Abstract
While the global down-regulation of microRNAs (miRNAs) is a common feature of human tumors, its genetic basis is largely undefined. To explore this question, we analyzed the consequences of conditional Dicer1 mutation (Dicer1 “floxed” or Dicer1fl) on several mouse models of cancer. Here we show Dicer1 functions as a haploinsufficient tumor suppressor gene. Deletion of a single copy of Dicer1 in tumors from Dicer1fl/+ animals led to reduced survival compared with controls. These tumors exhibited impaired miRNA processing but failed to lose the wild-type Dicer1 allele. Moreover, tumors from Dicer1fl/fl animals always maintained one functional Dicer1 allele. Consistent with selection against full loss of Dicer1 expression, enforced Dicer1 deletion caused inhibition of tumorigenesis. Analysis of human cancer genome copy number data reveals frequent deletion of DICER1. Importantly, however, the gene has not been reported to undergo homozygous deletion, suggesting that DICER1 is haploinsufficient in human cancer. These findings suggest Dicer1 may be an important haploinsufficient tumor suppressor gene and, furthermore, that other factors controlling miRNA biogenesis may also function in this manner.
Keywords: MicroRNAs, cancer, Dicer, tumor suppressor, mouse models, haploinsufficiency
MicroRNAs (miRNAs) are short, noncoding RNAs that function to suppress post-transcriptionally the expression of target mRNAs, predominately via inhibition of translation. Such translational inhibition relies on imperfect base-pairing between the miRNA and the target transcript, with the interaction at nucleotides 2–8 (or the “seed” region) of the miRNA being required for translational repression. Computational prediction of miRNA targets based on seed regions and sequence conservation has revealed a widespread potential for miRNA-mediated transcript regulation, with hundreds of putative mRNA targets for an individual miRNA (Bartel 2004).
In line with their broad-based effects, miRNAs have been proposed to function as oncogenes or tumor suppressor genes based on their inhibition of a variety of tumor-suppressive and oncogenic mRNAs, respectively (Plasterk 2006; Ventura and Jacks 2009). In particular, three distinct mechanisms have been posited. First, oncogenic miRNAs can undergo gain of function in tumors. This has been most clearly demonstrated for the miR-17∼92 cluster, whose amplification in B-cell lymphomas promotes their development, potentially through its control of B-cell differentiation (He et al. 2005; Koralov et al. 2008; Ventura et al. 2008). Furthermore, tumor-suppressive miRNAs could undergo loss of function in tumors. This has been shown for several miRNAs, including the let-7 family, whose expression can limit lung tumorigenesis through inhibition of oncogenes like the Ras family and HMGA2 (Esquela-Kerscher et al. 2008; Kumar et al. 2008). In particular, let-7 family members are in sites of frequent deletion in human tumors, and their processing is inhibited by the oncogenic Lin-28 proteins (Heo et al. 2008; Newman et al. 2008; Viswanathan et al. 2008; Chang et al. 2009). Finally, oncogenes can acquire mutations to remove miRNA-binding sites in tumors. This has been described for HMGA2, whose translocation promotes lipoma development by releasing the transcript from let-7-mediated tumor suppression (Mayr et al. 2007).
We reported a global down-regulation of miRNAs in several types of human and murine cancer (Lu et al. 2005). From this initial study, it was unclear whether this widespread loss of miRNAs was merely a consequence of tumor development or was functionally related to the disease process. We demonstrated previously that this global loss of miRNAs was functionally relevant to oncogenesis, as impairment of miRNA maturation enhanced transformation in both cancer cells and a K-Ras-driven model of lung cancer (Kumar et al. 2007). While these studies provide a framework to explain inhibition of miRNA biogenesis in cancer, the genetic basis of impaired miRNA processing in human cancer has been largely undefined. For a subset of miRNAs, widespread silencing occurs at the transcriptional level via the c-Myc oncogene (Chang et al. 2008). However, it has also been shown that such broad reductions in miRNAs can occur post-transcriptionally, since changes in miRNA levels frequently occur without changes in the levels of the primary miRNA transcript (Thomson et al. 2006). Recently, it was shown that mutations in the miRNA processing component TARBP2 occur frequently in mismatch repair-deficient colon cancer, and that these mutations promote tumorigenesis by impaired processing of miRNAs (Melo et al. 2009). While interesting, these limited cases do not resolve the common global reduction of miRNAs in human cancers. Moreover, the precise genetics of such changes in tumors is poorly defined, especially as no components of the miRNA processing pathway have been reported to be completely deleted in human tumors. This is not surprising, since it has been shown that germline deletion of miRNA processing components Dicer1 and Dgcr8 in mice fails to produce viable progeny (Bernstein et al. 2003; Wang et al. 2007). Thus, conditional deletion of miRNA processing components provides a powerful means of examining the role of miRNAs in tumorigenesis.
