Abstract
Our previous work has shown that efficient evasion from type I interferon responses by human cytomegalovirus (hCMV) requires expression of the 72-kDa immediate-early 1 (IE1) protein. It has been suggested that IE1 inhibits interferon signaling through intranuclear sequestration of the signal transducer and activator of transcription 2 (STAT2) protein. Here we show that physical association and subnuclear colocalization of IE1 and STAT2 depend on short acidic and serine/proline-rich low-complexity motifs in the carboxy-terminal region of the 491-amino-acid viral polypeptide. These motifs compose an essential core (amino acids 373 to 420) and an adjacent ancillary site (amino acids 421 to 445) for STAT2 interaction that are predicted to form part of a natively unstructured domain. The presence of presumably “disordered” carboxy-terminal domains enriched in low-complexity motifs is evolutionarily highly conserved across all examined mammalian IE1 orthologs, and the murine cytomegalovirus IE1 protein appears to interact with STAT2 just like the human counterpart. A recombinant hCMV specifically mutated in the IE1 core STAT2 binding site displays hypersensitivity to alpha interferon, delayed early viral protein accumulation, and attenuated growth in fibroblasts. However, replication of this mutant virus is specifically restored by knockdown of STAT2 expression. Interestingly, complex formation with STAT2 proved to be entirely separable from disruption of nuclear domain 10 (ND10), another key activity of IE1. Finally, our results demonstrate that IE1 counteracts the antiviral interferon response and promotes viral replication by at least two distinct mechanisms, one depending on sequestration of STAT2 and the other one likely involving ND10 interaction.
Human cytomegalovirus (hCMV) is an extremely widespread opportunistic pathogen causing morbidity and mortality in hundreds of thousands of children and adults each year (48). Within the ∼230,000-bp hCMV genome, the major immediate-early (IE) gene is believed to have a decisive role in acute infection and reactivation from viral latency. Through differential splicing, polyadenylation, and promoter usage, this viral genomic region produces multiple mRNAs. Although a variety of protein products expressed from these mRNAs have been identified (4, 55), the UL123-coded 72-kDa nuclear phosphoprotein IE1 and the UL122-coded 86-kDa nuclear phosphoprotein IE2 are the most abundant and important. They share 85 amino-terminal amino acids corresponding to major IE exons 2 and 3 but have distinct carboxy-terminal parts, encoded by exon 4 (IE1) or exon 5 (IE2). Both proteins have long been recognized as promiscuous transcriptional regulators. IE2 is the principal activator of the hCMV lytic cycle and is essential for productive viral replication (26, 40). The role of IE1 in hCMV infection is less clear than that of IE2. Whereas IE1-null viruses replicate efficiently in fibroblasts at high input multiplicities, the absence of IE1 results in inefficient hCMV early gene expression and attenuated viral growth under “single-hit” conditions (20, 22, 47). Consistent with its role in transcriptional activation, IE1 has been reported to interact with several transcriptional coactivators (25, 30, 41, 56, 84) and a histone deacetylase (28, 51). A small fraction of IE1 is also found covalently conjugated to the small ubiquitin-like modifier 1 (SUMO-1) (28, 36, 49, 50, 67). Furthermore, the viral protein has the remarkable ability to localize to both the chromatin (33) and the interchromatinic matrix-associated nuclear domain 10 (ND10) compartments of the cell nucleus. IE1 binds to at least one constituent of ND10, namely, the promyelocytic leukemia (PML) protein, and disrupts these structures upon ectopic expression or at early times after hCMV infection (2, 32, 78). It has recently been demonstrated that individual ND10 components, including PML, Sp100, and Daxx, mediate an intrinsic immune response against hCMV and other herpesviruses (18, 19, 60, 61, 71, 72, 79). This observation supports the idea that ND10-resident proteins are part of a cellular antiviral defense mechanism which is inactivated by virus-encoded proteins, including IE1 (3, 71, 72; reviewed in references 17, 42, and 73). The IE1 nucleotide and protein sequences are evolutionarily conserved among primate CMVs. In contrast, rodent CMVs have positional IE1 orthologs that share no obvious amino acid sequence similarity with the hCMV counterpart. Nonetheless, the hCMV and murine CMV (mCMV) IE1 proteins exhibit discrete as well as common functional activities (reviewed in references 7 and 43).
In addition to its proposed role in antagonizing ND10-related cellular defense mechanisms, hCMV IE1 also inactivates a crucial branch of the host's inducible innate immune system. We have demonstrated that the viral protein inhibits type I interferon (IFN) signaling, conferring a substantial degree of protection against the antiviral effects of IFN-α and IFN-β upon hCMV (53). Remarkably, IE1 interferes with one of the final steps in the Janus kinase-signal transducer and activator of transcription (Jak-STAT) cascade, which links the cytoplasmic membrane-bound type I IFN receptor to the promoters of IFN-stimulated genes (ISGs), many of which encode antiviral proteins or RNAs (reviewed in reference 62). In particular, the viral protein prevents sequence-specific promoter binding and transcriptional activation by the IFN-stimulated gene factor (ISGF3) complex, which forms in the presence of IFN-α or -β (53). ISGF3 is composed of three proteins: STAT1, STAT2, and IFN regulatory factor 9 (IRF9, also known as p48 or ISGF3γ), and IE1 was found to be physically associated with two of these components (STAT1 and STAT2) (28, 53). However, STAT2 appears to be the viral protein's primary target in the trimeric complex (S. Krauss, S. Meinel, I. Tschertner, C. Paulus, and M. Nevels, unpublished data). Despite the fact that STAT2 binding is expected to contribute substantially to the overall function of IE1 during hCMV infection, we have only just begun to understand the structural basis for and relevance of this interaction (28).
Here we define the structural requirements in the hCMV IE1 protein that contribute to physical and functional interaction with STAT2. We present evidence that STAT2 binding is evolutionarily conserved between mammalian CMV IE1 proteins. Moreover, we show for the first time that STAT2 binding and ND10 disruption are genetically separable activities of IE1 and that STAT2 interaction is one of at least two distinct mechanisms by which the viral protein antagonizes the antiviral IFN response to promote viral replication.
MATERIALS AND METHODS
Cell culture and virus infections.
All cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were cultured at 37°C in a humidified 5% CO2 atmosphere. Human fetal diploid lung fibroblasts (MRC-5) were obtained from the European Collection of Cell Cultures, and early-passage cells (15 to 25 population doublings before senescence) were used in all experiments. The fibrosarcoma cell line 2fTGH, which has been used extensively to study Jak-STAT signaling pathways, was obtained from George Stark (Case Western Reserve University, Cleveland, OH) (54) and maintained in medium containing 250 μg/ml hygromycin. H1299 cells have been described (46). Where applicable, cells were treated with 250 to 1,000 U/ml recombinant IFN-α (PBL Biomedical Laboratories) for various durations.
All infections were carried out on confluent MRC-5 cells with a bacterial artificial chromosome (BAC)-derived wild-type isolate (TNwt; from Hua Zhu, University of Medicine and Dentistry of New Jersey) (40) or with IE1-mutated variants (TNdlIE1 and TNdlIE1AD1-S/P) or a revertant (TNdlIE1rev) of the hCMV Towne strain at input multiplicities of 0.01 to 3 PFU per cell or equivalent infectious genome numbers. Virus was grown on MRC-5 fibroblasts (TNwt and TNdlIE1rev) or IE1-expressing derivatives (TNdlIE1 and TNdlIE1AD1-S/P), and TNwt and TNdlIE1rev titers were determined by standard plaque assay on MRC-5 cells. TNdlIE1 and TNdlIE1AD1-S/P infectious units equivalent to TNwt titers were calculated from relative intracellular genome numbers. To this end, cells were infected with various dilutions of virus stock, and particle adsorption was terminated after 2 h by a 1-min incubation in citrate wash buffer (40 mM citrate [pH 3.0], 10 mM KCl, 135 mM NaCl) followed by two wash steps in medium. DNA was extracted from cells at 8 h postinfection using a DNeasy blood and tissue kit from Qiagen. For viral replication analysis, the same kit was employed to purify DNA from infected culture supernatants. The DNA was used to template real-time PCRs performed in a Roche LightCycler 1.5 employing the hCMV-specific primers 294 and 295 (directed against the UL54 promoter) (Table 1) and a LightCycler FastStart DNA MasterPlus SYBR green I kit (Roche), following the manufacturer's instructions exactly. DNA levels were calculated using the efficiency-corrected relative quantification strategy described in Roche Applied Science Technical Note no. LC 13/2001. The hCMV clinical strains Coz and Par were isolated from blood and urine, respectively, by Stephen Spector (University of California at San Diego). A 5.8-kb fragment covering the major IE transcription units of Coz or Par was amplified by high-fidelity PCR from virion DNA using primers 588 and 589 (Table 1), cloned in vector pCR4-TOPO (Invitrogen), and sequenced (Princeton University Syn/Seq Facility).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′) | Use |
|---|---|---|
| 140 | GCGGCGCATGCGATCTCCACGCGAATCTCGGGTAC | hCMV IE1-ΔAD1-S/P mutagenesis |
| 145 | CGCGGCAAGCTTGGATCCACCATGGAGTCCTCTGCC | hCMV IE1 mutagenesis, BAC analysis |
| 146 | GGCGCGAAGCTTGAATTCTTACTGGTCAGCCTTG | hCMV IE1 mutagenesis, BAC analysis |
| 155 | GAGTCCGAAGCCGAACTGCAG | hCMV IE1 cloning |
| 156 | GCGGCGCGGCCGCGATCTCCACGCGAATCTCG | hCMV IE1 cloning |
| 205 | CTGACGGATCCGAGCCTTTCGAGGAGATGA | hCMV IE1ΔN cloning |
| 206 | CTGACGAATTCTTACTGGTCAGCCTTGCTT | hCMV IE1ΔN cloning |
| 207 | CTGACGGATCCATGGAGCCCGCCGCACCCAGTT | mIE1 cloning |
| 208 | CTGACGAATTCTCACTTCTTGCTCTTCTTCTTG | mIE1 cloning |
| 294 | CACCAAAGACACGTCGTT | hCMV genome quantification |
| 295 | GTCCTTTGCGACCAGAAT | hCMV genome quantification |
| 326 | TAGACGGATCCACCATGGAGTCCTCTGCCAAGAG | hCMV IE1 cloning |
| 327 | TAGACGAATTCTTACTGGTCAGCCTTGCTTC | hCMV IE1 cloning |
| 386 | TCCTAGTGTGGATGACCTACGGTACACTTTGGCCACCGCTGGTG | hCMV IE1-ΔAD1 mutagenesis |
| 387 | CACCAGCGGTGGCCAAAGTGTACCGTAGGTCATCCACACTAGGA | hCMV IE1-ΔAD1 mutagenesis |
| 388 | CACTTTGGCCACCGCTGGTGCCGCGACTATCCCTCTGTCCTCAG | hCMV IE1-ΔS/P mutagenesis |
| 389 | CTGAGGACAGAGGGATAGTCGCGGCACCAGCGGTGGCCAAAGTG | hCMV IE1-ΔS/P mutagenesis |
| 390 | TCTGTCCTCAGTAATTGTGGCTACTGTGTCTGTCAAGTCTGAGC | hCMV IE1-ΔAD2 mutagenesis |
| 391 | GCTCAGACTTGACAGACACAGTAGCCACAATTACTGAGGACAGA | hCMV IE1-ΔAD2 mutagenesis |
| 392 | GGAGGACACTGTGTCTGTCAAGGGAGGCAAGAGCACCCACCCTA | hCMV IE1-ΔAD3 mutagenesis |
| 393 | TAGGGTGGGTGCTCTTGCCTCCCTTGACAGACACAGTGTCCTCC | hCMV IE1-ΔAD3 mutagenesis |
| 394 | TCCTAGTGTGGATGACCTACGGGAGAACAGTGATCAGGAAGAAA | hCMV IE1-ΔAD1-S/P mutagenesis |
| 395 | TTTCTTCCTGATCACTGTTCTCCCGTAGGTCATCCACACTAGGA | hCMV IE1-ΔAD1-S/P mutagenesis |
| 401 | TGAGGAAGATGCTATTGTAGCCAGCTCCTCTGATTCTCTGGTGT | hCMV IE1-Δ387-394 mutagenesis |
| 402 | ACACCAGAGAATCAGAGGAGCTGGCTACAATAGCATCTTCCTCA | hCMV IE1-Δ387-394 mutagenesis |
| 422 | GCGGCGCTAGCCATGCGGCTCACCTCGTCAATCTTG | hCMV IE1-ΔAD1-S/P mutagenesis |
| 588 | GCGCCGGAATTCCAAAAGTCCTAGGTCGGCGTCGACG | hCMV major IE (Coz, Par) cloning |
| 589 | GCGCCGGGATCCGCATACGCGATATCTGGCGATAGCG | hCMV major IE (Coz, Par) cloning |
Protein production and glutathione S-transferase (GST) pull-down assays.
