV are only approximately 12% identical, however, RRV is much more closely related to JMHV, a rhadinovirus isolated from the Japanese macaque or snow monkey (Macaca fuscata). Our analysis predicted the existence of 9 conserved hairpin loci, all of which map to the region harboring the already-known RRV miRNAs (Fig. 2B; see also the alignments in data file S2 in the supplemental material). Four of the conserved hairpin pairs correspond to known RRV pre-miRNAs (rrv-miR-rR1-3, -4, -6, and -7 [black diamonds in Fig. 2B]) aligned with candidate pre-miRNAs in JMHV (gray diamonds in Fig. 2B). The remaining 5 candidates represent novel predictions of RRV as well as JMHV pre-miRNAs (Fig. 2B, open diamonds). Of course, given the ∼87% sequence identity observed in whole-genome alignments of RRV and JMHV, as with the subtype alignments of EBV and KSHV, one may argue that these predictions may well represent false positives. However, when we investigated the regions harboring the miRNA clusters in pairwise alignments, we observed that these regions are significantly more divergent than the rest of the genome, showing only approximately 67% sequence identity (see Fig. S1 in the supplemental material). In contrast, the corresponding regions are between 88% and 98% identical in the EBV or KSHV subtype alignments. Consequently, the detection of conserved hairpins within the generally less conserved background should be more significant in the case of RRV and JMHV. We therefore strongly suspect that some or all of the novel predictions represent authentic conserved pre-miRNAs. To what extent this is the case, however, will have to await experimental validation. Closer inspection of the VMir prediction data for both viruses furthermore reveals the presence of additional high-scoring candidates which map to the regions of the conserved miRNAs but which did not register in our conservation analysis (see data file S1 in the supplemental material), suggesting the existence of novel nonconserved miRNAs in RRV as well as JMHV.
With the exception of the above cases, our analysis did not suggest the existence of conserved pre-miRNAs in the gammaherpesvirus family. However, as shown in Fig. 2C (see also data file S1 in the supplemental material), the primary VMir analysis predicted the existence of large clusters of high-scoring pre-miRNA candidates located in non-protein-encoding regions of equine herpesvirus 2 (EHV-2), ovine herpesvirus 2 (OvHV-2), and alcelaphine herpesvirus 1 (AHV-1). Furthermore, in each of these viruses the predicted cluster is located immediately downstream of the polymerase gene (indicated by arrows in Fig. 2), an arrangement which is highly reminiscent of the location of the BamHI rightward transcript (BART) miRNA clusters in EBV and rLCV. Thus, while the positions of these miRNAs are conserved in the EHV-2, OvHV-2, and AHV-1 genomes, their sequences are not.
Experimental validation of novel conserved and nonconserved lymphocryptovirus miRNAs.
The known EBV and rLCV miRNAs are transcribed in direct (relative to the RefSeq genome sequence) orientation from two discrete genomic locations: either the BHRF1 locus or the region encompassed by the EBV-encoded BARTs and the homologous region in rLCV. Indeed, all of the novel pre-miRNAs predicted during our conservation-based analysis mapped to the region harboring the known miRNA clusters (Fig. 2A; see also data file S2 in the supplemental material). Furthermore, our initial VMir analysis had predicted the existence of several unique pre-miRNAs located within or close to these clusters (see data file S1 in the supplemental material), and we thus suspected the existence of novel conserved as well as nonconserved rLCV and EBV miRNAs. Regardless of their conservation status, we therefore investigated all predicted hairpins which mapped to the miRNA cluster regions (i.e., the genomic segments between ORFs BHLF1 and BFLF2 [BHRF1 cluster] or BILF2 and BALF5 [BART cluster]) for their ability to produce mature miRNAs. While reviewing the data from our conservation-based analysis, we also noted that one putative conserved, reverse-oriented rLCV hairpin (MR1588 [see data file S2 in the supplemental material]) aligned with an EBV hairpin (MR1392) which represented the reverse complement of ebv-miR-BART9. No putative rLCV homologue had been predicted, since the minimal free energy structure of the reverse complement rLCV sequence does not represent a hairpin. However, suboptimal folding within an energy range of 0.5 kcal/mol revealed an alternative hairpin structure with high similarity to known pre-miRNA stem-loops, and we therefore also designed oligonucleotide probes to detect potential miRNAs produced from this hairpin.