Results and Discussion
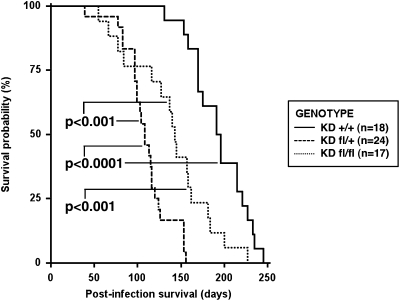
We previously reported results using the KrasLSL-G12D strain, which was either heterozygous or homozygous for a conditional allele of Dicer1 (hereafter referred to as KDfl/+ and KDfl/fl) (Jackson et al. 2001; Harfe et al. 2005). After intranasal infection with adenovirus expressing Cre (Ad-Cre), these compound mutant animals developed significantly more lung tumors compared with KrasLSL-G12D mice that were wild type for Dicer1 (hereafter referred to KD+/+); moreover, Dicer1 mutant mice had a modest shift in histological grade, further enforcing its role as a tumor suppressor (Kumar et al. 2007). To extend these observations of Dicer1-mediated tumor suppression, we characterized survival of KD+/+, KDfl/+, and KDfl/fl animals after intranasal Ad-Cre infection. Both KDfl/+ and KDfl/fl animals had significantly reduced survival relative to KD+/+ mice (Fig. 1). Surprisingly, we found that the survival of the KDfl/+ cohort was less than that of KDfl/fl animals. To extend these findings to additional cancer types, we used a mouse model of soft-tissue sarcoma generated through intramuscular infection with Ad-Cre of KrasLSL-G12D; Trp53fl/fl mice (Kirsch et al. 2007). When we compared survival of KrasLSL-G12D; Trp53fl/fl mice either wild type or heterozygous or homozygous for the conditional allele of Dicer1 (hereafter referred to as KPD+/+, KPDfl/+, and KPDfl/fl) intramuscularly infected with Ad-Cre, we observed a reduction in survival in only the KPDfl/+ cohort (Supplemental Fig. 1). Overall, these results indicate that induction of heterozygosity for a Dicer1 mutation in tumors can enhance tumor development.
Figure 1.
Dicer1 mutation reduces post-infection survival in a genetically engineered mouse model of K-Rasdriven lung cancer. KrasLSL-G12D mice, either wild type or heterozygous or homozygous conditional for Dicer1 (KD+/+, KDfl/+, and KDfl/fl, respectively), were intranasally infected with Ad-Cre, and survival was assessed. Median survival was 194 d for KD+/+ mice, 108 d for KDfl/+ mice, and 143 d for KDfl/fl mice. Statistical significance was assessed by the log-rank test.