Plasmid pGEX-IE1 was constructed by excising the 72-kDa hCMV IE1 coding sequence from pcDNA-IE1 (50) via consecutive treatment with HindIII, Klenow polymerase, and EcoRI. The resulting DNA fragment was inserted into the SmaI and EcoRI sites of vector pGEX-KG (23). For the construction of pGEX-IE2, the cDNA encoding the 86-kDa hCMV IE2 protein was derived from pEGFP-IE2 (see below) via BglII/EcoRI digestion and inserted into the BamHI and EcoRI sites of pGEX-KG. The IE1ΔN cDNA lacking nucleotides 4 to 411 from the 5′ end of the full-length hCMV IE1 coding sequence was PCR amplified from plasmid template pEGFP-IE1 (50) using primers 205 and 206 (Table 1). The PCR product was inserted into vector pGEX-KG via BamHI and EcoRI sites to generate pGEX-IE1ΔN. The IE1ΔC sequence, comprising nucleotides 1 to 1038 from the 5′ end of the hCMV IE1 gene, was derived from pEGFP-IE1 by BglII digestion. The large BglII fragment was subsequently inserted into the BamHI site of pGEX-KG to generate pGEX-IE1ΔC. The full-length cDNA of wild-type mIE1 was PCR amplified from pEMBL19-pp89 (a gift from Martin Messerle, Hannover Medical School) (77) using primers 207 and 208 (Table 1). The resulting PCR product was inserted into pGEX-KG via BamHI and EcoRI sites. The identity of each recombinant plasmid and error-free PCR amplification was confirmed by restriction digestion and DNA sequencing (Geneart).
A single colony of the expression strain Escherichia coli M15(pRep4) (Qiagen) transformed with the recombinant pGEX-KG derivatives or empty vector was grown in a shaker overnight at 37°C in Luria-Bertani (LB) medium containing ampicillin (50 μg/ml) and kanamycin (25 μg/ml). The culture was diluted 1:100 in fresh prewarmed medium and further grown for 1 h at 30°C to an optical density at 600 nm of 0.6. After that, gene expression was induced by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.5 mM. Following a 16-h incubation at 25°C, cells were harvested by centrifugation. After one wash step in ice-cold purification buffer (50 mM Tris-HCl [pH 7.6], 100 mM NaCl, 0.5% Triton X-100) and one freeze-thaw cycle, bacteria were resuspended in 1/50 culture volume of purification buffer with complete mini-protease inhibitor cocktail tablets (Roche). This was followed by sonification (three times for 2 min each; duty cycle, 70%; output control, 5) in a Branson Sonifier 450 and centrifugation for 30 min at 27,000 × g. From the supernatant, GST fusion proteins were affinity purified in a batch procedure using glutathione Sepharose 4B (GE Healthcare) according to the manufacturer's instructions. The purification steps were monitored by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis followed by Coomassie brilliant blue staining of the protein bands and/or by Western blot analysis.
For binding assays, total extracts from confluent MRC-5 cells were prepared in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.6], 10% glycerol, 100 mM NaCl, 0.5% Triton X-100, complete mini-protease inhibitor cocktail tablets) with short sonification. For each sample, the extract from one 15-cm dish of confluent MRC-5 cells (approximately 1.5 × 107 cells) was mixed with 7.5 μg of full-length GST or GST fusion protein bound to glutathione Sepharose in purification buffer and incubated for 90 min at 4°C with constant rotation. After that, the Sepharose beads were washed four times in GST wash buffer (50 mM Tris-HCl [pH 7.6], 100 mM NaCl, 0.1% Triton X-100), suspended in 2× sample buffer (100 mM Tris-HCl [pH 6.8], 200 mM β-mercaptoethanol, 0.2% bromphenol blue, 20% glycerol, 4% SDS), and heated for 5 min at 95°C. Finally, STAT2 binding was analyzed by SDS-polyacrylamide gel electrophoresis followed by Western blotting.
Immunofluorescence microscopy.
Total RNA purified from MRC-5 fibroblasts infected with hCMV (Towne) was reverse transcribed, and the IE1-specific cDNA was PCR amplified using primers 326 and 327 (Table 1). The resulting PCR product was subjected to EcoRI and BamHI restriction digestion. The IE1-coding DNA fragment was subsequently inserted, in frame with the enhanced green fluorescent protein (EGFP) coding sequence, into the EcoRI and BglII sites of vector pEGFP-C1 (BD Biosciences-Clontech), resulting in plasmid pEGFP-TNIE1. The correct wild-type sequence of the IE1 insert was verified (Geneart). Plasmids pEGFP-IE1ΔN and pEGFP-mIE1 were constructed by subcloning BamHI/EcoRI fragments from pGEX-IE1ΔN and pGEX-mIE1, respectively, into the BglII and EcoRI sites of pEGFP-C1. For pEGFP-ΙΕ1ΔC, the large BglII fragment from pEGFP-IE1 (50) was inserted into the BglII and BamHI sites of pEGFP-C1. The IE2 cDNA from pCGN-IE2 (85) was inserted into vector pcDNA3 via KpnI to generate pcDNA-IE2. From pcDNA-IE2, a HindIII/EcoRI fragment was inserted into the same sites of pEGFP-C1 to generate pEGFP-IE2. A fusion PCR strategy (27) was employed to introduce the internal deletions ΔAD1, ΔS/P, ΔAD1-S/P, Δ387-394, ΔAD2, and ΔAD3 into the IE1 coding sequence. Phusion polymerase (New England Biolabs) and the following primers were used for PCR amplification from template pEGFP-TNIE1: 145, 146 (outer primers used in all PCRs), 386 to 395, 401, and 402 (mutation-specific inner primers) (Table 1). Final PCR products were inserted via HindIII and BamHI sites into vector pEGFP-C1, and error-free amplification of the entire IE1 mutant sequences was confirmed (Geneart). Full-length IE1 and internal deletions ΔAD1, ΔS/P, ΔAD1-S/P, ΔAD2, and ΔAD3 were subsequently transferred to pcDNA-HA-N (a gift from Ron Hay, University of Dundee) (70) via BamHI and EcoRI sites following PCR amplification primed by oligonucleotides 145 and 146 (Table 1) and using the respective pEGFP derivatives as templates. A pSG5-derived plasmid, which encodes a hemagglutinin (HA)-tagged IE1 protein lacking the region spanning AD2 and AD3 (amino acids 421 to 475) (28), was provided by Jin-Hyun Ahn (Sungkyunkwan University, Suwon, South Korea).
Approximately 2 × 105 2fTGH cells were plated on coverslips in six-well dishes and transfected with 10 μg of plasmid DNA by calcium phosphate precipitation 24 h thereafter. At 48 h posttransfection, cells were fixed with methanol for 15 min at −20°C, and immunofluorescence staining was performed following a previously published protocol (52). An anti-STAT2 rabbit polyclonal antibody (H-190) or an anti-PML mouse monoclonal antibody (5E10; from Roel van Driel, University of Amsterdam) (68) and an anti-EGFP mouse monoclonal antibody (MAB3580) or an anti-IE1 rabbit polyclonal antibody (rbIE1-1) were used for primary protein detection (Table 2). To generate antibody rbIE1-1, GST-IE1ΔN protein was affinity purified on glutathione Sepharose 4B beads (see above) and eluted with reduced glutathione. A New Zealand White rabbit was immunized three times with 200 μg each of GST-IE1ΔN protein (Davids Biotechnology), and the reactivity of the resulting serum, compared to that of a preimmune serum, was tested by Western blotting and immunofluorescence (data not shown). The target epitopes of antibody rbIE1-1 were roughly mapped to the region between amino acids 138 and 404 in the hCMV IE1 protein (data not shown). Primary antibody incubation was followed by a combination of secondary conjugates, each used at a 1:1,000 dilution (2 μg/ml): Alexa Fluor 594 goat anti-mouse immunoglobulin G (IgG) (heavy plus light chain [H+L]) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) or Alexa Fluor 488 goat anti-mouse IgG (H+L) and Alexa Fluor 594 goat anti-rabbit IgG (H+L) (all highly cross-adsorbed and from Invitrogen). 4′,6-Diamidino-2-phenylindole (DAPI) from Roche was added to the secondary antibody solution at a final concentration of 0.2 μg/ml. Coverslips were mounted on glass slides using ProLong Gold (Invitrogen). Slides were analyzed and images were acquired using a Leica DMRX epifluorescence microscope equipped with a digital camera system (Retiga; Q-Imaging). Images were cropped and processed using Image-Pro Plus 6.2 (Q-Imaging), Meta Imaging series 4.5 (Universal Imaging Corporation), and Adobe Photoshop CS (version 8.0) software.
TABLE 2.
Primary antibodies used in this study
| Antigen | Antibody |
|||||
|---|---|---|---|---|---|---|
| Name | Source | Typea | IFb | Amt (μg) used for IPc | WB dilutiond | |
| EGFP | MAB3580 | Millipore-Chemicon | M | 1:500 | NAe | NA |
| GAPDH | ab9485 | Abcam | R | NA | NA | 1:4,000 |
| HA tag | HA-7 | Sigma-Aldrich | M | NA | 40 | NA |
| hCMV IE1 | MAB810 | Millipore-Chemicon | M | 1:100 | NA | NA |
| hCMV IE1 | rbIE1-1 | This work | R | 1:200 | NA | NA |
| hCMV IE1 | 1B12 | T. Shenk (85) | M | 1:2 | 2.5 | 1:10 |
| hCMV IE2 | 1218 | J. Nelson | R | 1:3,000 | NA | NA |
| hCMV IE2 | 3A9 | T. Shenk (52) | M | NA | NA | 1:10 |
| hCMV ppUL44 | 10D8 | GeneTex | M | NA | NA | 1:2,000 |
| hCMV pp28 | 10B4-29 | T. Shenk (65) | M | NA | NA | 1:15 |
| Nonspecific | IgG (mouse serum) | Sigma-Aldrich | R | NA | 2.5 | NA |
| Nonspecific | IgG (rabbit serum) | Sigma-Aldrich | M | NA | 2.5 | NA |
| PML | 5E10 | R. van Driel (68) | M | 1:2 | NA | NA |
| STAT2 | H-190 | Santa Cruz | R | 1:100 | 2.5 | 1:500 |
R, rabbit polyclonal; M, mouse monoclonal.
Dilution in phosphate-buffered saline with 0.05% Tween 20 and 0.02% IgG from human serum (Sigma-Aldrich). IF, immunofluorescence.
Amount used for one sample.
Dilution in phosphate-buffered saline. WB, Western blotting.
NA, not applicable.
Immunoprecipitation and Western blotting.