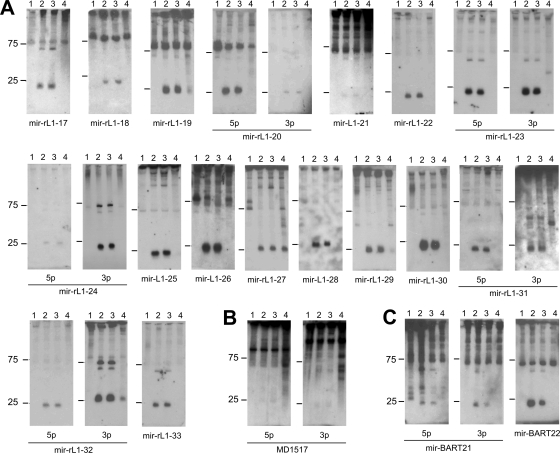
We then carried out Northern blot assays using RNA from the two rLCV-infected B-cell lines 260-98 and 211-98, two Burkitt's lymphoma lines infected with either a type I or a type II strain of EBV (Raji and Jijoye, respectively), an EBV type I-infected epithelial cell line derived from nasopharyngeal carcinoma (C666-1), and the EBV- and rLCV-negative Burkitt's lymphoma cell line BJAB. As shown in Fig. 3A, we were able to confirm the existence of 22 novel rLCV-encoded mature miRNAs which are derived from 17 distinct pre-miRNA hairpins. The pre-miRNAs include all but one candidate from our initial conservation-based analysis as well as the hairpin representing the reverse complement of the rLCV hairpin MR1588 (rlcv-miR-rL1-25 in Fig. 3). Several of the probes designed to detect rLCV miRNAs also showed cross-reactivity with RNA from EBV-infected C666-1 cells, suggesting extensive sequence conservation of the mature molecules. Compared among each other, there was significant variability in the relative expression levels of the various rLCV miRNAs, similar to what has been previously observed for the BART miRNAs (8, 15). However, the levels of expression of individual miRNAs in both of the rLCV-infected cell lines investigated here were remarkably similar (compare lanes 2 and 3 in Fig. 3A). Besides the 22 miRNAs shown in Fig. 3A, we also detected faint but distinct signals of the size expected for a mature miRNA with probes complementary to both arms of the nonconserved rLCV hairpin MD1517 (Fig. 3B). Detection of these bands was reproducible, and the signals were observed only in rLCV-positive cells. However, as we were unable to clone mature miRNAs from this hairpin (see below), we cannot exclude the possibility that the signals result from cross-reactivity of the probes with a cellular RNA species, especially given the relatively high GC content (∼80%) of the hairpin sequences (see data file S1 in the supplemental material). We thus refrain from designating this hairpin a pre-miRNA but include the Northern blot here for completeness.
FIG. 3.
Northern blot confirmation of predicted rLCV and EBV miRNAs. For the confirmation of predicted novel rLCV (A and B) and EBV (C) miRNAs, total RNA isolated from infected or uninfected cells was analyzed with probes specific for the indicated miRNAs. For the rLCV candidate hairpin MD1517 blot shown in panel B, we were unable to derive mature miRNA sequences by cloning of small RNAs (see text for details). (A and B) Lanes 1, BJAB (rLCV/EBV-negative B cells); lanes 2, 211-98 (rLCV-infected rhesus macaque cell line); lanes 3, 260-98 (rLCV-infected rhesus macaque cell line); lanes 4, C666-1 (EBV-positive epithelial cells). (C) Lanes 1, BJAB (rLCV/EBV-negative B cells); lanes 2, C666-1 (EBV-positive epithelial cells); lanes 3, Jijoye (EBV-positive B cells); lanes 4, Raji (EBV-positive B cells).
In addition to the novel rLCV miRNAs, our Northern blot assays also identified three EBV miRNAs (originating from two discrete hairpins, ebv-miR-BART21 and -22) which had been unknown at the time of our analysis but which in the meantime have been reported by another group (38). The authors of the study cloned the miRNAs from NPC tissue and also demonstrated their expression in Jijoye cells; the data presented in Fig. 3C thus confirm the findings by Zhu et al. and additionally show that the miRNAs are not expressed (or only very weakly expressed) in Raji cells (lanes 4 in Fig. 3C), similarly to what has been previously shown for the other EBV miRNAs mapping to the BART cluster (8).