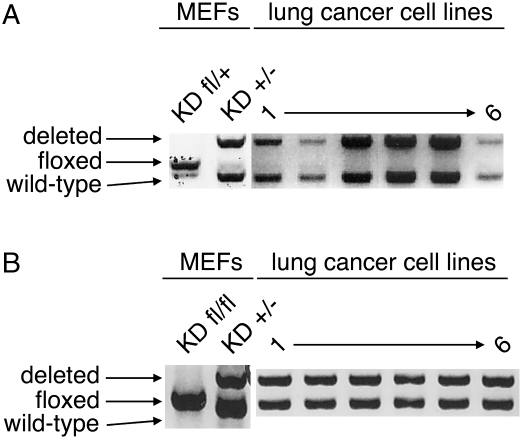
These findings stand in opposition to traditional tumor suppressor genes, in which a homozygous mutation provides a direct advantage to tumor cells while a heterozygous mutation promotes tumorigenesis only after subsequent mutation or inactivation of the remaining wild-type allele. In particular, the survival disadvantage of KDfl/+ tumors to KDfl/fl suggests active selection against complete loss of Dicer1 in these tumors. To characterize precisely the mutation status of Dicer1 conditionally mutant tumors, we generated lung cancer cell lines from KPDfl/+ animals and examined the Dicer1 locus for further deletion. As shown in Figure 2, we observed recombination of the conditional allele of Dicer1 and retention of the wild-type allele, generating KPD+/− lung cancer cells (Fig. 2A). Similar maintenance of the wild-type allele was found in sarcoma cell lines from KPDfl/+ mice (Supplemental Fig. 2A).
Figure 2.
Dicer1 undergoes hemizygous loss in lung tumors. (A) DNA was prepared from KrasLSL-G12D mouse embryonic fibroblasts (MEFs) either heterozygous conditional or mutant for Dicer1 (KDfl/+ and KD+/−) and lung cancer cell lines from KPDfl/+ mice, and the Dicer1 locus was examined by PCR. (B) DNA was prepared from KrasLSL-G12D mouse embryonic fibroblasts either homozygous conditional or heterozygous mutant for Dicer1 (KDfl/fl and KD+/−) and lung cancer cell lines from KPDfl/fl mice, and the Dicer1 locus was examined by PCR.
We further examined Dicer1 recombination in lung cancer cell lines from KPDfl/fl animals. In these cell lines, we again found evidence of Cre-Lox recombination. However, the recombination was incomplete, generating KPDfl/− lung cancer cells (Fig. 2B); this incomplete loss of Dicer1 was also observed in sarcoma cell lines from KPDfl/fl mice (Supplemental Fig. 2A). Thus, it is likely that KDfl/fl lung tumors undergo selection against complete Dicer1 recombination during tumor progression. To determine if complete loss of Dicer1 product occurs, Dicer1 protein levels were examined in lung cancer cells and sarcomas either wild type or mutant for Dicer1. Although we observed a general reduction in Dicer1 protein in Dicer1 mutant cancer cell lines, all retained some expression of Dicer1 (Supplemental Fig. 2B). In addition, resequencing of the Dicer1 ORF in lung cancer and sarcoma cell lines from KPDfl/+ and KPDfl/fl mice revealed no further mutations of the retained Dicer1 allele (data not shown), suggesting that complete loss of Dicer1 function is not achieved through second site mutation. In sum, these data indicate that only partial loss of Dicer1 occurs in these genetically engineered mouse models during tumorigenesis.
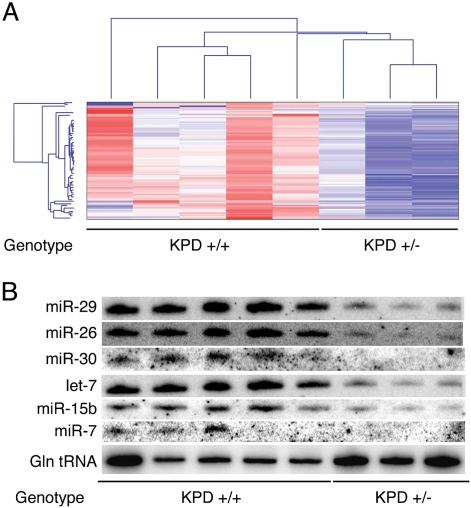
Based on the protumorigenic effect of hemizygous mutation of Dicer1 in these tumor models, we went on to assess the consequence of partial loss of Dicer1 on global miRNA expression. Comparing miRNA profiles in Dicer1 wild-type and heterozygous mutant lung cancer cells, there was a global decrease in steady-state miRNA levels in KPD+/− cells (Fig. 3A; Supplemental Table 1). A small RNA Northern blot analysis verified these reductions in miRNA levels (Fig. 3B). Thus, a Dicer1 mutation is capable of significantly altering miRNA levels, even when incomplete.
Figure 3.