Approximately 48 h following transfection of H1299 cells by calcium phosphate precipitation, whole-cell extracts were prepared in lysis buffer (50 mM Tris [pH 8.0], 10% glycerol, 150 mM NaCl, 0.5% Triton X-100, protease inhibitor cocktail set III [Calbiochem]). Preclearing was performed for 1 h with protein A-agarose (Sigma-Aldrich). Following centrifugation (1 min, 1,000 × g, 4°C), extracts were incubated with monoclonal anti-HA-agarose (clone HA-7; Sigma-Aldrich) for 90 min at 4°C. The agarose beads were subsequently washed four times in immunoprecipitation wash buffer (50 mM Tris [pH 8.0], 0.1% Igepal CA-630, 150 mM NaCl). For immunoprecipitations from hCMV-infected cells, Dynabeads Protein A M-280 (Invitrogen) were used, following the manufacturer's instructions exactly. After the last wash step, beads were resuspended in 2× sample buffer and heated for 5 to 8 min at 95°C. Polyacrylamide-SDS gel electrophoresis and Western blotting were performed as described previously (50). For a list of primary antibodies, see Table 2.
Mutagenesis of hCMV BACs.
An IE1 cDNA lacking the exon 4 nucleotides corresponding to amino acids 373 to 420 (AD1 and S/P) was generated using a fusion PCR strategy (27) on template pGS284-MIE (E.-M. Hauer and C. Paulus, unpublished data) with primer pairs 395/140, 394/422, and 140/422. The final PCR product was inserted into vector pCR4-TOPO to generate plasmid pCR4-MIEdlIE1AD1-S/P. Subsequently, a 3.2-kb NheI/SphI fragment from pCR4-MIEdlIE1AD1-S/P covering the mutant IE1 sequence was transferred to pGS284, a derivative of the positive-selection suicide vector pCVD442 carrying ampicillin resistance and sacB genes (66). The resulting transfer plasmid, pGS284-MIEdlIE1AD1-S/P, was used for homologous recombination with the hCMV BAC pTNIE1kanlacZ (E.-M. Hauer and C. Paulus, unpublished data), in which the major IE exon 4 is replaced by a kanamycin resistance (kan) and lacZ cassette (see Fig. S3A in the supplemental material). The BAC also carries a chloramphenicol resistance gene. For allelic exchange via conjugation, the recA+ E. coli strain GS500 (66) containing pTNIE1kanlacZ and E. coli S17λpir transformed with the pGS284-MIEdlIE1AD1-S/P donor plasmid were cross-streaked on LB plates (without antibiotics) and incubated overnight at 37°C. To select for cointegrates, 10 ml LB broth containing ampicillin and chloramphenicol was inoculated with bacteria from intersection regions, and cells were grown at 37°C overnight. On the next day, 5 μl of the stationary culture was again inoculated in 10 ml LB medium with ampicillin plus chloramphenicol and incubated for another 8 h at 37°C. This was followed by dilution in 10 ml chloramphenicol containing LB broth and incubation for 15 h at 37°C. Finally, a small culture volume was incubated for 48 h at 30°C on LB plates containing 15 μg/ml chloramphenicol, 20 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 200 mg/ml isopropyl-β-d-thiogalactopyranoside, and 5% sucrose. This step allowed selection against the sacB-containing donor vector and nonresolved cointegrates and facilitated identification of positive bacterial clones via blue-white screening. White colonies were replica plated on LB plates containing either ampicillin, kanamycin, or chloramphenicol and incubated at 37°C. Clones resistant to chloramphenicol but not ampicillin or kanamycin were selected, and BAC DNA purified from these cells was further examined by analytical PCR (see Fig. S3B in the supplemental material), restriction enzyme digestion analysis (see Fig. S3B in the supplemental material), and DNA sequencing (Geneart). From two verified BAC clones, pTNdlIE1AD1-S/P-1 and pTNdlIE1AD1-S/P-2, infectious virus was reconstituted by transfection of MRC-5 fibroblasts as described previously (83). The pTNdlIE1-1, pTNdlIE1-2, pTNdlIE1rev-1, and pTNdlIE1rev-2 hCMV BACs were generated in an analogous fashion (see Fig. S3A in the supplemental material), and their construction and reconstitution to infectious virus will be described in detail elsewhere (C. Paulus, T. Knoblach, C. Winterling, and M. Nevels, unpublished data).
RNA interference.
Short interfering RNA (siRNA) duplexes directed against human STAT2 transcripts (sSTAT2-1, r[AAGAUUCUGCAGCAUUUCCCACUCC]; sSTAT2-2, r[AUAUCCGAAGCAGAAGACAUCCUGC]) were purchased from Invitrogen (12937-64). An EGFP-targeted negative control siRNA without related sequences in the human genome (sEGFP, r[GCAAGCUGACCCUGAAGUUC]dTdT) was provided by IBA. MRC-5 cells on 12-well dishes were transfected with 30 nM of individual siRNAs using the reagent Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. At 48 h following transfection, cells were subjected to hCMV infection. Quantitations of viral DNA and protein analyses were performed 4 days postinfection.
Nucleotide sequence accession numbers.
The IE1 nucleotide and amino acid sequences from hCMV strains Coz (GQ244522) and Par (GQ244523) have been deposited in the GenBank database (National Center for Biotechnology Information).
RESULTS
Residues in the carboxy-terminal third of the IE1 protein are required for STAT2 interaction.
Our previous work has shown that complex formation between hCMV IE1 and STAT2 can be reproduced by combining a purified GST-IE1 fusion protein expressed in E. coli with total human cell extracts in an in vitro capture assay (53). To narrow down the STAT2 interaction region(s) in the viral polypeptide, we purified bacterially expressed wild-type GST-IE1 and truncated derivatives lacking large parts (137 or 145 amino acids) from the amino (IE1ΔN) and carboxy (IE1ΔC) termini of the full-length protein, respectively. We also prepared unfused GST as well as GST fusions of the hCMV 86-kDa IE2 protein, sharing 85 amino-terminal residues with IE1, and of the mCMV IE1 ortholog (mIE1) (Fig. 1A). The mIE1 gene product is functionally related to hCMV IE1, but the two proteins exhibit only very limited primary sequence similarity (7, 31).
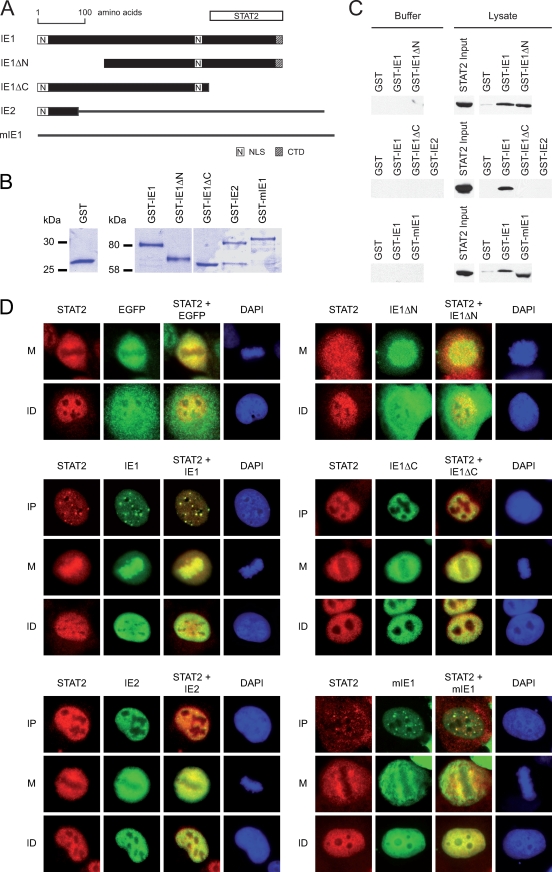
FIG. 1.
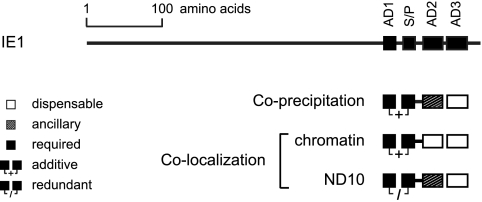
Mapping the STAT2 interaction domain to the carboxy-terminal region of the hCMV IE1 protein. (A) Schematic overview of the tested wild-type and mutant CMV major IE proteins (drawn to scale). Black bars correspond to identical sequences in the respective IE1 and IE2 proteins. Black lines symbolize unique sequences in the IE2 and mIE1 proteins. The STAT2 region interacting within IE1, deduced from the experiments whose results are shown in panels C and D, is shown as a white bar at the top of the diagram. NLS, IE1 nuclear localization signal (locations of IE2- and mIE1-specific nuclear localization signals are not shown); CTD, chromatin-tethering domain. (B) Purified proteins used for GST pull-down assays. Following affinity purification, the indicated GST and GST-IE proteins were separated in 10% polyacrylamide-SDS gels and stained with Coomassie brilliant blue. (C) Results of pull-down binding assays. Identical volumes of glutathione Sepharose beads carrying equal amounts of GST or the indicated GST-IE fusion proteins were reacted with lysis buffer or whole-cell lysate from MRC-5 fibroblasts. Samples were separated in 10% polyacrylamide-SDS gels and Western blotted. Input and copurified STAT2 was detected using antibody H-190. In the GST-mIE1 lane (lower right panel), STAT2 (113 kDa) was detected at an apparently lower molecular weight compared to the other lanes. This is likely caused by comigrating excess GST-mIE1 (∼115 kDa) affecting STAT2 mobility. (D) Nuclear distribution of ectopically expressed EGFP-IE proteins and endogenous STAT2 in 2fTGH cells. After plasmid transfection and short selection in G418 (300 μg/ml), cells were treated with 1,000 U IFN-α for 1 h and fixed with methanol. Then samples were simultaneously reacted with primary antibodies against STAT2 (H-190) and EGFP (MAB3580), followed by incubation with a rabbit-specific Alexa Fluor 594 conjugate, a mouse-specific Alexa Fluor 488 conjugate, and DAPI. Representative single and merged fluorescent stainings of three different subcellular localization patterns are shown: mitotic (M), interphase with nuclear punctate (and diffuse) IE protein distribution (IP), and interphase with nuclear diffuse protein staining (ID). Since EGFP and EGFP-IE1ΔN do not localize to nuclear dots, only M and ID patterns are shown in the upper two sets of images. Magnification, ∼×500.
Sufficient amounts of GST and GST-IE fusion proteins were expressed and purified (Fig. 1B), and the identity of each recombinant protein was confirmed by Western blotting (data not shown). Subsequently, glutathione Sepharose beads carrying comparable amounts of full-length proteins were reacted with equal volumes of cell extract prepared from MRC-5 cells (Fig. 1C). Little or no STAT2 was captured when only GST or binding buffer was used for the assays. As expected, wild-type GST-IE1 pulled down readily detectable amounts of STAT2. Similar results were obtained for the GST-IE1ΔN protein indicating that this mutant displays wild-type characteristics with respect to STAT2 binding. For GST-IE1ΔC and GST-IE2, no physical association with STAT2 was detectable, implying that these proteins lack the pertinent interacting domain(s). Surprisingly, however, robust complex formation between mIE1 and human STAT2 was reproducibly observed (Fig. 1C).