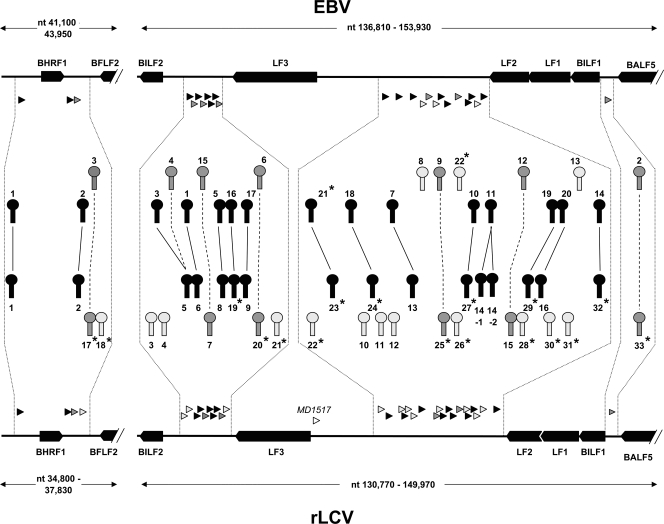
Figure 4 depicts the genomic location and conservation status of all EBV and rLCV pre-miRNAs and summarizes the result of our analysis. Pre-miRNAs of homologous origin are connected by lines, with dotted or solid lines indicating conservation of seed sequences (see next paragraph). Detailed alignments of all pre-miRNAs are provided in data file S3 in the supplemental material. In addition to the 20 authentic pre-miRNAs which had registered during our primary conservation-based analysis, Fig. 4 also indicates a common ancestry for rlcv-miR-17 and ebv-miR-BHRF1-3, as well as rlcv-miR-20 and ebv-miR-BART6 (see also alignments in data file S3 in the supplemental material). While these pre-miRNAs are in fact distantly related, the homology was too weak to be picked up by the stringent BLAST procedure of our original screen and emerged only when the pre-miRNAs were compared using relaxed-stringency parameters (see Materials and Methods for details).
FIG. 4.
Genomic locations of EBV- and rLCV-encoded miRNAs. Shown is a map of the two genomic regions encoding EBV and rLCV miRNAs. Open reading frames at each of the loci are shown as black arrows, and the locations of pre-miRNAs are indicated by triangles underneath the EBV and above the rLCV backbone. Pre-miRNA hairpin loci are depicted in higher magnification toward the center: light gray hairpins drawn proximal to the backbone symbolize nonconserved pre-miRNAs, and dark gray or black hairpins depict homologous hairpins. Solid lines connecting black hairpins indicate pairs in which the seed region of at least one mature miRNA has been conserved, whereas dark gray hairpins connected by dotted lines indicate orthologues in which the seed regions have diverged. Novel miRNAs identified in this study are marked with an asterisk.
Out of the 17 novel rLCV pre-miRNAs identified in this study, two map to the BHRF1 locus. Whereas miR-rL1-17 is a distant orthologue of miR-BHRF1-3, miR-rL1-18 does not show any recognizable sequence homology. The remaining miRNAs are located in the region homologous to the EBV BART locus, where a total of 19 hairpins show evidence of common ancestry. Interestingly, ebv-miR-BART3 and -BART4 can both be aligned with rlcv-miR-rL1-5 (see data file S3 in the supplemental material), indicating duplication and diversification of a common ancestor. Likewise, the pre-miRNA hairpin originally identified as rlcv-miR-rL1-14 (designated rlcv-miR-rL1-14-1 in Fig. 4) appears to have undergone a relatively recent duplication event, with its paralogue (rlcv-miR-rL1-14-2 in Fig. 4; see also data file S3 in the supplemental material) having not yet significantly diverged. In fact, the sequences of the two hairpins are identical except for 2 nucleotide exchanges within their terminal loop sequences. Thus, both stem-loops may give rise to the mature miRNAs rlcv-miR-rL1-14-5p and -3p identified by Cai and colleagues (8).
Determination of 5′ ends of mature miRNAs.
Our initial analysis of miRNA conservation was based on the alignment of hairpin sequences and structures, thus generally identifying pre-miRNAs which share a common ancestor. If miRNAs produced from these precursors recognize evolutionarily conserved target sequences, one would expect that especially the regions encoding the mature seed sequences should be conserved. To judge seed conservation, we thus sought to determine the 5′ ends of the mature miRNAs. Due to variability in the precise cleavage sites chosen by Drosha and Dicer, as well as frequent inaccuracies in the prediction of terminal loop size and position, the computational identification of these termini is notoriously difficult. Therefore, to experimentally determine the exact 5′ termini of the novel miRNAs identified here, we isolated small RNA moieties from EBV-infected C666-1 or rLCV-infected 211-98 cells, ligated them to linkers, and subjected them to a standard cloning protocol. From the resulting library, we then amplified and sequenced the 5′ ends of the mature miRNAs, using primers complementary to the linker and the 3′ end of the miRNA. Primer design was chosen such that the amplified sequences contained at least 4 specific nucleotides not covered by the primers, and at least 3 independent clones were sequenced to determine the 5′ ends. Not surprisingly, we observed 5′-end heterogeneity for some of the mature miRNAs, indicating variable Drosha and/or Dicer cleavage. In these cases, additional clones were sequenced to determine the major miRNA species. We were able to determine 5′-end sequences for all miRNAs shown in Fig. 3A but repeatedly failed to obtain clones for hairpin MD1517, indicating that the bands detected on Northern blots as shown in Fig. 3B may indeed represent false-positive signals resulting from cross-hybridization. A summary of the results from our sequencing of rLCV miRNAs and the frequency of obtained clones are given in Table 1 (note that we did not experimentally determine the precise 3′ termini but only estimated their position, assuming an average miRNA length of 22 nt). The termini of the three novel EBV miRNAs were the same as those determined by Zhu et al. (38) and are thus not reproduced here.