Hemizygous deletion of Dicer1 causes a global reduction in steady-state miRNA levels. (A) miRNA profiling and hierarchical clustering were performed on lung cancer cell lines either wild type (KPD+/+) or heterozygous for Dicer1 (KPD+/− and KPDfl/−). (B) Small RNA Northern blotting analysis of miRNAs and glutamine tRNA was performed on lung cancer cell lines wild type or heterozygous for Dicer1 as above.
While the genetic analysis of the Dicer1 locus in our mouse cancer models suggested that Dicer1 was functioning as a haploinsufficient tumor suppression gene, it was still formally possible that Dicer1 mutant tumors eventually underwent complete inactivation of Dicer1 via an alternative mechanism like epigenetic silencing. Thus, we assessed the consequences of complete loss of Dicer1 in the genetically engineered mouse models. First, to promote complete loss of Dicer1 in the lung, we infected KDfl/+ and KDfl/fl mice with a lentivirus expressing Cre (Lenti-Cre). In contrast to the recombinant adenoviruses, which do not integrate in the genome of the infected cells and are eventually lost, lentiviral integration is expected to allow for stable, longer-term expression of Cre and, more likely, complete recombination at the Dicer1 locus. Infection of mice with Lenti-Cre led to a striking decrease in tumor burden in KDfl/fl compared with KDfl/+ mice (Supplemental Fig. 3A–C). This result contrasts sharply with previous studies in which KDfl/+ and KDfl/fl mice created a similar tumor burden after Ad-Cre infection. Notably, when we isolated DNA from the tumors that did arise in KDfl/fl mice and assessed Dicer1 loss, all tumors were once again found to have undergone incomplete recombination, suggesting that there is selection against total Dicer1 loss in lung tumors (Supplemental Fig. 3D).
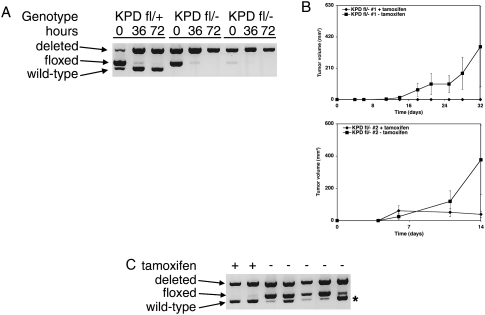
To further examine the effects of complete Dicer1 loss, we transduced a set of sarcoma cell lines with CreERT2 to permit tamoxifen-dependent Cre activity. Treatment with 4-hydroxytamoxifen (4-OHT) led to efficient deletion of Dicer1 in vitro (Fig. 4A). To assess the effect of complete Dicer1 loss on tumorigenesis, we transplanted untreated KPDfl/− sarcoma cells expressing CreERT2 into immune-competent hosts, and recombination was induced by systemic tamoxifen administration. Compared with control treatments, tamoxifen administration substantially slowed tumor growth in KPDfl/− sarcomas (Fig. 4B). Of note, this system allowed for Cre-mediated deletion of Dicer1, as the tumors that developed in tamoxifen-treated animals had extensive Dicer1 recombination (Fig. 4C). Importantly, tumor suppression was not merely an effect of tamoxifen- or Cre-mediated toxicity, as tamoxifen treatment of animals injected with KPD+/+ sarcoma cells expressing CreERT2 did not impair tumor growth (Supplemental Fig. 4). Taken together, these studies demonstrate that complete deletion of Dicer1 is deleterious to tumor development, strongly suggesting that Dicer1 is a haploinsufficient tumor suppressor. Of note, in a separate study using these cancer cell lines, A Ravi, MS Kumar, C Chin, T Jacks, and PS Sharp (in prep.) have shown that full loss of Dicer1 function can be tolerated, but that Dicer1-null cells have impaired proliferative capacity.
Figure 4.