To verify the results from the in vitro binding assays in intact cells, we performed transfection experiments with plasmids expressing EGFP-tagged wild-type IE1, IE1ΔN, IE1ΔC, IE2, and mIE1 proteins. Our prior work has demonstrated that binding of IE1 to STAT2 correlates with specific intranuclear colocalization patterns (53). Thus, the nuclear distribution of the individual viral IE proteins in spatial relation to endogenous STAT2 was examined by double-labeling indirect immunofluorescence microscopy (Fig. 1D). In agreement with previous observations (53), the majority (≥90%) of interphase cells displayed a rather diffuse nuclear staining (excluding nucleoli) for both IE1 and STAT2. However, intensive colocalization of STAT2 with EGFP-IE1 but not EGFP alone at punctate nuclear structures was observed in a subset (≤10%) of cells. The fact that most of the IE1/STAT2-containing dots stained positive for the PML protein (data not shown) identified these structures as ND10, which is a well-established target of IE1 (reviewed in references 42 and 73). In our experimental setting, STAT2 was recruited to ND10 only in the presence of the hCMV protein, not in IE1-negative cells. Likewise, STAT2 was relocalized to nuclear dots by mIE1, although costaining with the cellular protein was less pronounced than with hCMV IE1. Similar to IE1 and mIE1, the IE1ΔN, IE1ΔC, and IE2 proteins also localized to the nucleus, although nuclear targeting of IE1ΔN was less efficient than that of the wild-type, most likely due to one missing nuclear localization signal (amino acids 1 to 24) (Fig. 1A) (35, 78). Moreover, no clear dot pattern was observed for IE1ΔN, IE1ΔC, and IE2, reflecting the lack of critical residues for efficient ND10 targeting (2, 29, 35, 36, 49, 78). Notably, occasional small IE1ΔC- and IE2-positive nuclear dots did not costain for STAT2. In mitotic (metaphase) cells, wild-type IE1 and, to a lesser extent, IE1ΔN were found to be enriched at condensed chromatin (Fig. 1D). In contrast, mIE1, IE1ΔC, and IE2 failed to localize to mitotic chromatin, consistent with the fact that these proteins do not contain the previously defined IE1 chromatin-tethering domain (amino acids 476 to 491) (Fig. 1A) (58, 78). Importantly, STAT2 displayed exclusion from metaphase chromatin in nontransfected cells but was recruited to mitotic chromosomes by IE1 and IE1ΔN, in accordance with the results from our binding experiments (Fig. 1C).
These results indicate that residues in the carboxy-terminal 145 amino acids of the hCMV IE1 protein are required for complex formation with STAT2. In contrast, the amino-terminal 137 amino acids of IE1, including the region shared with IE2, do not contribute significantly to this interaction. Interestingly, STAT2 binding seems to be evolutionarily conserved between the hCMV and mCMV IE1 orthologs.
The STAT2 binding region of IE1 is characterized by evolutionarily conserved LC motifs and a predicted disordered structure.
As yet, no experimental data on the three-dimensional structures of the CMV major IE proteins are available. To explore the structural architecture of the STAT2-interacting carboxy-terminal 150 residues of the IE1 protein in silico, we used the Simple Modular Architecture Research Tool (SMART) (37, 63) and tools available on the PredictProtein server (http://www.predictprotein.org/) (59). Based on the SEG algorithm developed by Wootton and Federhen (80), both SMART and PredictProtein identified four low-complexity (LC) regions of strong compositional bias in the hCMV IE1 (Towne) sequence (Fig. 2A): (i) a 14-residue (positions 373 to 386) acidic region referred to here as AD1, (ii) a 15-amino-acid serine- and proline-rich region between positions 395 and 409 (S/P), (iii) a 25-residue (positions 421 to 445) acidic region (AD2), and (iv) another 25-residue acidic element between amino acids 451 and 475 (AD3). AD2 and AD3 together have been designated the acidic domain of IE1 (e.g., see references 28 and 58). Interestingly, the SEG program predicted the same LC domain architecture in the carboxy-terminal parts of IE1 proteins from a variety of different laboratory-adapted and clinical hCMV strains, including 13 sequences available through GenBank (National Center for Biotechnology Information) and two sequences from virus isolates (Coz and Par) first described in this work (Fig. 2A). In some cases, AD1, S/P, and the amino acids between these two regions were recognized as one continuous LC domain (hCMV type II versus type I) (see Fig. S1A in the supplemental material). Furthermore, the number (two to four), approximate lengths, and relative positions of carboxy-terminal LC motifs were remarkably highly conserved between hCMV IE1 and the respective orthologs of primate and nonprimate CMVs (see Fig. S1A in the supplemental material). On the other hand, such motifs were rarely present in protein regions outside the carboxy-terminal domains, and the few LC sequences identified there were not positionally conserved between the orthologs. Intriguingly, even the carboxy-terminal regions of rat CMV IE1 and mIE1 were specifically enriched in LC motifs (see Fig. S1A in the supplemental material).
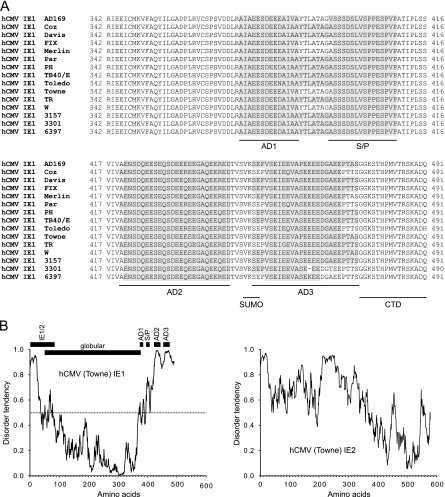
FIG. 2.
Evolutionarily conserved LC motifs and predicted structural disorder in the STAT2 binding region of IE1. (A) Alignment of the carboxy-terminal 150 (149 in strain 3301) amino acids from IE1 proteins of 15 clinical or laboratory-adapted hCMV strains derived from GenBank (National Center for Biotechnology Information): NP_040060 (AD169), GQ244522 (Coz), AAR31419 (Davis), AC146907 (FIX), AAR31666 (Merlin), GQ244523 (Par), AC146904 (PH), AAR31361 (TB40/E), AAR31504 (Toledo), AAR31448 (Towne), AC146906 (TR), AAR31332 (W), AAR31390 (3157), AAR31303 (3301), AAR31477 (6397). The sequence blocks composing the four conserved LC motifs (AD1, S/P, AD2, and AD3) identified by the SEG program are indicated. The locations of the SUMO-1 modification site (SUMO) and chromatin-tethering domain (CTD) are also shown. (B) LC motifs in the STAT2 binding region of IE1 belong to a predicted natively unstructured domain. The IUPred program was used to predict and plot the tendency of intrinsic disorder in the IE1 protein from hCMV (Towne). For comparison, the disorder prediction for the Towne strain IE2 protein is also shown. For IE1, the locations of the 85 amino-terminal residues shared with IE2, a globular central domain (amino acids 50 to 378) predicted by the “structured domains” application of IUPred, and the four LC motifs described for panel A are presented relative to the disorder profile. The chosen IUPred algorithm (“long disorder”) allows prediction of context-independent global disorder that encompasses at least 30 consecutive residues of predicted disorder while the sequential neighborhood of 100 residues is considered. The disorder tendency score can range between 0 and 1, and scores above 0.5 (dotted horizontal line in the left panel) indicate disorder. For additional data, see Fig. S1 in the supplemental material.
Compositionally biased amino acid sequences are regarded as hallmarks of intrinsically unstructured or disordered protein domains which have no single well-defined tertiary structure in their native, functional state (reviewed in reference 15). Frequently, disordered domains function via binding to a structured partner, thereby undergoing a disorder-to-order transition. Several in silico predictors of protein disorder have been developed (reviewed in reference 14), including IUPred (http://iupred.enzim.hu) (13). IUPred recognizes disordered regions from the primary sequence based on the assumption that globular proteins are composed of amino acids which have the potential to form a large number of favorable interactions; in contrast, intrinsically unstructured proteins adopt no stable structure, because their amino acid composition does not allow sufficient favorable interactions to form. Using IUPred, we identified disordered structures at the amino- and carboxy-terminal ends of the hCMV IE1 protein (amino acids 1 to ∼50 and ∼380 to 491, respectively) with very high confidence, while the central region, between amino acids 50 and 378, was predicted to constitute a structured globular domain (Fig. 2B). Very similar results were obtained with other computational tools for structural disorder prediction, including DisEMBL (http://dis.embl.de) (38) (data not shown). Strikingly, the location of the putative disordered region at the IE1 carboxy terminus and its peak scores almost exactly overlap the AD1, S/P, AD2, and AD3 LC motifs, suggesting a structural relation. Furthermore, the strong tendency for structurally disordered carboxy-terminal domains is highly conserved among IE1 proteins of all human and animal CMVs examined, including chimpanzee, rhesus macaque, African green monkey, rat, and mouse virus strains. In fact, extremely high disorder tendency scores were determined for the extended carboxy-terminal regions of mIE1 and rat CMV IE1 proteins (see Fig. S1B in the supplemental material). By comparison, hCMV IE2 is predicted to be rather globally disordered with a relatively structured carboxy-terminal region (Fig. 2B).
Thus, despite limited primary sequence similarity, IE1 proteins of CMVs from diverse mammalian species share the presence of LC motifs and a putatively unfolded carboxy-terminal domain. The predicted structural disorder of this domain has likely evolved to enable binding to multiple host cell proteins, including STAT2.
The LC elements in the IE1 protein constitute core and accessory interaction sites for STAT2.
To examine the relative contributions of each carboxy-terminal LC element to STAT2 binding, we constructed and cloned IE1 mutant cDNAs with individual deletions of the AD1, AD2, AD3, and S/P sequences (Fig. 3A). An AD1-S/P double deletion was also generated. Wild-type and mutant IE1 cDNAs were transiently expressed in H1299 cells, and coimmunoprecipitation assays were performed (Fig. 3B). The results show that the ΔAD1 and ΔS/P mutants were severely (≥80% and ≥60%, respectively) compromised for binding to STAT2 (Fig. 3B and C). Combined mutation of AD1 and S/P (ΔAD1-S/P) resulted in less than 3% of wild-type binding activity (Fig. 3C). In contrast, loss of AD2 or AD3 affected IE1-STAT2 complex formation only moderately (by <40%). In fact, the ΔAD3 mutant exhibited a measurable binding defect only when the amounts of coprecipitated STAT2 were normalized to the slightly varying input IE1 protein levels (Fig. 3C; note that this normalization is relevant only if the overall amounts of IE1 are limiting for complex formation with STAT2, which may not be the case given the high abundance of the overexpressed viral protein).
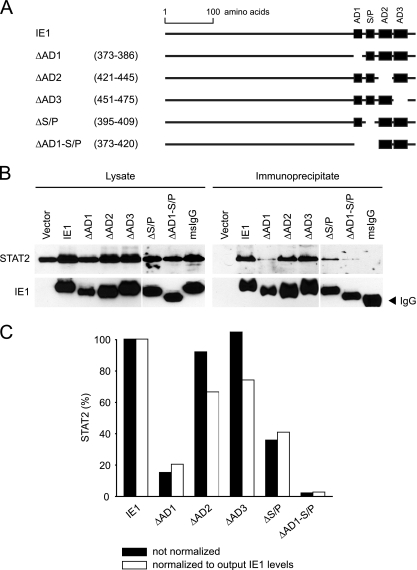
FIG. 3.
LC motifs in the carboxy-terminal IE1 domain specify distinct core and ancillary interfaces for STAT2 binding. (A) Schematic presenting relative positions of carboxy-terminal LC motifs in the full-length IE1 polypeptide and IE1 mutant proteins used in binding and colocalization analyses. The amino acid coordinates of each deletion are provided in parentheses. (B) Results from IE1-STAT2 coimmunoprecipitation experiments. Following cotransfection of H1299 cells with pcDNA-HA-N (vector) or the indicated IE1-expressing derivatives and pcDNA-STAT2-Flag (provided by Curt Horvath, Mount Sinai School of Medicine, New York, NY), cell extracts were subjected to immunoprecipitation using anti-HA agarose or agarose coated with nonspecific mouse (ms) IgG (Table 2). Proteins were separated in 10% polyacrylamide-SDS gels, Western blotted, and detected with primary antibodies H-190 (STAT2) and 1B12 (IE1; from Tom Shenk, Princeton University). Input lysate (5% of material used for immunoprecipitation) and immunoprecipitated output protein levels are shown. The data represent one of two independent experiments with similar outcomes. (C) The output STAT2 bands from B were quantified using Scion Image 4.0.3 (Scion Corporation) and normalized to immunoprecipitated IE1. Both normalized and nonnormalized results are plotted.