TABLE 1.
Sequences of novel rLCV miRNAs
| Name | Sequence | Frequency |
|---|---|---|
| rlcv-miR-rL1-17 | UGCUUCGCCCUCUCCAUCAUAA | 4 |
| rlcv-miR-rL1-18 | UUAGCCCCUCCCCAUCAUCUUG | 4 |
| UAGCCCCUCCCCAUCAUCUUG | 2 | |
| rlcv-miR-rL1-19 | UAUAGAUAGCGUGGGUGUGUGA | 6 |
| AUAGAUAGCGUGGGUGUGUGA | 2 | |
| rlcv-miR-rL1-20-5p | UAAAGGGUAGUGUGGUUCACAG | 4 |
| rlcv-miR-rL1-20-3p | UGUGGAUCUAACUUCCCUUAGU | 3 |
| rlcv-miR-rL1-21 | AGGCCUCUUCUCACAAUUCUAA | 3 |
| GCCUCUUCUCACAAUUCUAA | 1 | |
| rlcv-miR-rL1-22 | UCACCGUUUCAUCCCCACGAUU | 3 |
| rlcv-miR-rL1-23-5p | UCACUAGUGCUGGCACCUAAGA | 3 |
| rlcv-miR-rL1-23-3p | UUAGUUGUCUGCACUGGAGAGU | 3 |
| rlcv-miR-rL1-24-5p | CUCAAGUUCUCAUUUCCAAUAC | 4 |
| rlcv-miR-rL1-24-3p | UAUCGGAGAUAGGACUUGAUAC | 4 |
| rlcv-miR-rL1-25 | UGACAAUUUAAUGGGUCUAGUA | 3 |
| rlcv-miR-rL1-26 | UACGAUUCUCCGGGUUUAGCAG | 3 |
| rlcv-miR-rL1-27 | CACAUAACCAUGGAGGUGGUUG | 4 |
| ACAUAACCAUGGAGGUGGUUG | 1 | |
| rlcv-miR-rL1-28 | GAGGAAAGUAUCGCCUUCUAGA | 3 |
| rlcv-miR-rL1-29 | UUUUGUUUGCUUGGGACUGCAG | 3 |
| rlcv-miR-rL1-30 | UUUCGUUUUCGAAGGUCGGCUG | 3 |
| UUCGUUUUCGAAGGUCGGCUG | 1 | |
| rlcv-miR-rL1-31-5p | UCGCUUACGCCGACCGCGUAAC | 3 |
| GUCGCUUACGCCGACCGCGUAAC | 1 | |
| rlcv-miR-rL1-31-3p | UAGGCGUGUCGUCGUAAGCAGC | 3 |
| rlcv-miR-rL1-32-5p | UAACUUACUACUGCCGCUUUAC | 3 |
| GUAACUUACUACUGCCGCUUUAC | 1 | |
| rlcv-miR-rL1-32-3p | UAAAUGCGAGCAGUAGUAGGCG | 3 |
| rlcv-miR-rL1-33 | CAUUGUCUUCAUUUGCCCUUGC | 3 |
Figure 5 shows the predicted structures of the 17 novel rLCV pre-miRNA stem-loops. Given known requirements for Drosha processing (17, 36) and the experimentally determined position of the mature miRNAs, for three hairpins (rlcv-miR-rL1-23, -25, and -28) both the minimal free energy structures and the energetically reasonable alternative stem-loop predictions that are more likely to represent the authentic Drosha substrates are shown.
FIG. 5.
Predicted hairpin structures of novel rLCV pre-miRNAs. The structures of rLCV pre-miRNAs were predicted by minimal free energy folding using the RNAfold algorithm (16). The positions of mature miRNAs are indicated by shaded residues (note that only the 5′ termini of mature miRNAs were determined experimentally, whereas the 3′ ends are given based on the assumption that the miRNA is of the average [22-nt] size). Free energy values are given underneath each hairpin. For rlcv-miR-rL1-23, -25, and -28, alternative predictions which are likely to represent the authentic stem-loop structures recognized and cleaved by Drosha are shown underneath the minimal energy structures.