Complete deletion of Dicer1 inhibits sarcoma development. (A) KPDfl/+ and KPDfl/− sarcoma cell lines were infected with MSCV.CreERT2.puro. Cre-mediated recombination was induced by treatment with 4-OHT for defined time points. DNA was prepared, and recombination of the Dicer1 locus was assessed by PCR. (B) Two independent KPDfl/− sarcoma cell lines infected with MSCV.CreERT2.puro were injected subcutaneously into C57Bl6/129SV F1 animals. Animals were treated with or without tamoxifen by intraperitoneal injection, and tumor growth was measured over time. Values are mean ± SEM (n = 8 each). (C) Tumors were isolated from KPDfl/− sarcoma cell line transplants treated with or without tamoxifen. DNA was prepared, and recombination of the Dicer1 locus was assessed by PCR. Asterisk corresponds to the wild-type Dicer1 locus from the C57Bl6/129SV F1 host animals.
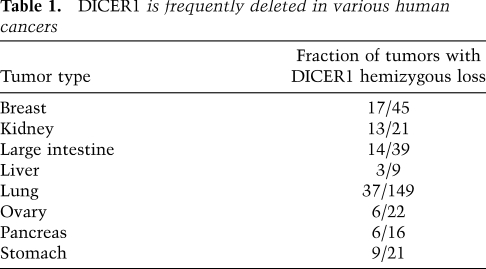
While these findings indicate that partial loss of Dicer1 promotes tumor development, it was not clear whether a comparable situation occurs in human cancer. To explore this question, we assessed DICER1 copy number data from the Cancer Genome Project at the Sanger Institute (Forbes et al. 2008). In these data sets, there was frequent loss of one allele of DICER1 (via hemizygous deletion) in several different tumor types (Table 1); similar results were obtained from analysis of copy number data sets of glioblastomas and ovarian cancers from The Cancer Genome Atlas (TCGA) (data not shown). Although the deletions of DICER1 seen in human cancer were generally broad (>1 Mb), there were never high-level amplifications of the DICER1 locus in these tumor sets, suggesting these losses were not due to random genome instability. More importantly, homozygous deletion of DICER1 was never reported, in line with our findings of Dicer1 functioning as a haploinsufficient tumor suppressor (Hill et al. 2009; Melo et al. 2009).
Table 1.
DICER1 is frequently deleted in various human cancers
Our results suggest a major cause for the global loss of miRNAs in human cancer (Lu et al. 2005), via partial loss of function of the miRNA processing machinery in human tumors. Although the frequent DICER1 single-copy deletion noted in human cancers provides a relevant mechanism of impairing miRNA biogenesis, additional mechanisms may occur. In particular, in light of the frequent mutation of TARBP2 in mismatch repair-deficient colon cancer (Hill et al. 2009) and heterozygous germline point mutations in DICER1 in patients with pleuropulmonary blastoma (Melo et al. 2009), it is possible that point mutations of DICER1 or other components of the miRNA processing machinery can occur in different types of human cancer. Indeed, this has been described for DICER1 and DROSHA in a small number of ovarian cancer cell lines (Merritt et al. 2008). Importantly, while this study describes Dicer1 as a haploinsufficient tumor suppressor in the context of K-Ras-driven cancer, the analysis of miRNA processing machinery mutations has found changes in cancers of a variety of genotypes, suggesting a broad role for Dicer1 as a tumor suppressor.
Beyond such mutational analyses, these results represent an expansion of the list of haploinsufficient tumor suppressors to include components of the miRNA processing machinery. The breadth of tumor suppressor genes that function via haploinsufficiency is only beginning to be appreciated (Santarosa and Ashworth 2004). In fact, the traditional reliance on complete loss of tumor suppressor genes by heterozygous mutation and subsequent loss of heterozygosity (LOH) is likely to overlook factors whose effects do not require complete loss, such as p27Kip1 and Dmp1 (Inoue et al. 2001; Muraoka et al. 2002). Moreover, such analyses are certain to ignore genes like Dicer1, for which partial loss is advantageous to tumors while complete loss is disadvantageous. As genome-wide studies begin to explore the functional role of chromosomal deletions in human cancer, it will be important to consider haploinsufficiency of deleted genes in such contexts.
Materials and methods
Mice
KrasLSL-G12D; Dicer1+/+, fl/+,fl/fl animals were generated as described previously (Kumar et al. (2007). KrasLSL-G12D; Trp53fl/fl animals were bred with KrasLSL-G12D; Dicer1fl/fl animals to produce KrasLSL-G12D; Trp53fl/+; Dicer1fl/+ animals. These animals were backcrossed to KrasLSL-G12D; Trp53fl/fl animals to produce KrasLSL-G12D; Trp53fl/fl; Dicer1fl/+ animals, which were interbred to produce the experimental cohort.