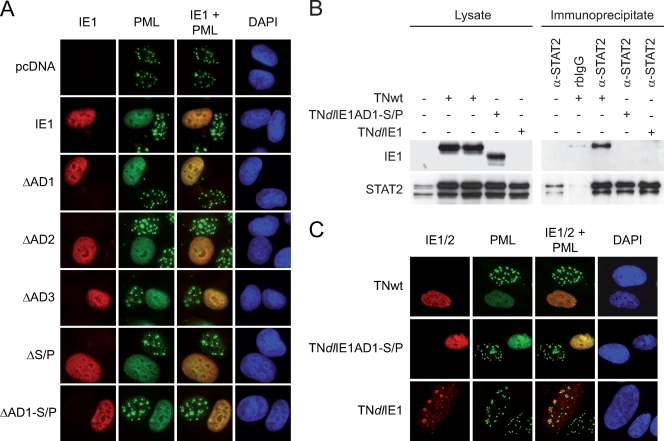
In addition to the binding assays, double-labeling fluorescence microscopy was performed to study the spatial distribution of the mutant IE1 proteins relative to endogenous STAT2 (Fig. 4). Consistent with the binding data, the ΔAD2 and ΔAD3 mutants displayed characteristics similar to those of the wild-type regarding colocalization with STAT2 at ND10 and mitotic chromatin, although fewer ND10s stained double positive for ΔAD2 and STAT2 than for ΔAD3/STAT2 and wild-type IE1/STAT2 (Fig. 4B). Surprisingly, deletion of either AD1 or S/P did not significantly affect colocalization with STAT2 at ND10. However, both mutations individually diminished sequestration of STAT2 at mitotic chromatin. As in the coimmunoprecipitations, the most severe phenotype was observed for the ΔAD1-S/P protein, which was entirely inactive for STAT2 recruitment to either ND10 or chromatin despite the fact that this mutant localized efficiently to both nuclear compartments (Fig. 4A). Deletion of the short sequence (amino acids 387 to 394) between the AD1 and S/P elements did not detectably affect IE1-STAT2 subnuclear codistribution (data not shown), verifying that the two LC elements are the critical determinants in this interaction.
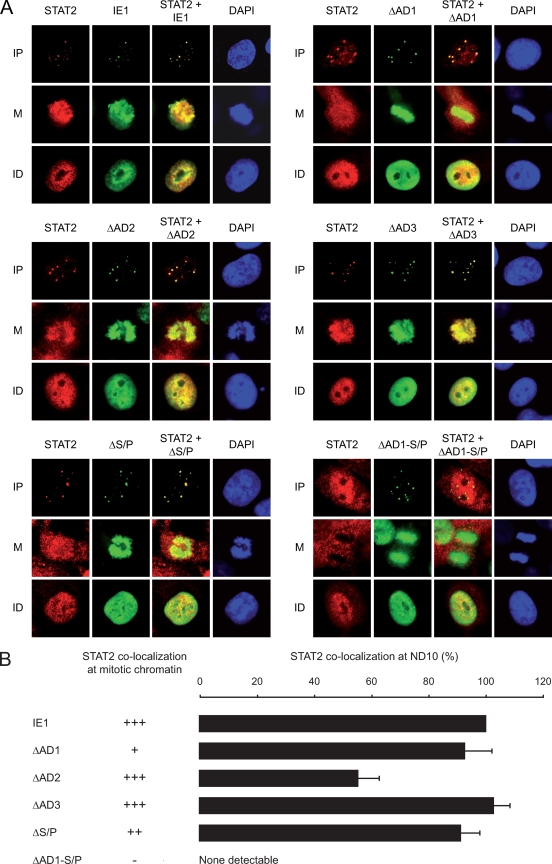
FIG. 4.
Relative contributions of IE1 LC motifs to subnuclear colocalization with STAT2. (A) Nuclear distribution of ectopically expressed wild-type and mutant EGFP-IE1 proteins and endogenous STAT2 in 2fTGH cells. After plasmid transfection and short selection in G418 (300 μg/ml), cells were treated with 1,000 U IFN-α for 1 h and fixed with methanol. Then samples were simultaneously reacted with primary antibodies against STAT2 (H-190) and EGFP (MAB3580), followed by incubation with a rabbit-specific Alexa Fluor 594 conjugate, a mouse-specific Alexa Fluor 488 conjugate, and DAPI. Representative single and merged fluorescent stainings of three different subcellular localization patterns are shown: mitotic (M), interphase with predominantly nuclear punctate IE1 distribution (IP), and interphase with nuclear diffuse protein staining (ID). Magnification, ∼×500. (B) Quantitation of IE1-dependent STAT2 sequestration at ND10. At least 100 nuclei with punctate staining of wild-type or mutant IE1 were analyzed for codistribution of STAT2 by immunofluorescence microscopy, as shown in panel A. Results are percentages of full-length-IE1 activity (set to 100%). For comparison, semiquantitative results from immunofluorescent colocalizations between IE1 and STAT2 at mitotic chromatin are also shown: +++, wild-type activity; ++, moderately reduced activity; +, severely reduced activity; −, no detectable activity.
Curiously, Huh et al. recently identified a region exactly comprising the AD2 and AD3 motifs (amino acids 421 to 475) as being critical for IE1-STAT2 physical interaction (28). To compare the relative contributions of AD2-AD3 and AD1-S/P to STAT2 interaction in our binding and colocalization assays, we transfected H1299 cells with plasmids expressing full-length IE1 or one of the two deletion mutants (see Fig. S2 in the supplemental material). The coimmunoprecipitations confirmed that the ΔAD1-S/P IE1 protein is severely defective for STAT2 binding, whereas the Δ421-475 mutant exhibited binding characteristics resembling wild-type in our hands (see Fig. S2B in the supplemental material). Moreover, in contrast to IE1 lacking AD1-S/P, the Δ421-475 protein colocalized with STAT2 at condensed chromatin or ND10 in most mitotic or interphase cells, respectively (see Fig. S2C in the supplemental material). After all, quantitative image analysis (analogous to Fig. 4B) revealed a ∼20% reduction in IE1-STAT2 colocalization efficiency at ND10 associated with the Δ421-475 mutation compared to the wild-type (data not shown).
These results demonstrate that mutation of individual carboxy-terminal LC motifs, including combined deletion of AD1 and S/P, does not significantly affect protein stability, nuclear import, or subnuclear targeting of IE1. However, the presence of AD1 and S/P elements is critical for IE1-STAT2 interaction. The two proximal LC motifs cooperate to form a core interface for STAT2 binding, while the adjacent AD2 element appears to have an ancillary function in this interaction (Fig. 5). The fact that colocalization with STAT2 at chromosomes and at ND10 does not cosegregate in each IE1 LC mutant might indicate that the viral protein forms distinct STAT2-containing complexes in the chromatin and interchromatin compartments of the cell nucleus.
FIG. 5.
Summary of results from IE1-STAT2 interaction analyses. The relative contributions of each carboxy-terminal LC motif to physical association and subcellular colocalization between IE1 and STAT2, as deduced from the data in Fig. 3 and Fig. 4, are schematically depicted.
STAT2 binding and ND10 disruption are genetically separable activities of IE1.
The experiments whose results are shown in Fig. 4 had already indicated that none of the four conserved LC motifs in the IE1 carboxy-terminal domain is required for condensed chromatin association or ND10 targeting by the viral protein. In addition, we examined all our IE1 internal deletion mutants by immunofluorescence microscopy following transient transfection for their capacity to disrupt ND10 integrity (Fig. 6A). As expected, the wild-type viral protein was able to efficiently trigger relocalization of the ND10 principal marker protein PML from a punctate to a rather homogenous nuclear distribution in most transfected cells, while the dot structures remained intact in all IE1-negative interphase cells. The effect of each mutant tested (ΔAD1, ΔS/P, ΔAD2, ΔAD3, and ΔAD1-S/P) on ND10 integrity was indiscernible from that of the full-length IE1 protein. In fact, even the ΔAD1-S/P mutant, which proved to be completely defective for STAT2 interaction (Fig. 3 and 4), disrupted ND10 with wild-type characteristics (Fig. 6A). Therefore, we conclude that the failure of some IE1 variant proteins to bind to and colocalize with STAT2 efficiently does not result from global structural perturbations induced by the mutations but rather specifically reflects the absence of critical STAT2 binding motifs. Moreover, these results demonstrate that STAT2 interaction and ND10 disruption are genetically separable, distinct activities of the hCMV IE1 protein.
FIG. 6.
STAT2 binding and ND10 disruption are separate activities of IE1. (A) Immunofluorescence analysis examining effects of ectopically expressed wild-type and mutant IE1 proteins on subnuclear distribution of endogenous PML (a major structural component of ND10). After transfection of 2fTGH cells with pcDNA-HA-N derivatives or empty vector and methanol fixation, samples were simultaneously reacted with primary antibodies against IE1 (rbIE1-1) and PML (5E10), followed by incubation with a rabbit-specific Alexa Fluor 594 conjugate, a mouse-specific Alexa Fluor 488 conjugate, and DAPI. Representative single and merged fluorescent stainings of IE1-positive and nontransfected nuclei are shown. Magnification, ∼×500. (B) Results from IE1-STAT2 coimmunoprecipitation experiments. Following infection of MRC-5 cells with TNwt, TNdlIE1AD1-S/P-2, or TNdlIE1-2 viruses (1 PFU/cell, 16 h), cell extracts were subjected to immunoprecipitation using an antibody directed against STAT2 (H-190) or nonspecific rabbit IgG (rbIgG) (Table 2). Proteins were separated in 12% polyacrylamide-SDS gels and Western blotted. STAT2 and IE1 input lysates (5% of material used for immunoprecipitation) and immunoprecipitated output protein levels are shown. (C) Immunofluorescence analysis examining effects of wild-type and mutant viruses on subnuclear distribution of endogenous PML. At 8 h after infection of MRC-5 cells with TNwt, TNdlIE1AD1-S/P-2, or TNdlIE1-2 viruses (1 PFU/cell), samples were simultaneously reacted with primary antibodies against either IE1 (rbIE1-1) (TNwt and TNdlIE1AD1-S/P) or IE2 (1218; a gift from Jay Nelson, Oregon Health and Science University) (TNdlIE1) and PML (5E10), followed by incubation with a rabbit-specific Alexa Fluor 594 conjugate, a mouse-specific Alexa Fluor 488 conjugate, and DAPI. Representative single and merged fluorescent stainings of IE1/IE2-positive and -negative nuclei are shown. Magnification, ∼×500.
To confirm these results in the context of an authentic hCMV infection, we constructed a mutant virus that specifically lacks the STAT2 core binding site (AD1-S/P) of IE1 using a BAC-based allelic exchange strategy (see Fig. S3A in the supplemental material). IE1 expressed from the resulting mutant virus, TNdlIE1AD1-S/P, was compared with results obtained with matching wild-type and IE1-null viruses for STAT2 binding following infection of human fibroblasts (Fig. 6B). In concordance with our transfection assays (Fig. 3), the ΔAD1-S/P protein, although stably expressed from the viral genome, completely failed to bind to endogenous human STAT2 under conditions that allowed binding to full-length IE1 (Fig. 6B). Furthermore, neither the efficiency nor the temporal kinetics of ND10 disruption was detectably different between the wild-type virus and TNdlIE1AD1-S/P, whereas ND10 remained intact after infection with TNdlIE1 (Fig. 6C and data not shown). These results strongly support our view that binding to STAT2 and interaction with ND10 are two separable and, most likely, unrelated activities of the hCMV IE1 protein.
