Abstract
mTOR is a central regulator of cellular growth and metabolism. Using metabolic profiling and numerous small-molecule probes, we investigated whether mTOR affects immediate control over cellular metabolism by posttranslational mechanisms. Inhibiting the FKBP12/rapamycin-sensitive subset of mTOR functions in leukemic cells enhanced aerobic glycolysis and decreased uncoupled mitochondrial respiration within 25 min. mTOR is in a complex with the mitochondrial outer-membrane protein Bcl-xl and VDAC1. Bcl-xl, but not VDAC1, is a kinase substrate for mTOR in vitro, and mTOR regulates the association of Bcl-xl with mTOR. Inhibition of mTOR not only enhances aerobic glycolysis, but also induces a state of increased dependence on aerobic glycolysis in leukemic cells, as shown by the synergy between the glycolytic inhibitor 2-deoxyglucose and rapamycin in decreasing cell viability.
Keywords: metabolomics, mitochondria, chemical biology
mTOR functions as a multichannel processor in a cellular nutrient-sensing network by receiving multiple inputs derived from distinct environmental cues and directing different outputs to appropriate downstream effectors. Upstream regulators of mTOR are primarily mitogens, nutrients, and energy (1). mTOR exists in 2 separate protein complexes: the mitogen-, nutrient-, and rapamycin-sensitive mTOR complex 1 (mTORC1) and the mitogen-sensitive and nutrient- and rapamycin-insensitive mTORC2 (2). mTOR regulates key cellular processes, including mRNA translation, ribosome biogenesis, autophagy, and metabolism. The most extensively studied targets of mTOR are the translation regulators S6K1 and 4E-BP1 (3). Yeast cells treated with rapamycin mount an immediate (within 20 min) and widespread transcriptional response that results in metabolic reprogramming characteristic of the diauxic shift (4). Mammalian cells do not display an immediate and widespread transcriptional response to mTOR inhibition by rapamycin (5). Inhibition of mTORC1 using either rapamycin or RNA-mediated interference of proteins involved in the signaling network decreases mitochondrial respiration and levels of fully uncoupled respiration, independent of S6K1 and 4EBP1 (6). Recently, rapamycin was reported to decrease the transcription of genes involved in mitochondrial oxidative function by disrupting a complex involving TORC1, YY1, and PGC-1α, thereby preventing the coactivation of YY1 by PGC-1α (7). The authors suggested this transcriptional mechanism as the means by which mTOR controls mitochondrial function; however, treatment with rapamycin does not result in decreased mitochondrial content (6) even after 8 h.
Here we describe an immediate change in mitochondrial function following the inhibition of mTOR. Using global physiological profiling, we show that inhibition of mTOR has immediate effects on carbon and mitochondrial metabolism in a leukemic cell line. Rapamycin-treated leukemic cells display reduced mitochondrial function, resulting in energy production via enhanced aerobic glycolysis in preference over mitochondrial respiration. We discovered that the mTOR-mediated control of mitochondrial metabolism occurs via a complex comprising Bcl-xl and VDAC1 at the mitochondrial outer membrane.
Results and Discussion
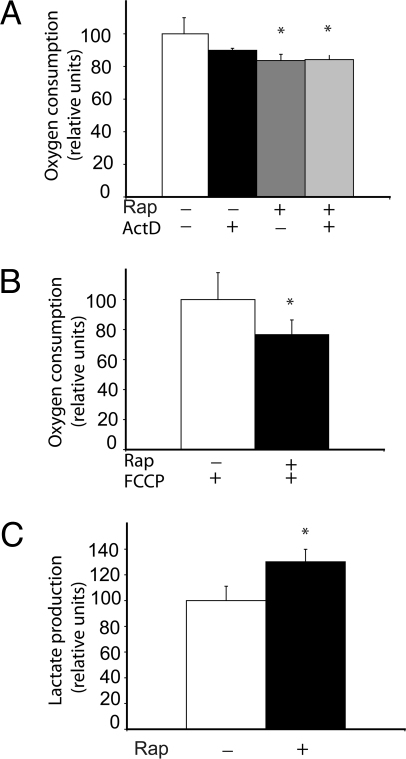
Inhibition of mTOR by rapamycin decreased oxygen consumption and mitochondrial capacity (as determined by adding a mitochondrial uncoupler immediately after treatment with rapamycin) of Jurkat cells within 30 min (Fig. 1B). The rapamycin-induced decrease in oxygen consumption persisted even when cells were cotreated with the transcriptional inhibitor actinomycin D (Fig. 1A), indicating that this effect did not require transcription. Pyruvate, the end product of glycolysis, can be converted either into acetyl CoA by mitochondrial pyruvate dehydrogenase or into lactate by cytoplasmic lactate dehydrogenase. Inhibition of mTOR by rapamycin increased lactate production in Jurkat cells within 25 min (Fig. 1C). We observed no significant changes in glucose consumption at this time point.