Intranasal, intratracheal, and intramuscular infection
Mice were infected intranasally, intratracheally, and intramuscularly with Ad-Cre and Lenti-Cre as described (Jackson et al. 2001; Kirsch et al. 2007; Kumar et al. 2008). Lung and tumor areas were determined using Bioquant Image Analysis software as described previously (Kumar et al. 2007).
Cell line injection and tamoxifen treatment
C57Bl6/129SV F1 animals were subcutaneously injected with 2.5 × 104 sarcoma cells. Mice were subsequently treated with corn oil with and without tamoxifen as described previously (Ventura et al. 2007). Tumors were measured as described previously (Kumar et al. 2007).
Cell culture
Primary sarcoma and lung cancer cell lines were generated by dissection of tumors from mice infected with Ad-Cre, digestion with trypsin, and plating and propagation in serum-containing media. KrasLSL-G12D; Dicer1fl/+,fl/fl mouse embryonic fibroblasts were generated as described previously (Tuveson et al. 2004). Sarcoma cell lines were infected with MSCV.CreERT2.puro and treated with 4-OHT for defined time points as described previously (Ventura et al. 2007). Cells were maintained using standard conditions.
Recombination analysis
DNA was prepared from tumors and cancer cell lines, and Dicer1 and Kras recombination was assessed by PCR using previously described primers (Harfe et al. 2005). Kras primer information is provided in the Supplemental Material.
Northern blotting
Small RNA Northern blotting was performed as described previously (Ventura et al. 2004). Probe information is provided in the Supplemental Material.
Acknowledgments
We thank H. Zhang and D. Crowley for experimental assistance, and D.G. McFadden and E. Meylan for critical review of the manuscript. This work was supported by grant 2-PO1-CA42063 from the National Cancer Institute, and partially by Cancer Center Support grant P30-CA14051 from the National Cancer Institute. Research was approved by the Committee for Animal Care, was conducted in compliance with the Animal Welfare Act regulations and other federal statutes relating to animals and experiments involving animals, and adheres to the principles set forth in the Guide for Care and Use of Laboratory Animals (National Research Council, 1996) (institutional animal welfare assurance no. A-3125-01). M.S.K. was an NSF Graduate Research Fellow. T.R.G. and T.J. are investigators of the Howard Hughes Medical Institute. T.J. is a Ludwig Scholar. M.S.K. conceived the project. M.S.K., R.E.P., C.Y.C., C.C., J.L., A.L. Dooley, and D.G.K. carried out all experiments described. T.R.G. and T.J. supervised the experimental work and interpretation of data. The manuscript was prepared by M.S.K. and T.J.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1848209.
Supplemental material is available at http://www.genesdev.org.
References
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA, et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci. 2009;106:3384–3389. doi: 10.1073/pnas.0808300106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The catalogue of somatic mutations in cancer (COSMIC) Curr Protoc Hum Genet. 2008;57:10.11.1–10.11.26. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokamp KA, Suarez BK, Whelan AJ, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes & Dev. 2001;15:2934–2939. doi: 10.1101/gad.929901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, Nielsen GP, Quade BJ, Chaber CJ, Schultz CP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci. 2008;105:3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka RS, Lenferink AE, Law B, Hamilton E, Brantley DM, Roebuck LR, Arteaga CL. ErbB2/Neu-induced, cyclin D1-dependent transformation is accelerated in p27-haploinsufficient mammary epithelial cells but impaired in p27-null cells. Mol Cell Biol. 2002;22:2204–2219. doi: 10.1128/MCB.22.7.2204-2219.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Santarosa M, Ashworth A. Haploinsufficiency for tumour suppressor genes: When you don't need to go all the way. Biochim Biophys Acta. 2004;1654:105–122. doi: 10.1016/j.bbcan.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP, Jackson EL, Chang S, Mercer KL, Grochow R, Hock H, Crowley D, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]