STAT2 interaction-dependent and -independent activities of IE1 contribute to type I IFN resistance and efficient replication of hCMV.
Our prior work has shown that an IE1-null mutant hCMV (CR208) (22) lacking all of major IE exon 4 is hypersensitive to exogenous human IFN-α compared to the corresponding wild-type virus (53). Moreover, neutralization of endogenously produced IFN-β partially complemented the low-multiplicity-dependent growth phenotype of the IE1-deficient virus. These findings suggest that IE1 promotes hCMV replication at low input multiplicities, at least in part, by antagonizing the antiviral type I IFN response (53).
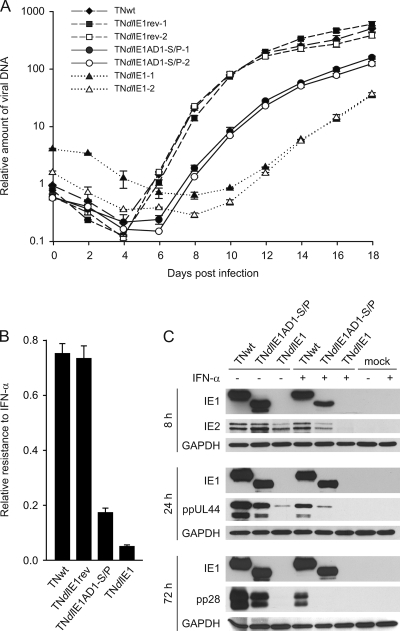
In order to investigate to what extent hCMV IFN resistance depends on the IE1-STAT2 interaction, we compared the replication efficiencies of TNwt, TNdlIE1rev, TNdlIE1AD1-S/P, and TNdlIE1 viruses in the presence and absence of exogenously added IFN-α. Multistep growth analyses after low-multiplicity infection revealed that, as expected, TNdlIE1 strains were severely attenuated in terms of replication kinetics and peak titers compared to the parental virus (Fig. 7A). TNdlIE1AD1-S/P viruses displayed an intermediate phenotype between revertant and TNdlIE1 viruses in these analyses. The differentially attenuated phenotypes of ΔAD1-S/P and IE1-null mutants likely result, at least in part, from varying sensitivities to IFN-β produced from the infected fibroblasts. In support of this view, the TNdlIE1AD1-S/P mutant also exhibited an intermediate phenotype between revertant and IE1-deficient viruses when exogenous IFN-α was added to high-multiplicity infections (Fig. 7B; also, see Fig. S4 in the supplemental material). To obtain further support for these findings, we performed a series of Western blotting experiments (Fig. 7C). In the absence of exogenous IFN-α, IE1 and IE2 steady-state protein levels were comparable between wild-type and TNdlIE1AD1-S/P viruses, and slightly less IE2 was found in the TNdlIE1 infections. However, TNdlIE1 produced markedly reduced levels of early (ppUL44) and late (pp28) viral proteins compared to TNwt, confirming the fact that IE1 activates early gene expression (20, 22). In comparison, accumulation of pUL44 and pp28 was much less severely affected in the TNdlIE1AD1-S/P mutant. When TNwt-infected cells were treated with IFN-α, ppUL44 and pp28 but not IE2 accumulated to significantly lower levels than in nontreated infections, reflecting partial sensitivity of hCMV early gene expression to exogenous type I IFN. However, protein (including IE2) production from TNdlIE1 was hardly detectable, indicating that without IE1, hCMV is not able to initiate replication in the presence of large amounts of IFN-α. Again, TNdlIE1AD1-S/P showed intermediate characteristics between the wild-type and IE1-null phenotypes in this experiment (Fig. 7C).
FIG. 7.
Effects of STAT2 core binding site mutation in IE1 on productive replication and IFN resistance of hCMV. (A) Multistep analysis of replication by wild-type and IE1 mutant viruses. Following inoculation of MRC-5 cells with the indicated virus clones at 0.01 PFU/cell, viral replication was monitored at 2-day intervals by real-time-PCR-based relative quantification of hCMV DNA from culture supernatants. Values are means and standard deviations from two independent infections, each quantified in duplicate. (B) IE1-dependent relative resistance of hCMV to exogenous IFN-α. MRC-5 cells were pretreated for 24 h with 250 U of IFN-α or mock-treated and infected with TNwt, TNdlIE1rev-2, TNdlIE1AD1-S/P-2, or TNdlIE1-2 at an input multiplicity of 3 PFU/cell. Viral replication was assessed by real-time-PCR-based relative quantification of hCMV DNA from culture supernatants, and the ratios between titers from IFN-α-treated and nontreated infections were determined 9 days postinfection. Values are means and standard deviations from two independent infections, each quantified in duplicate. For additional data, see Fig. S4 in the supplemental material. (C) Accumulation of viral proteins as a function of postinfection time, presence of exogenous IFN-α, and IE1 status. MRC-5 cells were pretreated for 24 h with 1,000 U of IFN-α or mock-treated and infected with TNwt, TNdlIE1AD1-S/P-2, or TNdlIE1-2 at an input multiplicity of ∼1 PFU/cell. Whole-cell protein extracts were prepared at very early (8-h), early (24-h) and late (72-h) times postinfection, separated in 12% polyacrylamide-SDS gels, and Western blotted. Viral proteins were detected with the following primary antibodies: 1B12 (IE1), 3A9 (IE2; a gift from Tom Shenk, Princeton University), 10D8 (ppUL44), and 10B4-29 (pp28; also from Tom Shenk). Glyceraldehyde-phosphate dehydrogenase (GAPDH) (antibody ab9485) served as a cellular loading control.
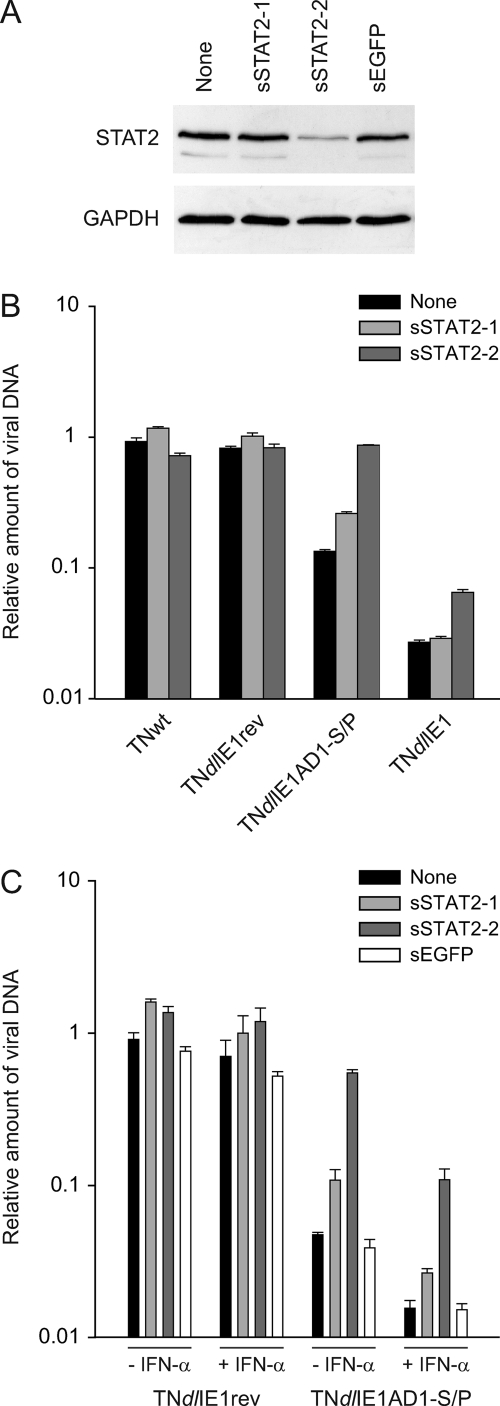
To further substantiate the contribution of STAT2 interaction to the attenuated phenotype of the TNdlIE1AD1-S/P virus, we monitored viral replication following siRNA-mediated knockdown of STAT2 gene expression (Fig. 8). Two siRNAs designed to target the human STAT2 mRNA (sSTAT2-1 and sSTAT2-2) and a negative control siRNA directed against EGFP (sEGFP) were tested for their effects on STAT2 protein levels (Fig. 8A). GAPDH was used as a control protein in these assays. As expected, the EGFP-specific siRNA had no detectable effect on STAT2 or GAPDH levels. Of the two STAT2-directed siRNAs, only one duplex (sSTAT2-2) caused considerable (>80%) STAT2 knockdown, whereas the other one (sSTAT2-1) had little if any effect (Fig. 8A). Consistently, transfection of sSTAT2-2 but not sSTAT2-1 or sEGFP restored the ∼10-fold defect in viral titers associated with the ΔAD1-S/P mutation to wild-type levels (Fig. 8B and C). At the same time, sSTAT2-2 partially rescued defective replication of the TNdlIE1 mutant, and neither one of the two STAT2 siRNAs had any obvious effect on TNwt and TNdlIE1rev virus titers (Fig. 8B and C). Interestingly, STAT2 knockdown by sSTAT2-2 transfection also more than compensated the negative effect of exogenous IFN-α on TNdlIE1AD1-S/P titers (Fig. 8C). This likely indicates that, in the absence of IE1-STAT2 interaction, hCMV replication is hypersensitive not only to exogenous but also to endogenous type I IFN.
FIG. 8.
Knockdown of STAT2 expression restores the viral replication defect associated with the ΔAD1-S/P mutation. (A) Specific reduction in STAT2 protein levels by siRNA-mediated gene silencing. MRC-5 cells were mock transfected (None) or transfected with the indicated siRNA duplexes, followed by infection with TNdlIE1AD1-S/P-2 (3 PFU/cell). Four days postinfection, whole-cell protein extracts were prepared, separated in 10% polyacrylamide-SDS gels, and Western blotted. STAT2 and GAPDH proteins were detected using primary antibodies H-190 and ab9485, respectively. (B) STAT2 knockdown selectively rescues defective replication by IE1 mutant viruses. MRC-5 cells were mock transfected (None) or transfected with the indicated siRNA duplexes, followed by infection with TNwt, TNdlIE1rev-2, TNdlIE1AD1-S/P-2, or TNdlIE1-2 (3 PFU/cell). Four days postinfection, viral replication was assessed by real-time PCR-based relative quantification of hCMV DNA from culture supernatants. Values are means and standard deviations from two independent infections, each quantified in duplicate. (C) IFN dependence of STAT2 knockdown effects on IE1 mutant virus replication. MRC-5 cells were mock transfected (None) or transfected with the indicated siRNA duplexes. At 24 h after transfection, cells were pretreated with 250 U of IFN-α for 24 h or left untreated, followed by infection with TNdlIE1rev-2 or TNdlIE1AD1-S/P-2 (3 PFU/cell). Four days postinfection, viral replication was assessed by real-time PCR-based relative quantification of hCMV DNA from culture supernatants. Values are means and standard deviations from two independent infections, each quantified in duplicate.
Taken together, our results strongly support the view that the relative resistance of hCMV gene expression and replication to type I IFNs crucially depends on the IE1 protein and that interaction between the viral protein and STAT2 via the AD1 and S/P LC motifs contributes significantly to this phenotype. Nonetheless, additional mechanisms independent of STAT2 binding must exist that account for the role of IE1 in counteracting the antiviral IFN response and in promoting viral replication.