Fig. 1.
Effect of rapamycin on mitochondria and glycolysis. (A) Jurkat cells were treated with 100 nM rapamycin in the presence or absence of the transcription inhibitor actinomycin D for 45 min. (B) Levels of maximally uncoupled respiration were measured after treatment with rapamycin or vehicle for 30 min, followed by treatment with 1 μm FCCP for 15 min. (C) Levels of lactic acid in the extracellular medium were measured after cells were treated with rapamycin for 45 min. *, P < .05. All experiments were performed in biological triplicate.
We next measured the global changes in relative levels of intracellular metabolites after a 25-min treatment of Jurkat cells with rapamycin. This time point was selected for 2 reasons: (i) after 25 min, cellular responses to mTOR inhibition should still be largely independent of transcriptional or translational effects, and (ii) this was the shortest feasible time point given our experimental methods. The measurement of relative levels of 197 intracellular small-molecule metabolites provided a global snapshot of the physiological status of rapamycin-treated cells. Table 1 lists the metabolites that changed significantly after the 25-min treatment.
Table 1.
Metabolic profiling of Jurkat cells treated with rapamycin
| % change (P < 0.05) | |
|---|---|
| Amino acid | |
| Arginine | 42.2 |
| Phenylalanine | 33.7 |
| Histidine | 33.1 |
| Leucine/isoleucine | 23.4 |
| Tyrosine | 23.1 |
| Proline | 22.9 |
| Methionine | 21.4 |
| Valine | 19.7 |
| Tryptophan | 16.3 |
| Glutamic acid | −5.9 |
| Glutamine | −6.6 |
| Hydroxyproline | −23.2 |
| Threonine | −26.2 |
| Homoserine | −28 |
| Taurine | −30.7 |
| Serine | −30.7 |
| Asparagine | −31.3 |
| Mitochondrial metabolism | |
| Fumaric acid/maleic acid | −19.9 |
| Malic acid | −28.5 |
| Alpha-keto-glutarate | −30.4 |
| Succinic acid | −37.1 |
| Fumaric acid/maleic acid | −46.3 |
| Orotic acid | −99.6 |
| Acetyl-CoA | 57 |
| Malonyl-CoA | 47 |
| Glucose metabolism | |
| F1P/F6P/G1P/G6P | 85.3 |
| Ribose-5-P/ribulose-5-P | 49.2 |
| Lactic acid | 40.8 |
| UDP-glucose | −27.7 |
| Lipid metabolism | |
| Glycerol | 55.2 |
| Carnitine | −26.1 |
| Nucleotides | |
| ATP | 66.3 |
| UTP | 125.5 |
| CDP | 70.1 |
| GTP | 78.1 |
| UDP | 86.6 |
| CTP | 134 |
| GDP | 52 |
| ADP | 79 |
Jurkat cells were treated with 100 nM rapamycin for 25 min. Intracellular metabolites were extracted and profiled using LC-MS/MS. The percent changes in relative levels of peak areas of metabolites that differed significantly (P < 0.05) after rapamycin treatment are displayed. Profiling experiments were performed in 5 biological replicates. Bold indicates an increase, and italic indicates a decrease, in relative levels.