In Fig. 6, we present alignments of all orthologous pairs of mature rLCV and EBV miRNAs as detected during our analysis (note that for the sake of completeness we have also reproduced the 9 orthologous pairs already identified by Cai et al. [8], marked with an asterisk in Fig. 6). Alignments of the pre-miRNA hairpin sequences and their structures are given in data file S3 in the supplemental material. Seed conservation of mature miRNAs is also indicated in Fig. 4, where solid lines connect conserved hairpins for which the seed sequence of at least one mature miRNA has been conserved whereas dotted lines indicate pre-miRNAs which harbor no conserved seed region or which produce miRNAs from different arms of the orthologous stem-loops in EBV and rLCV. Taken together, EBV and rLCV are now known to encode a total of 44 and 43 mature miRNAs, produced from 25 and 34 discrete pre-miRNA stem-loops, respectively. According to our analysis, 39 (or ∼85%) of all mature EBV and 31 (∼72%) of the mature rLCV miRNAs map to 22 pre-miRNA hairpins which show signs of common ancestry in the two viruses. Of the mature miRNAs, a total of 16 have conserved their seed sequences, which corresponds to approximately 35% of all miRNAs encoded by EBV and rLCV. Considering the 22 orthologous pre-miRNA hairpins, 14 of these harbor at least one miRNA with a conserved seed sequence.
FIG. 6.
Alignment of novel mature rLCV miRNAs and their EBV orthologues. Shown are alignments of all mature EBV and rLCV miRNA sequences which map to homologous pre-miRNA hairpins, as detected in our study. In cases where no mature miRNA is produced from the homologous arm of the aligned hairpin, genomic sequences are shown. Alignments of miRNAs which have been previously identified as being conserved (8) are marked with an asterisk. Sequences of mature miRNAs as determined in this or previous studies (8, 18, 22) are shown shaded gray. Dark gray shading indicates sequence variations (Table 1 shows variations observed in this study) which may arise due to variable Drosha and/or Dicer cleavage during miRNA maturation or result from differential end processing after cleavage.
DISCUSSION
The vast majority of the currently known viral miRNAs are encoded by members of the herpesvirus family. However, while the propensity to encode miRNAs appears to be a conserved feature of herpesviruses, only a few examples of evolutionarily conserved miRNAs have been reported so far (8, 33). Here, we have performed a global analysis of miRNA conservation among all 14 fully sequenced gammaherpesvirus genomes currently deposited in GenBank, using an approach which combines ab initio computational prediction with sequence/structure alignments to identify conserved candidates. Our analysis is thus not limited to the contingent of presently known viral miRNAs but also includes gammaherpesviruses which hitherto have not been analyzed for the presence of such molecules, as well as candidate miRNAs which may have evaded detection in previous studies aimed at identifying viral miRNAs in KSHV-, EBV-, rLCV-, RRV-, or MHV-68-infected cells. The results of our study indicate that evolutionary conservation of gammaherpesvirus miRNAs is indeed rare. In general, we did not find evidence of miRNA conservation among genomes which exhibit less than 60% overall sequence identity. For example, OvHV-2 and AHV-1 show approximately 26% sequence identity and are furthermore both predicted to encode significant numbers of miRNAs, but none of the candidate miRNAs show recognizable sequence homology. Strikingly, however, despite the absence of primary sequence conservation, the predicted miRNA clusters of OvHV-2 and AHV-1 as well as EHV-2 are found at the same genomic location as the BART miRNA cluster of EBV and the orthologous cluster of rLCV, i.e., in a region immediately downstream of the DNA polymerase gene which is largely devoid of open reading frames. In contrast, the miRNAs of KSHV and RRV (and probably also JMHV) map to a region which is the positional equivalent of the BHRF miRNA cluster in EBV and rLCV. It is certainly possible that OvHV-2, AHV-1, and EHV-2 encode additional miRNAs at a location similar to that in KSHV and RRV, although their numbers are expected to be smaller than those expressed from the large cluster downstream of the polymerase gene (Fig. 2C). Likewise, whereas we did not observe large clusters of predicted miRNAs in the genomes of the lymphocryptovirus CaHV-3 or the rhadinoviruses bovine herpesvirus 4 (BoHV-4) and herpesvirus saimiri (HVS), each of these viruses harbors at least a few candidates at the positions equivalent to those of the EBV- and/or KSHV-encoded miRNAs (see data file S1 in the supplemental material). Whether these predictions represent bona fide miRNAs, however, has to await experimental confirmation.