DISCUSSION
IFN-α and -β are extremely potent antiviral cytokines that are rapidly synthesized and secreted upon exposure of vertebrates to a broad variety of pathogens. The IFNs transduce through well-characterized intermediates, resulting in the induction of numerous ISG-coded effector proteins or RNAs that combat viral replication in the infected host (reviewed in references 39 and 57). Presumably due to the strong selective pressures of IFN-related host responses, most if not all vertebrate viruses have evolved cleverly devised counteractive strategies. In hCMV and mCMV, several viral functions have been identified that interfere with multiple distinct steps in IFN synthesis, IFN-dependent signaling, and ISG effector function (1, 5, 6, 9-11, 28, 34, 44, 45, 52, 53, 75, 76, 86; reviewed in reference 12). Among the viral factors involved in evasion from type I IFN responses, the hCMV IE1 protein is unique in its ability to counteract a terminal step in Jak-STAT signaling through targeting of nuclear STAT2 (53). The present work was designed to define the physical requirements in the viral protein necessary for binding to STAT2 and to clarify the significance of this interaction for IE1 function in hCMV IFN signaling evasion and productive replication.
The present work confirms that the hCMV IE1 protein forms stable complexes with human STAT2 in transfected and hCMV-infected cells. At first glance, this may appear contradictory or even irrelevant in the light of recent reports showing that STAT2 is efficiently degraded via the proteasome during mCMV and hCMV replication (34, 86). However, STAT2 proteolysis was found to vary with the virus strain and input multiplicity used for infection (34). Moreover, reduced STAT2 protein levels were evident only in the late stages (48 to 72 h), not in the early stages (4 to 24 h), of the hCMV life cycle (34). Our interaction assays were consistently performed in noninfected cells or at 8 to 16 h following hCMV infection (Fig. 3, 4, and 6). In fact, at 16 h postinfection we observed a marked increase rather than a decrease in STAT2 steady-state protein levels compared to mock-infected cells (Fig. 6B). In mCMV, M27 was identified as the protein that targets STAT2 for degradation (86). However, UL27, the M27 ortholog in hCMV, is apparently not involved in destabilization of STAT2 (34). Thus, the viral protein(s) targeting STAT2 for degradation during hCMV replication remains to be determined. IE1 has not been implicated in proteolytic processes, and our data do not indicate a role for this protein in STAT2 degradation. Consequently, two distinct hCMV gene products may target the same host cell factor, although a perhaps more indirect role of IE1 in STAT2 degradation during the late phase of infection cannot yet be ruled out.
Our initial results from coaffinity purification and subnuclear colocalization experiments indicated that the carboxy-terminal 145 amino acids of hCMV IE1 but not residues in the amino-terminal region, including the 85-amino-acid domain shared by IE2 and IE1, are involved in STAT2 interaction (Fig. 1). This observation is in agreement with recent data from Ahn and colleagues (28) and identifies STAT2 as the first and so far only cellular or viral protein associating with the IE1 carboxy terminus via noncovalent interaction. The only other protein known to interact with IE1 in this region is SUMO-1, which covalently attaches to lysine 450 in the viral protein (49, 50, 67, 82) and may negatively regulate STAT2 binding (28).
Protein-protein interactions have been traditionally associated with stable three-dimensional structures in each binding partner. More recently, it has been recognized that even entirely unstructured polypeptides can undergo specific protein interactions (reviewed in reference 16). High-confidence predictions by different algorithms suggest that the carboxy-terminal ∼120 amino acids of IE1 form a natively disordered domain (Fig. 2B and data not shown). Intrinsically disordered regions are thought to exist in a flexible and dynamic state, allowing proteins to adopt multiple conformations and thereby maximizing their interaction capabilities. In fact, many disordered proteins or protein domains fold on binding to structured biological targets (coupled folding and binding [81]). Thus, it is conceivable that a primarily unstructured carboxy-terminal IE1 domain may become locked into a static conformation as a consequence of STAT2 binding.
A common signature of intrinsic disorder is the presence of amino acid compositional bias with a low proportion of bulky hydrophobic amino acids, which would normally form the core of a folded globular protein, and a high proportion of polar and charged amino acids (15, 16). Based on the popular SEG program by Wootton and Federhen (80), the present work identifies four short (14- to 25-amino-acid) elements of pronounced compositional bias (LC motifs) in the carboxy-terminal region of the hCMV IE1 polypeptide (AD1, S/P, AD2, and AD3) (Fig. 2A). The proximal two adjacent LC motifs (AD1 and S/P), which are recognized as one continuous LC element in some virus strains (see Fig. S1 in the supplemental material), turned out to be most important for IE1-STAT2 interaction. In fact, deletion of either one of the two sequences affected binding and colocalization between IE1 and STAT2 significantly (Fig. 3 and 4). However, simultaneous mutation of both AD1 and S/P resulted in complete loss of subnuclear costaining and barely detectable or no complex formation (Fig. 3, 4, and 6). Thus, we propose that the AD1 and S/P motifs act in an additive manner to mediate IE1-STAT2 interaction. On the other hand, mutation of the AD2 motif had only a small, albeit reproducibly detectable, negative effect on IE1-STAT2 binding, and AD3 did not contribute appreciably to the interaction (Fig. 3 and 4). Likewise, simultaneous deletion of AD2 and AD3 affected IE1-STAT2 interaction only marginally in our hands (see Fig. S2 in the supplemental material). This contrasts with the report by Huh et al., who identified a region exactly comprising the AD2 and AD3 motifs (amino acids 421 to 475) as being critical for IE1-STAT2 binding (28). It is difficult to reconcile their findings with our own results, since IE1 proteins from the same virus strain (Towne) and similar cells (human fibroblasts) were used in the two studies. One curious side observation of the present paper concerns the fact that IE1-dependent sequestration of STAT2 at mitotic chromosomes or ND10 did not exactly cosegregate (Fig. 4 and 5). In fact, the AD1 and S/P elements were found to contribute additively to localization of STAT2 at condensed chromatin, while at the same time they were mutually redundant regarding STAT2 targeting to ND10. In addition, AD2 and AD3 were individually dispensable for sequestering STAT2 at chromatin but AD2 measurably affected the efficiency of ND10 localization by the signaling protein. Interestingly, it has been questioned whether deposition of STAT2 at metaphase chromosomes in hCMV-infected cells depends at all on IE1, since the Δ421-475 mutant virus claimed to be all deficient for STAT2 binding still relocalized the cellular protein to metaphase chromatin (28). However, this virus retains intact AD1 and S/P elements that, according to our work, direct STAT2 to chromosomes. Collectively, these data suggest that IE1 might form distinct STAT2-containing complexes, involving somewhat differing cofactors, in the chromatin and interchromosomal (ND10) compartments of the cell nucleus. Alternatively, IE1 might target different STAT2 complexes during the interphase and mitotic stages of the cell cycle. STAT2 does not appear to exist as a monomer or homodimer but usually forms either a trimeric complex (ISGF3) with STAT1 and IRF9 or heterodimers with STAT1, STAT3, or STAT6 (24, 69; reviewed in references 62 and 64). Notably, ISGF3 as well as the three known STAT2 heterodimers are all activated by type I IFNs. Moreover, we have shown that IE1 physically interacts with STAT1 but fails to associate with IRF9 (53; Krauss et al., unpublished). Thus, one attractive speculation is that IE1 may independently interact with at least two different STAT2 heterodimers through slightly varying interfaces in its carboxy-terminal domain.
Intriguingly, we identified predicted structurally disordered carboxy-terminal domains with two to five LC elements in the IE1 orthologs of all examined mammalian CMVs from mice to humans (see Fig. S1 in the supplemental material). In the two rodent CMVs tested (mouse and rat), the presumably disordered regions appeared to be even longer and more distinctive than those in the human and nonhuman primate species. This was not necessarily expected given the very limited amino acid sequence similarity between primate and rodent CMV IE1 proteins. In fact, a recent analysis identified only 22% identical and another 20% related residues over a low-stringency alignment between the hCMV IE1 and mIE1 polypeptide sequences, which may not be statistically significant (7). However, based on our structural predictions for the IE1 carboxy-terminal domains, it appears likely that interaction with STAT2 is an evolutionarily conserved activity of CMV IE1 proteins. Indeed, our results indicate that hCMV IE1 and mIE1 bind to STAT2 with comparable efficiencies in vitro (Fig. 1B and C). Moreover, mIE1 may sequester STAT2 at ND10, similar to its human counterpart (Fig. 1D). Relocalization of STAT2 to the chromatin compartment could not be studied with the mouse ortholog, since mIE1 does not detectably localize to mitotic chromosomes (Fig. 1D) (43). The hCMV and mCMV IE1 gene products are expressed from positionally conserved viral genomic regions, and they were generally believed to exert analogous functions during infection by their respective viruses. However, this view has been challenged by the fact that hCMV IE1 and mIE1 deletion viruses do not seem to share the same growth phenotype in cell culture (21) and may not target the same set of host cell proteins besides PML (reviewed in references 7 and 43). Thus, STAT2 may turn out to be one of the few cellular target proteins shared by the hCMV and mCMV IE1 proteins.
In this study, we also describe the construction and initial characterization of an IE1 mutant virus (TNdlIE1AD1-S/P) which specifically lacks the AD1-S/P core STAT2 binding interface and thus fails to interact with STAT2 (Fig. 6 and 7; also, see Fig. S3 in the supplemental material). Especially at low input multiplicities, this mutant displayed delayed viral early protein accumulation and attenuated replication in human fibroblasts compared to wild-type virus. This phenotype resembles the one originally described for IE1-null viruses (20, 22, 47) and can be efficiently rescued by selective knockdown of STAT2 expression (Fig. 8). However, a matching virus lacking the entire IE1-specific coding sequence (TNdlIE1) showed a much more severe replication defect than TNdlIE1AD1-S/P under the exact same infection conditions. Therefore, we conclude that STAT2 binding contributes to IE1 function in the hCMV lytic cycle but that other (STAT2-independent) activities of the viral protein may have an at least equally important role. The growth defect of an IE1-deficient mutant in human fibroblasts has been ascribed in part to the virus' inability to efficiently counteract the antiviral effects of endogenously produced IFN-β (53). Moreover, several IE1 mutant viruses have been shown to be excessively sensitive to IFN-α or IFN-β added exogenously to cells prior to infection (28, 53). Similarly, TNdlIE1AD1-S/P proved to be hypersensitive to exogenous IFN-α compared to the corresponding revertant virus. However, TNdlIE1 was even more sensitive to IFN-α pretreatment, again indicating that although IE1-STAT2 interaction is important for the relative type I IFN resistance of hCMV, additional IE1-dependent activities certainly contribute. Interestingly, ND10 and individual components of these structures have been implicated in intrinsic as well as IFN-mediated innate defense mechanisms against herpesviruses (8, 74; reviewed in references 17 and 73). Notably, neither colocalization with PML nor the timing and efficiency of ND10 disruption were affected in the ΔAD1-S/P mutant indicating that STAT2 and ND10 interaction are entirely separable, most likely independent activities of IE1 (Fig. 6A and C). This observation is consistent with previous studies that used transient transfection to map the PML relocalizing activity to segments in the amino-terminal (amino acids 53 to 69; reference 35) and central (amino acids 105 to 345 [29, 36, 49, 78]) parts of the viral protein. Consequently, it is very tempting to speculate that, besides STAT2 inhibition, targeting of ND10 by IE1 may be another crucial factor that allows hCMV gene expression and replication in the presence of type I IFNs (see Fig. S5 in the supplemental material). Experiments to investigate this possibility are under way.
Supplementary Material
Acknowledgments
We thank Eva-Maria Hauer, Theresa Knoblach, Sandra Meinel, Ines Tschertner, and Carla Winterling for experimental help. We are also grateful to the following colleagues for providing important reagents: Jin-Hyun Ahn (Suwon, Korea), Roel van Driel (Amsterdam, The Netherlands), Ron Hay (Dundee, United Kingdom), Curt Horvath (New York, NY), Martin Messerle (Hannover, Germany), Jay Nelson (Portland, OR), Tom Shenk (Princeton, NJ), Stephen Spector (San Diego, CA), George Stark (Cleveland, OH), and Hua Zhu (Newark, DE). Finally, we thank Hans Wolf (Regensburg, Germany) for continuous support.