In agreement with the hypothesis that inhibition of mTOR shifts glucose metabolism away from mitochondrial respiration, we observed an immediate buildup of intracellular lactic acid. This was accompanied by an accumulation of mono-phosphorylated carbohydrates (glucose and fructose-6 phosphate), including ribose-5-phosphate, which is an intermediate in the glycolysis-derived pentose-phosphate pathway. Of the other glucose metabolites measured, we found a decrease in the relative levels of UDP-glucose, an intermediate in glycogen synthesis, and a general increase in levels of ribonucleotides, including ATP and ADP. We found decreased levels of all 6 mitochondrially metabolized organic acids measured following the inhibition of mTOR. Five of these are tri-carboxylic acid (TCA) cycle intermediates; the other, orotic acid, is synthesized in the mitochondria during purine metabolism. This striking decrease in mitochondrial organic acids, in conjunction with diminished oxygen consumption, is indicative of a decrease in mitochondrial metabolism. We noted an accumulation of acetyl CoA and malonyl CoA, which are substrates for lipid synthesis, along with a 55% increase in the relative level of glycerol, which results from the hydrolysis of lipid esters.
Among the amino acids, arginine showed the largest relative increase (42.2%) in intracellular pools. The other amino acids demonstrating significant increases were phenylalanine, histidine, leucine/isoleucine, tyrosine, proline, methionine, valine, and tryptophan. Asparagine showed the greatest decrease in relative levels (31.3%), and threonine and serine decreased by <10%. We believe that the observed increases in intracellular levels of certain amino acids following the inhibition of mTOR may be due largely to amino acid uptake via transporters, rather than to anabolic processes. Rapamycin has been reported to increase the influx of arginine into endothelial cells (8). It is noteworthy that the amino acids that were enhanced in response to mTOR inhibition can yield TCA cycle intermediates following the catabolism of their carbon skeletons (ie, arginine, histidine, and proline, yielding α-ketoglutarate; isoleucine, methionine, and valine, yielding succinyl-CoA; phenylalanine and tyrosine, yielding fumarate). These amino acids can replenish depleted levels of TCA cycle intermediates through the process of anaplerosis. Overall, the profile indicates an immediate and profound change in cellular metabolism caused by inhibition of the FKBP12/rapamycin-sensitive functions of mTOR, consistent with enhanced aerobic glycolysis.
We observed a synergistic interaction between inhibitors of glycolysis and mTOR in leukemic cells. The striking synergy between the glycolytic inhibitor 2-deoxyglucose (2DG) and rapamycin, as demonstrated by combination index (CI) values (9) significantly below 0.5 (Table 2), suggests that inhibition of mTOR in leukemia cells causes an increased dependence on glycolytic metabolism. These insights suggest that the use of small-molecule probes having activities that strongly distinguish different types of cancers (e.g., the bioenergetic-classifying ATP synthase inhibitors), along with an increased understanding of the bioenergetics of cancers in general, may be useful in identifying cancers that are especially sensitive to mTOR- and other bioenergetics-targeting drugs, and in developing effective combinations of drugs that individually affect cancer metabolism.
Table 2.
Glycolytic dependency induced by treatment with rapamycin
| Rapamycin (nM) | 2DG (mM) | CI |
|---|---|---|
| 5 | 1.5 | 0.46 |
| 5 | 2 | 0.071 |
| 5 | 2.5 | 0.006 |
| 5 | 5 | 9.51E-05 |
| 20 | 1.5 | 0.317 |
| 20 | 2 | 0.166 |
| 20 | 2.5 | 0.038 |
| 20 | 5 | 2.53E-05 |
| 40 | 1.5 | 0.056 |
| 40 | 2 | 0.181 |
| 40 | 2.5 | 0.112 |
| 40 | 5 | 0 |
The synergy of glycolytic inhibitor 2-deoxyglucose with rapamycin was measured by calculating the CI of various cotreatments. A CI < 1 indicates synergy.