While the inclusion of computationally predicted miRNAs has enabled us to perform a global analysis of gammaherpesvirus miRNA conservation, one has to be aware of the limitations of this approach. First and chief among these are the limitations inherent to all computational miRNA prediction methods. All ab initio pre-miRNA prediction algorithms face the challenge of striking an appropriate balance between accuracy and sensitivity of the predictions, as there are only a few structural features which distinguish a bona fide pre-miRNA stem-loop from other hairpin structures (15). For cellular miRNAs, elimination of candidates which are not evolutionarily conserved is the most widely used and efficient method to weed out false positives—such filters, for obvious reasons, could not be used here. In order to maintain a high degree of sensitivity, VMir was deliberately designed to err rather on the side of overpredicting candidates. We reasoned this to be greatly favorable for viral genomes, as the total number of such false positives remains manageable due to the comparatively small genome size. Under the moderate-stringency conditions used here (see Materials and Methods for details), we detect and retain greater than 90% of all known gammaherpesvirus miRNAs for our analysis, but the predictions will undoubtedly also contain false positives. This does not compromise our conclusion that conservation is a rare feature among gammaherpesvirus miRNAs, as our analysis is over- rather than undersensitive. However, researchers interested in the experimental confirmation of novel predicted miRNAs given in the data in the supplemental material should be aware of the likely presence of false positives. Of course, despite its comparatively high sensitivity our prediction method may also have missed some candidates. Most such false negatives are expected to arise due to inaccurate structure predictions. To keep the computational load manageable, VMir (like most other prediction methods) considers only the lowest free energy structure. Even though pre-miRNA hairpins generally exhibit low free energy values (4), this may result in some genuine precursors being missed. One such example is rlcv-miR-rL1-25, which presented a favorable structure only upon suboptimal folding. While there may be similar cases of conserved or nonconserved miRNAs, the fact that VMir predictions were made for nearly all known gammaherpesvirus miRNAs indicates that their numbers are likely to be low. Lastly, an additional potential source for false negatives may result from the necessity of eliminating candidates which overlap with open reading frames. In these cases, it would be impossible to decide to what extent sequence conservation is due to the preservation of miRNA function or protein coding capacity. However, as only three (one in KSHV and two in RRV) of all currently known gammaherpesvirus miRNAs are located in protein coding regions, we are confident that most authentic miRNAs were present within the contingent of predicted candidates.
While our analysis had predicted the presence of novel conserved as well as nonconserved miRNAs encoded by rLCV, EBV, RRV, and JMHV, we have performed experimental verification of the rLCV and EBV candidates only. In addition to the 17 novel rLCV and 2 EBV pre-miRNAs shown in Fig. 3, our conservation analysis also includes 7 EBV miRNAs (ebv-miR-BART15 to -22 [15]) which had not been identified in the study by Cai et al. (8) and which therefore have not been investigated with regard to their conservation status before. The overall results of our study of EBV and rLCV as shown in Fig. 4 provide a detailed picture of the evolution of miRNA clusters in these two viruses. Taken together, 22 (and therefore almost three times as many EBV and rLCV pre-miRNAs as previously thought) show signs of evolutionary conservation. However, not in every case does common ancestry also translate into conservation of mature miRNAs and especially conservation of miRNA seed regions. Taken together, roughly one-third of all mature EBV and rLCV miRNAs have conserved their seed sequences. Interestingly, although 7 of the 9 EBV miRNAs designated “star” species (miRNA*) map to hairpins which have homologues in EBV, none of the mature miRNAs* are conserved (Fig. 6; note that for ebv-miR-BART3-5p we have followed the original nomenclature of Cai et al. [8], who have isolated nearly equal numbers of 5p and 3p clones, whereas Landgraf et al. [18] designate ebv-miR-BART3-5p an miRNA*). At the same time, 6 out of the 7 major mature miRNAs produced from the same hairpins have conserved their seeds, indicating that they are subject to significantly higher evolutionary pressure than are the miRNAs*.
There are a few cases of homologous EBV and rLCV hairpins which undergo differential processing, leading to the production of miRNAs which are expected to be functionally different. This is most obvious in the case of ebv-miR-BHRF1-3/rlcv-miR-rL1-17 and ebv-miR-BART15/rlcv-miR-rL1-7, which appear to exclusively produce mature miRNAs from opposite arms of the EBV or rLCV hairpins. As ebv-miR-BHRF1-3 has been reported to target the T-cell-attracting chemokine CXCL-11 (35), it seems that this function is not conserved in rLCV. Another case is rlcv-miR-rL1-19. Compared to the orthologous ebv-miR-BART16, 6 of the sequenced rlcv-miR-rL1-19 clones display an additional nucleotide in position 1 and therefore have an altered seed sequence. However, the 2 remaining clones have maintained the same 5′ terminus and seed as those of ebv-miR-BART16. Alternative processing thus leads to the production of two different mature miRNA species, one of which may conserve the function(s) of ebv-miR-BART16, whereas the other may have evolved additional or different targets.