This work was funded in part by the Sixth Framework Programme of the European Union (“TargetHerpes,” LSHG-CT-2006-037517).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 7 October 2009.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J. H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. [DOI] [PubMed] [Google Scholar]
- 4.Awasthi, S., J. A. Isler, and J. C. Alwine. 2004. Analysis of splice variants of the immediate-early 1 region of human cytomegalovirus. J. Virol. 78:8191-8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browne, E. P., and T. Shenk. 2003. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc. Natl. Acad. Sci. USA 100:11439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budt, M., L. Niederstadt, R. S. Valchanova, S. Jonjic, and W. Brune. 2009. Specific inhibition of the PKR-mediated antiviral response by the murine cytomegalovirus proteins m142 and m143. J. Virol. 83:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busche, A., A. Angulo, P. Kay-Jackson, P. Ghazal, and M. Messerle. 2008. Phenotypes of major immediate-early gene mutants of mouse cytomegalovirus. Med. Microbiol. Immunol. 197:233-240. [DOI] [PubMed] [Google Scholar]
- 8.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Child, S. J., M. Hakki, K. L. De Niro, and A. P. Geballe. 2004. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J. Virol. 78:197-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Child, S. J., S. Jarrahian, V. M. Harper, and A. P. Geballe. 2002. Complementation of vaccinia virus lacking the double-stranded RNA-binding protein gene E3L by human cytomegalovirus. J. Virol. 76:4912-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chin, K. C., and P. Cresswell. 2001. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 98:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFilippis, V. R. 2007. Induction and evasion of the type I interferon response by cytomegaloviruses. Adv. Exp. Med. Biol. 598:309-324. [DOI] [PubMed] [Google Scholar]
- 13.Dosztanyi, Z., V. Csizmok, P. Tompa, and I. Simon. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433-3434. [DOI] [PubMed] [Google Scholar]
- 14.Dosztanyi, Z., and P. Tompa. 2008. Prediction of protein disorder. Methods Mol. Biol. 426:103-115. [DOI] [PubMed] [Google Scholar]
- 15.Dunker, A. K., I. Silman, V. N. Uversky, and J. L. Sussman. 2008. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 18:756-764. [DOI] [PubMed] [Google Scholar]
- 16.Dyson, H. J., and P. E. Wright. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6:197-208. [DOI] [PubMed] [Google Scholar]
- 17.Everett, R. D., and M. K. Chelbi-Alix. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819-830. [DOI] [PubMed] [Google Scholar]
- 18.Everett, R. D., C. Parada, P. Gripon, H. Sirma, and A. Orr. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gawn, J. M., and R. F. Greaves. 2002. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J. Virol. 76:4441-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghazal, P., A. E. Visser, M. Gustems, R. Garcia, E. M. Borst, K. Sullivan, M. Messerle, and A. Angulo. 2005. Elimination of ie1 significantly attenuates murine cytomegalovirus virulence but does not alter replicative capacity in cell culture. J. Virol. 79:7182-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 24.Gupta, S., M. Jiang, and A. B. Pernis. 1999. IFN-alpha activates Stat6 and leads to the formation of Stat2:Stat6 complexes in B cells. J. Immunol. 163:3834-3841. [PubMed] [Google Scholar]
- 25.Hayhurst, G. P., L. A. Bryant, R. C. Caswell, S. M. Walker, and J. H. Sinclair. 1995. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J. Virol. 69:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heider, J. A., W. A. Bresnahan, and T. E. Shenk. 2002. Construction of a rationally designed human cytomegalovirus variant encoding a temperature-sensitive immediate-early 2 protein. Proc. Natl. Acad. Sci. USA 99:3141-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 28.Huh, Y. H., Y. E. Kim, E. T. Kim, J. J. Park, M. J. Song, H. Zhu, G. S. Hayward, and J. H. Ahn. 2008. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J. Virol. 82:10444-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, R. A., A. D. Yurochko, E. E. Poma, L. Zhu, and E. S. Huang. 1999. Domain mapping of the human cytomegalovirus IE1-72 and cellular p107 protein-protein interaction and the possible functional consequences. J. Gen. Virol. 80:1293-1303. [DOI] [PubMed] [Google Scholar]
- 31.Keil, G. M., A. Ebeling-Keil, and U. H. Koszinowski. 1987. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J. Virol. 61:1901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korioth, F., G. G. Maul, B. Plachter, T. Stamminger, and J. Frey. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155-158. [DOI] [PubMed] [Google Scholar]
- 33.Lafemina, R. L., M. C. Pizzorno, J. D. Mosca, and G. S. Hayward. 1989. Expression of the acidic nuclear immediate-early protein (IE1) of human cytomegalovirus in stable cell lines and its preferential association with metaphase chromosomes. Virology 172:584-600. [DOI] [PubMed] [Google Scholar]
- 34.Le, V. T., M. Trilling, M. Wilborn, H. Hengel, and A. Zimmermann. 2008. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J. Gen. Virol. 89:2416-2426. [DOI] [PubMed] [Google Scholar]
- 35.Lee, H. R., Y. H. Huh, Y. E. Kim, K. Lee, S. Kim, and J. H. Ahn. 2007. N-terminal determinants of human cytomegalovirus IE1 protein in nuclear targeting and disrupting PML-associated subnuclear structures. Biochem. Biophys. Res. Commun. 356:499-504. [DOI] [PubMed] [Google Scholar]
- 36.Lee, H. R., D. J. Kim, J. M. Lee, C. Y. Choi, B. Y. Ahn, G. S. Hayward, and J. H. Ahn. 2004. Ability of the human cytomegalovirus IE1 protein to modulate SUMOylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linding, R., L. J. Jensen, F. Diella, P. Bork, T. J. Gibson, and R. B. Russell. 2003. Protein disorder prediction: implications for structural proteomics. Structure 11:1453-1459. [DOI] [PubMed] [Google Scholar]
- 39.Loo, Y. M., and M. Gale, Jr. 2007. Viral regulation and evasion of the host response. Curr. Top. Microbiol. Immunol. 316:295-313. [DOI] [PubMed] [Google Scholar]
- 40.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margolis, M. J., S. Pajovic, E. L. Wong, M. Wade, R. Jupp, J. A. Nelson, and J. C. Azizkhan. 1995. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J. Virol. 69:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maul, G. G. 2008. Initiation of cytomegalovirus infection at ND10. Curr. Top. Microbiol. Immunol. 325:117-132. [DOI] [PubMed] [Google Scholar]
- 43.Maul, G. G., and D. Negorev. 2008. Differences between mouse and human cytomegalovirus interactions with their respective hosts at immediate early times of the replication cycle. Med. Microbiol. Immunol. 197:241-249. [DOI] [PubMed] [Google Scholar]
- 44.Miller, D. M., B. M. Rahill, J. M. Boss, M. D. Lairmore, J. E. Durbin, J. W. Waldman, and D. D. Sedmak. 1998. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J. Exp. Med. 187:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller, D. M., Y. Zhang, B. M. Rahill, W. J. Waldman, and D. D. Sedmak. 1999. Human cytomegalovirus inhibits IFN-alpha-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-alpha signal transduction. J. Immunol. 162:6107-6113. [PubMed] [Google Scholar]
- 46.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, S. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, et al. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7:171-180. [PubMed] [Google Scholar]
- 47.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2773. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, and M. A. Martin (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 49.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nevels, M., W. Brune, and T. Shenk. 2004. SUMOylation of the human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 78:7803-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 101:17234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nitzsche, A., C. Paulus, and M. Nevels. 2008. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol. 82:11167-11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paulus, C., S. Krauss, and M. Nevels. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. USA 103:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pellegrini, S., J. John, M. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pizzorno, M. C., M. A. Mullen, Y. N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poma, E. E., T. F. Kowalik, L. Zhu, J. H. Sinclair, and E. S. Huang. 1996. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J. Virol. 70:7867-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1-47. [DOI] [PubMed] [Google Scholar]
- 58.Reinhardt, J., G. B. Smith, C. T. Himmelheber, J. Azizkhan-Clifford, and E. S. Mocarski. 2005. The carboxyl-terminal region of human cytomegalovirus IE1491aa contains an acidic domain that plays a regulatory role and a chromatin-tethering domain that is dispensable during viral replication. J. Virol. 79:225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rost, B., G. Yachdav, and J. Liu. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321-W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saffert, R. T., and R. F. Kalejta. 2007. Human cytomegalovirus gene expression is silenced by Daxx-mediated intrinsic immune defense in model latent infections established in vitro. J. Virol. 81:9109-9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80:3863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindler, C., and C. Plumlee. 2008. Interferons pen the JAK-STAT pathway. Semin. Cell Dev. Biol. 19:311-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sehgal, P. B. 2008. Paradigm shifts in the cell biology of STAT signaling. Semin. Cell Dev. Biol. 19:329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spengler, M. L., K. Kurapatwinski, A. R. Black, and J. Azizkhan-Clifford. 2002. SUMO-1 modification of human cytomegalovirus IE1/IE72. J. Virol. 76:2990-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773-784. [DOI] [PubMed] [Google Scholar]
- 69.Tang, X., J. S. Gao, Y. J. Guan, K. E. McLane, Z. L. Yuan, B. Ramratnam, and Y. E. Chin. 2007. Acetylation-dependent signal transduction for type I interferon receptor. Cell 131:93-105. [DOI] [PubMed] [Google Scholar]
- 70.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 71.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tavalai, N., P. Papior, S. Rechter, and T. Stamminger. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82:126-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tavalai, N., and T. Stamminger. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 1783:2207-2221. [DOI] [PubMed] [Google Scholar]
- 74.Taylor, J. L., D. Unverrich, W. J. O'Brien, and K. W. Wilcox. 2000. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J. Interferon Cytokine Res. 20:805-815. [DOI] [PubMed] [Google Scholar]
- 75.Taylor, R. T., and W. A. Bresnahan. 2005. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J. Virol. 79:3873-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valchanova, R. S., M. Picard-Maureau, M. Budt, and W. Brune. 2006. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J. Virol. 80:10181-10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Volkmer, H., C. Bertholet, S. Jonjic, R. Wittek, and U. H. Koszinowski. 1987. Cytolytic T lymphocyte recognition of the murine cytomegalovirus nonstructural immediate-early protein pp89 expressed by recombinant vaccinia virus. J. Exp. Med. 166:668-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilkinson, G. W., C. Kelly, J. H. Sinclair, and C. Rickards. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233-1245. [DOI] [PubMed] [Google Scholar]
- 79.Woodhall, D. L., I. J. Groves, M. B. Reeves, G. Wilkinson, and J. H. Sinclair. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate-early promoter. J. Biol. Chem. 281:37652-37660. [DOI] [PubMed] [Google Scholar]
- 80.Wootton, J. C., and S. Federhen. 1996. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 266:554-571. [DOI] [PubMed] [Google Scholar]
- 81.Wright, P. E., and H. J. Dyson. 2009. Linking folding and binding. Curr. Opin. Struct. Biol. 19:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu, Y., J. H. Ahn, M. Cheng, C. M. apRhys, C. J. Chiou, J. Zong, M. J. Matunis, and G. S. Hayward. 2001. Proteasome-independent disruption of PML oncogenic domains (PODs), but not covalent modification by SUMO-1, is required for human cytomegalovirus immediate-early protein IE1 to inhibit PML-mediated transcriptional repression. J. Virol. 75:10683-10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yurochko, A. D., M. W. Mayo, E. E. Poma, A. S. Baldwin, Jr., and E. S. Huang. 1997. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoters. J. Virol. 71:4638-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zimmermann, A., M. Trilling, M. Wagner, M. Wilborn, I. Bubic, S. Jonjic, U. Koszinowski, and H. Hengel. 2005. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-γ signaling and antiviral responses. J. Exp. Med. 201:1543-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.