The rapamycin-induced increase in lactate production and decrease in uncoupled respiration without detectable change in mitochondrial content led us to hypothesize that inhibiting mTOR diverts the metabolism of pyruvate away from the mitochondria by exerting control over the mitochondrial substrate availability. We investigated this by inhibiting VDAC proteins, which are situated in the outer mitochondrial membrane and are known to be key mediators of substrate transport into the mitochondria (10). We reasoned that if rapamycin affects substrate availability, then a global metabolic profile of VDAC inhibition might recapitulate important aspects of the rapamycin profile. Thus, we measured the global metabolic response of VDAC inhibition by treating Jurkat cells with the small molecule erastin for 25 min, the same treatment period we used for rapamycin. Erastin, a cell-permeable inhibitor of VDAC2 (11), has been used to study the immediate effects of perturbing mitochondrial outer membrane permeability. (A cell-permeable inhibitor, VDAC1, remains to be discovered.) Comparing the metabolic profiles of erastin and rapamycin perturbations reveals that erastin-mediated VDAC inhibition reproduces key facets of rapamycin-mediated mTOR inhibition, such as increases in the relative intracellular levels of lactate, glycerol, and upstream glycolytic intermediates (Fig. S1 A and B). The erastin treatment led to decreased levels of the TCA cycle intermediates succinic, fumaric, and malic acid. These results suggest that inhibition of substrate availability leads to the nonmitochondrial metabolism of pyruvate into lactate and the resulting accumulation of glycolytic intermediates, similar to rapamycin.
It has been previously demonstrated that mTOR colocalizes with the outer membrane of the mitochondria (12). The mechanism by which mTOR is targeted to the mitochondria is unclear. A member of the FK506-binding family of proteins, FKBP38, binds directly to the FRB domain of mTOR (13). FKBP38 is localized on the mitochondria and has been shown to anchor proteins like Bcl2 and Bcl-xl to this organelle (14). The FKBP38–mTOR interaction is not thought to be involved in the Rheb protein–mediated activation of mTOR (15, 16). mTOR might possibly be localized to the mitochondria via an FKBP38-mediated anchoring mechanism. The upstream signals to mitochondrial mTOR merit further investigation. mTOR has been demonstrated to be sensitive to mitochondrial dysfunction (12). Localization of mTOR to the mitochondria might directly sensitize the activity of mTOR to the redox status of the cell. Another signal to mitochondrial mTOR might be through the AMP-dependent protein kinase, an important sensor of energy in cells. AMPK is known to activate TSC2 (via the tumor suppressor LKB1) and to inhibit mTOR (17).
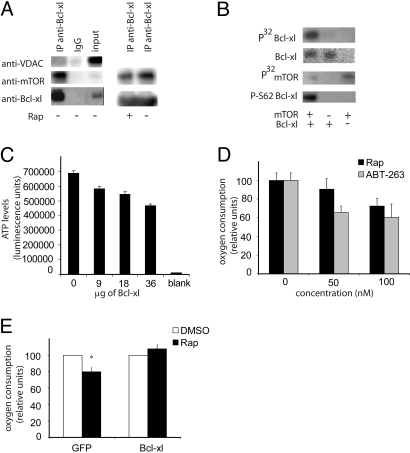
We investigated whether mTOR is in complex with key proteins that mediate substrate availability at the mitochondrial outer membrane, and found that mTOR coimmunoprecipitates with the outer mitochondrial membrane proteins VDAC1 and Bcl-xl (Fig. 2A). Bcl-xl, a key mediator of mitochondrial function and cellular apoptosis has been shown to bind to VDAC1 and to increase substrate permeability (18, 19). We explored whether these 2 proteins serve as kinase substrates for mTOR and discovered that Bcl-xl, but not VDAC1, is phosphorylated by mTOR in vitro (Fig. 2 B and C). Bcl-xl function has been shown to be sensitive to its phosphorylation state (20, 21). Phosphorylation of the serine-62 residue on Bcl-xl was recently shown to be a key regulator of Bcl-xl activity (21), but the immediate upstream kinase responsible for this phosphorylation remains mysterious. We found that Bcl-xl serine 62 was one of the sites phosphorylated by mTOR in vitro (Fig. 2B), suggesting that mTOR might be an important and direct modulator of Bcl-xl function in vivo.
Fig. 2.
mTOR affects mitochondrial metabolism via a complex with Bcl-xl. (A) Bcl-xl interacts with mTOR and VDAC1. Interaction of Bcl-xl with mTOR was decreased by treatment with rapamycin. (B) An in vitro kinase assay was performed using recombinant purified mTOR and Bcl-xl proteins and γP32-labeled ATP. Bcl-xl was detected using a Ponceau stain, and the phosphorylated serine 62 on Bcl-xl was detected using an antibody. (C) mTOR-mediated phosphorylation of Bcl-xl was monitored using recombinant, purified Bcl-xl protein at indicated concentrations, using an ATP-coupled luminescence assay, with 100 ng of mTOR protein used per reaction. (D) Jurkat cells were treated with rapamycin and ABT-263 at indicated concentrations, and oxygen consumption was measured. (E) Jurkat cells overexpressing either GFP or Bcl-xl were treated with DMSO vehicle or rapamycin, and oxygen consumption was measured. *, P < .05. All experiments were preformed in biological triplicate.