We expect that knowledge of the full complement of conserved and nonconserved miRNAs of EBV and rLCV will be helpful in the identification of functionally important viral or cellular miRNA targets. For example, ebv-miR-BART2 has been previously reported to negatively regulate the expression of the viral DNA polymerase encoded by BALF5 (3, 22). The BALF5 gene lacks a canonical polyadenylation signal, and its transcripts are thus extended through the region which encodes ebv-miR-BART2 on the opposite strand. As a result, ebv-miR-BART2 is perfectly complementary to the BALF5 transcripts and induces their cleavage in a small interfering RNA (siRNA)-like manner. While no equivalent miRNA in rLCV has been described before, we have identified rlcv-miR-rL1-33 as the evolutionary homologue of ebv-miR-BART2. Although the seed sequences of the mature miRNAs are not conserved, both miRNAs are nevertheless fully complementary to transcripts produced from the opposite strand of the parental genome. Interestingly, as in EBV, inspection of the sequences downstream of the BALF5 gene reveals the lack of a canonical polyadenylation signal in the rLCV genome, pointing toward a conserved function of ebv-miR-BART2 and rlcv-miR-rL1-33, despite the absence of seed conservation. In contrast, another function of ebv-miR-BART2 appears not to be conserved: ebv-miR-BART2 has also been reported to target MICB, a stress-induced natural killer cell ligand, thereby potentially helping virus-infected cells to evade the immune system (20). However, whereas the reported MICB target sequence is conserved in the rhesus macaque genome, the seed sequences of the viral miRNAs are not, and it thus seems unlikely that rlcv-miR-rL1-33 mediates negative regulation of MICB expression (it is of course possible that another rLCV miRNA counteracts MICB by targeting a different region of the transcript).
At first, it may seem that miRNAs which have diverged in their seed sequences also have acquired different target transcripts and thus have functionally diverged. However, the absence of seed conservation between homologous viral miRNAs should certainly not be taken as evidence of insignificant function, as it is reasonable to assume that at least some of these changes may reflect alterations which occur first in the target sequence, with the miRNA secondarily acquiring compensatory changes to maintain sequence complementarity. It would seem that this could be especially the case for cellular targets, as the host is not expected to gain any benefit from maintaining target site complementarity to a viral miRNA in the first place. Thus, the most significant evidence of a functionally important host target would be a site which has diverged in two host species but which is nevertheless targeted by homologous miRNAs having accumulated compensatory changes. Comparative genomic approaches to discover such or conventionally conserved target sites will require detailed knowledge of the full complement of miRNAs encoded by related viral species, as well as their evolutionary relationship. Our detailed study of EBV and rLCV miRNAs thus should greatly aid in identifying targets which have been functionally conserved throughout the evolution of these related viruses.
Supplementary Material
Acknowledgments
This work was funded, in part, by the Deutsche Forschungsgemeinschaft (GR 3316/1-1). The Heinrich Pette Institute is a member of the Leibniz Gemeinschaft (WGL) and is supported by the Free and Hanseatic City of Hamburg and the Federal Ministry of Health.
Footnotes
Published ahead of print on 4 November 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bartel, D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth, S., T. Pfuhl, A. Mamiani, C. Ehses, K. Roemer, E. Kremmer, C. Jaker, J. Hock, G. Meister, and F. A. Grasser. 2008. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 36:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnet, E., J. Wuyts, P. Rouze, and Y. Van de Peer. 2004. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20:2911-2917. [DOI] [PubMed] [Google Scholar]
- 5.Brodersen, P., and O. Voinnet. 2009. Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10:141-148. [DOI] [PubMed] [Google Scholar]
- 6.Buck, A. H., J. Santoyo-Lopez, K. A. Robertson, D. S. Kumar, M. Reczko, and P. Ghazal. 2007. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J. Virol. 81:13761-13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, X., A. Schafer, S. Lu, J. P. Bilello, R. C. Desrosiers, R. Edwards, N. Raab-Traub, and B. R. Cullen. 2006. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carthew, R. W., and E. J. Sontheimer. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, S. T., D. P. Huang, A. B. Hui, K. W. Lo, C. W. Ko, Y. S. Tsang, N. Wong, B. M. Whitney, and J. C. Lee. 1999. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int. J. Cancer 83:121-126. [DOI] [PubMed] [Google Scholar]
- 11.Dolken, L., J. Perot, V. Cognat, A. Alioua, M. John, J. Soutschek, Z. Ruzsics, U. Koszinowski, O. Voinnet, and S. Pfeffer. 2007. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J. Virol. 81:13771-13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottwein, E., N. Mukherjee, C. Sachse, C. Frenzel, W. H. Majoros, J. T. Chi, R. Braich, M. Manoharan, J. Soutschek, U. Ohler, and B. R. Cullen. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones, S. 2004. The microRNA registry. Nucleic Acids Res. 32:D109-D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34:D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofacker, I. L., W. Fontana, P. F. Stadler, S. Bonhoeffer, P. Tacker, and P. Schuster. 1994. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 125:167-188. [Google Scholar]
- 17.Kim, V. N., J. Han, and M. C. Siomi. 2009. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10:126-139. [DOI] [PubMed] [Google Scholar]
- 18.Landgraf, P., M. Rusu, R. Sheridan, A. Sewer, N. Iovino, A. Aravin, S. Pfeffer, A. Rice, A. O. Kamphorst, M. Landthaler, C. Lin, N. D. Socci, L. Hermida, V. Fulci, S. Chiaretti, R. Foa, J. Schliwka, U. Fuchs, A. Novosel, R. U. Muller, B. Schermer, U. Bissels, J. Inman, Q. Phan, M. Chien, D. B. Weir, R. Choksi, G. De Vita, D. Frezzetti, H. I. Trompeter, V. Hornung, G. Teng, G. Hartmann, M. Palkovits, R. Di Lauro, P. Wernet, G. Macino, C. E. Rogler, J. W. Nagle, J. Ju, F. N. Papavasiliou, T. Benzing, P. Lichter, W. Tam, M. J. Brownstein, A. Bosio, A. Borkhardt, J. J. Russo, C. Sander, M. Zavolan, and T. Tuschl. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall, V., T. Parks, R. Bagni, C. D. Wang, M. A. Samols, J. Hu, K. M. Wyvil, K. Aleman, R. F. Little, R. Yarchoan, R. Renne, and D. Whitby. 2007. Conservation of virally encoded microRNAs in Kaposi sarcoma-associated herpesvirus in primary effusion lymphoma cell lines and in patients with Kaposi sarcoma or multicentric Castleman disease. J. Infect. Dis. 195:645-659. [DOI] [PubMed] [Google Scholar]
- 20.Nachmani, D., N. Stern-Ginossar, R. Sarid, and O. Mandelboim. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376-385. [DOI] [PubMed] [Google Scholar]
- 21.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 22.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 23.Rivailler, P., A. Carville, A. Kaur, P. Rao, C. Quink, J. L. Kutok, S. Westmoreland, S. Klumpp, M. Simon, J. C. Aster, and F. Wang. 2004. Experimental rhesus lymphocryptovirus infection in immunosuppressed macaques: an animal model for Epstein-Barr virus pathogenesis in the immunosuppressed host. Blood 104:1482-1489. [DOI] [PubMed] [Google Scholar]
- 24.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schafer, A., X. Cai, J. P. Bilello, R. C. Desrosiers, and B. R. Cullen. 2007. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology 364:21-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo, G. J., C. J. Chen, and C. S. Sullivan. 2009. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology 383:183-187. [DOI] [PubMed] [Google Scholar]
- 27.Seo, G. J., L. H. Fink, B. O'Hara, W. J. Atwood, and C. S. Sullivan. 2008. Evolutionarily conserved function of a viral microRNA. J. Virol. 82:9823-9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skalsky, R. L., M. A. Samols, K. B. Plaisance, I. W. Boss, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836-12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sullivan, C. S., and A. Grundhoff. 2007. Identification of viral microRNAs. Methods Enzymol. 427:3-23. [DOI] [PubMed] [Google Scholar]
- 30.Sullivan, C. S., A. T. Grundhoff, S. Tevethia, J. M. Pipas, and D. Ganem. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682-686. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan, C. S., C. K. Sung, C. D. Pack, A. Grundhoff, A. E. Lukacher, T. L. Benjamin, and D. Ganem. 2009. Murine polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology 387:157-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umbach, J. L., and B. R. Cullen. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umbach, J. L., M. F. Kramer, I. Jurak, H. W. Karnowski, D. M. Coen, and B. R. Cullen. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visone, R., and C. M. Croce. 2009. MiRNAs and cancer. Am. J. Pathol. 174:1131-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia, T., A. O'Hara, I. Araujo, J. Barreto, E. Carvalho, J. B. Sapucaia, J. C. Ramos, E. Luz, C. Pedroso, M. Manrique, N. L. Toomey, C. Brites, D. P. Dittmer, and W. J. Harrington, Jr. 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng, Y., and B. R. Cullen. 2006. Recognition and cleavage of primary microRNA transcripts. Methods Mol. Biol. 342:49-56. [DOI] [PubMed] [Google Scholar]
- 37.Zhao, Y., Y. Yao, H. Xu, L. Lambeth, L. P. Smith, L. Kgosana, X. Wang, and V. Nair. 2009. A functional microRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J. Virol. 83:489-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, J. Y., T. Pfuhl, N. Motsch, S. Barth, J. Nicholls, F. Grasser, and G. Meister. 2009. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 83:3333-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.