The treatment of cells with rapamycin also caused the dissociation of mTOR from Bcl-xl (Fig. 2A). mTOR also might mediate its function via other members of the Bcl protein family; this requires further investigation. We tested whether the inhibition of Bcl proteins, like the inhibition of mTOR by rapamycin, also would result in an immediate decrease in mitochondrial respiration. This investigation was feasible by small-molecule perturbation using the small molecule ABT-263, a potent inhibitor of protein–protein interactions involving Bcl proteins (22). ABT-263 at 100 nM immediately inhibited mitochondrial respiration by 30% (Fig. 2D), indicating that, like rapamycin, ABT-263 has an immediate effect on mitochondrial function. We next tested the effect of rapamycin on mitochondrial respiration in cells overexpressing either Bcl-xl protein or GFP. Cells overexpressing Bcl-xl were less sensitive to rapamycin-induced inhibition of respiration even after 8 h of rapamycin treatment (Fig. 2E); thus, we hypothesize that mTOR controls mitochondrial metabolism in a Bcl-xl–dependent fashion.
In summary, we have identified a mitochondrial mTOR complex that appears to function as a cellular switch regulating glycolytic and respiratory metabolism. Inhibiting mTOR, which is now the basis of treatment of certain cancers, enhances aerobic glycolysis and induces a state of increased dependence on aerobic glycolysis in leukemic cells.
Materials and Methods
Metabolic Measurements.
Jurkat (clone E6–1) T cell leukemic cells were purchased from ATCC and were treated with 40 nM rapamycin (LC Laboratories), 7.3 μM erastin, or 100 nM ABT-263. Cells were filtered of growth media and washed twice with 50 mL of ice-cold PBS. Intracellular metabolites were extracted using 1 mL of 80% methanol/20% water and dried under nitrogen. The metabolite extracts were reconstituted in 100 μL of deionized water, and the relative levels of metabolites were measured by liquid chromatography–mass spectrometry (LC-MS) as described previously (23). The experiments were preformed in 5 biological replicates. The oxygen consumption of Jurkat cells was measured using a BD Biosciences oxygen biosensor system as described previously (6). Relative levels of extracellular lactate were measured using the Amplex Red kit (Invitrogen).
Immunoprecipitation and SDS/PAGE Analysis.
Cells were lysed with RIPA buffer (Pierce Biotechnology), containing 1 mM DTT and protease inhibitor mixture, for 30 min at 4 °C. From the cell extract, an anti-mTOR antibody (Cell Signal, no. 2972) was used for immunoprecipitation. Antibodies against Bclxl, VDAC1, and mTOR were obtained from Cell Signal. The antibody against phosphorylated serine 62 was purchased from Abcam. The immune complex was resolved using 4%–12% SDS/PAGE.
DNA Constructs and Electroporation.
Bcl-xl and GFP expression constructs were obtained from Dr. Tim Chambers (17). The electroporation of Jurkat cells was performed with an Amaxa electroporation kit, using cell line–specific protocols optimized by the manufacturer. GFP transfection experiments were performed in duplicate, and the efficiency of transfection was estimated to be ≈65%.
mTOR Kinase Assay.
Recombinantly purified, active mTOR protein was purchased from Millipore. The radiolabeled ATP-based kinase assay protocol was performed as described previously (3). For luminescence-based assays, the kinase assay was performed without the use of radiolabeled ATP. The Kinase Glo kit (Promega) was used to quantitate the kinase-dependent depletion of ATP.
Synergy Analysis.
Jurkat cells were treated with varying combinations of rapamycin and glycolytic inhibitors for 48 h, and intracellular ATP levels were measured using a kit purchased from Promega. The synergy calculations were performed using Calcusyn software (Biosoft).
Supplementary Material
Acknowledgments.
We thank Yendi Linares for providing technical support with the small-molecule synergy experiments; Xiang Wang for providing assay development and screening of chemical libraries; R. Wei for managing the metabolic profiling experiments and instrumentation; Brent Stockwell for providing the erastin; Timothy Lewis (Broad Institute Chemical Biology Platform) for synthesizing the ABT-263 compound; Marco Colombini, Gerhard Wagner, and Sebastian Hiller for providing advice and a gift of purified VDAC1 protein; and Tim Chambers for providing the Bcl-xl and GFP expression constructs. This research was supported by National Institute for General Medical Sciences Grant GM38627 (to S.L.S.). S.L.S. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflicts of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912074106/DCSupplemental.
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Guertin DA, Sabitini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 4.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;26:1840–1851. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng T, Golub TR, Sabatini DM. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol Cell Biol. 2002;22:5575–5584. doi: 10.1128/MCB.22.15.5575-5584.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schieke SM, et al. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 8.Visigalli R, et al. Rapamycin stimulates arginine influx through CAT2 transporters in human endothelial cells. Biochim Biophys Acta. 2007;6:1479–1487. doi: 10.1016/j.bbamem.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Chou T. Theoretical basis, experimental design and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 10.Colombini M. VDAC: The channel at the interface between mitochondria and the cytosol. Mol Cell Biochem. 2004;256:107–115. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- 11.Yagoda N, et al. RAS-RAF-MEK–dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–868. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai BN, Myers BR, Schreiber SL. FKBP12-rapamycin–associated protein associates with mitochondria and senses osmotic stress via mitochondrial dysfunction. Proc Natl Acad Sci USA. 2002;99:4319–4324. doi: 10.1073/pnas.261702698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai X, et al. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- 14.Uhlenbrock K. Reassessment of the role of FKBP38 in the Rheb/mTORC1 pathway. FEBS Lett. 2009;583:965–970. doi: 10.1016/j.febslet.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Sato T, Nakashima A, Guo L, Tamanoi F. Specific activation of mTORC1 by Rheb G-protein in vitro involves enhanced recruitment of its substrate protein. J Biol Chem. 2009;284:12783–12791. doi: 10.1074/jbc.M809207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang CB, Tai J, Chia J, Yoon HS. The flexible loop of Bcl-2 is required for molecular interaction with immunosuppressant FK-506 binding protein 38 (FKBP38) FEBS Lett. 2005;579:1469–1476. doi: 10.1016/j.febslet.2005.01.053. [DOI] [PubMed] [Google Scholar]
- 17.Corradetti MN, Inoki K, Bardeesy N, DePinho RA, Guan KL. Regulation of the TSC pathway by LKB1: Evidence of a molecular link between tuberous sclerosis complex and Peutz–Jeghers syndrome. Genes Dev 2004. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vander Heiden MG, Chandel NS, Schumacker PT, Thompson CB. Bcl-xL prevents cell death following growth factor withdrawal by facilitating mitochondrial ATP/ADP exchange. Mol Cell. 1999;3:159–167. doi: 10.1016/s1097-2765(00)80307-x. [DOI] [PubMed] [Google Scholar]
- 19.Malia TJ, Wagner G. NMR structural investigation of the mitochondrial outer membrane protein VDAC and its interaction with antiapoptotic Bcl-xL. Biochemistry. 2007;16:514–525. doi: 10.1021/bi061577h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grad JM, Zeng XR, Boise LH. Regulation of Bcl-xL: A little bit of this and a little bit of STAT. Curr Opin Oncol. 2000;12:543–549. doi: 10.1097/00001622-200011000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Upreti M, et al. Identification of the major phosphorylation site in Bcl-xL induced by microtubule inhibitors and analysis of its functional significance. J Biol Chem. 2008;283:35517–35525. doi: 10.1074/jbc.M805019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park CM, et al. Discovery of an orally bioavailable small-molecule inhibitor of prosurvival B-cell lymphoma 2 proteins. J Med Chem. 2008;51:6902–6915. doi: 10.1021/jm800669s. [DOI] [PubMed] [Google Scholar]
- 23.Lewis GD, et al. Metabolite profiling of blood from individuals undergoing planned myocardial infarction reveals early markers of myocardial injury. J Clin Invest. 2008;118:3503–3512. doi: 10.1172/JCI35111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.