Abstract
The mechanism(s) regulating the expression of the TBX2 gene, a key regulator of development, is poorly understood and thus limits an understanding of its function(s). Here we demonstrate that 12-O-tetradecanoylphorbol-13-acetate (TPA) induces TBX2 expression in normal human fibroblasts in a protein kinase C (PKC)-dependent and MAPK-independent manner. Our data further reveal that TPA activates transcription of TBX2 through activating MSK1, which leads to an increase in phosphorylated histone H3 and the recruitment of Sp1 to the TBX2 gene. In addition, TPA was shown to activate MSK1 in a PKC-dependent and MAPK-independent manner. This study is the first to provide evidence that phosphorylation of histone H3 leads to the transcriptional activation of the TBX2 gene and to link MSK1 to PKC.
TBX2, a member of the developmentally important T-box gene family, encodes a transcriptional repressor and is expressed in a wide variety of tissues and organs in developing embryos (1–4). Tbx2 has been shown to repress Nppa, a key molecule of heart development (5) and is essential for limb development and establishing digit identities (6, 7). Moreover, there is growing evidence that TBX2 may play a role in tumorigenesis, because its expression levels are up-regulated in several cancers (7–10), and it is able to override key cell cycle checkpoints when it is inappropriately expressed (11–13). The above reports would suggest that deregulated TBX2 levels have detrimental consequences to normal cellular function, and it is therefore critical to understand the mechanism(s) regulating TBX2 gene expression. However, while several pathways, including BMP, Shh, and stress signaling, have been associated with the regulation of TBX2 gene expression, the details of this regulation remain unknown (14, 6, 7).
Protein kinase C (PKC)2 represents an important multigene family of serine/threonine kinases that have broad and diverse functions in many physiological and pathological processes because they can be activated by various extracellular and intracellular signals (15, 16). The PKC family comprises at least 12 isoforms that are divided into three subfamilies: conventional or classical PKCs (cPKCs; α, β ½, and γ), novel PKCs (nPKCs; δ, ϵ, η, and θ), and atypical PKCs (aPKCs; ζ and λ/ι). PKC is a major intracellular target of 12-O-tetradecanoylphorbol-13-acetate (TPA), and it is well established that in response to TPA, PKCs can activate the Ras/Raf/MEK/ERK MAPK pathway. There is also accumulating evidence suggesting that TPA can activate p38 and JNK in a cell type-dependent manner (17, 18).
The nucleosome is the basic building unit of chromatin and consists of a protein core of a histone octamer (H2A, H2B, H3, and H4) with 146 base pairs of DNA wound around it. N-terminal tails of histones protrude outside of the nucleosome and are thus readily subject to a variety of covalent modifications such as acetylation, phosphorylation, methylation, ubiquitination, and ADP-ribosylation. These modifications have been proposed to cause an alteration of chromatin configuration and/or form a recognition motif allowing access of relevant proteins of transcription to regulate gene expression (19, 20). For example, phosphorylation of histone H3 has been associated with transcriptional activation of c-jun and c-fos immediate-early (IE) response genes in response to growth factors and phorbol esters (21). The list of genes identified that are associated with stimulus-induced phosphorylation of histone H3 is growing (22).
Here, we demonstrate that TPA induces TBX2 gene expression in normal human lung fibroblasts in a PKC-dependent and MAPK-independent manner. Our data further reveal that TPA-induced phosphorylation of histone H3 plays a critical role in regulating TBX2 gene expression. Moreover, we provide evidence to link PKC to the activation of the mitogen- and stress-activated kinase 1 (MSK1), which we show to be responsible for the TPA-induced phosphorylation of histone H3 seen in this study.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
WI-38 (ATCC CCL-75) normal human embryonic lung fibroblasts, their in vitro transformed counterparts WI-38 CT-1 (23) and SVWI-38 cell lines were maintained in Dulbecco's-modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were maintained at 37 °C in an atmosphere of 5% CO2. For TPA treatment, we used 100 nm TPA (Sigma). The PKC inhibitor GF109203 (20 μm), MEK-1 inhibitor PD98095 (25 μm), p38 inhibitor SB203580 (10 μm), and JNK inhibitor SP600125 (20 μm) (Calbiochem) were added 1 h prior to treatment with TPA.
Plasmids, Transfections, and Reporter Assays
The human TBX2 promoter luciferase reporter construct was generated by inserting the SacI(-1606)-HindIII(+32) fragment of the human TBX2 gene into the appropriately cleaved luciferase reporter vector pGL3-basic (Promega). The pRL-TK vector (Promega) was used as an internal control reporter to test for transfection efficiency. The 4xAP-1-luc construct, pCMV-c-Jun and pCMV-JunB expression vectors were kindly provided by Dr. Michael Birrer (National Institutes of Health). Cells were plated at 1.5 × 105 cells/ml in 6-well plates 1 day before transfection. Non-liposomal-mediated gene transfer was performed using FuGENE®6 (Roche Applied Science) according to manufacturer's instructions using 1 μg of DNA consisting of the reporter construct, expression vectors, and the internal control vector. Thirty hours after transfection, cells were analyzed for luciferase activity using the Dual-Luciferase® Reporter Assay (Promega) following manufacturer's instructions and quantified with a Luminoskan Ascent Luminometer (Thermo LabSystems).
Small Interfering RNA (siRNA)
Suppression of MSK1 cellular expression was achieved using siRNA that specifically targets MSK1 mRNA. WI-38 cells were transfected with 5 μm anti-MSK1 siRNA or control (non-silencing) siRNA (Qiagen) for 72 h, using HiPerfect reagent (Qiagen) according to the manufacturer's instructions. Cells were treated with TPA 8 h prior to harvesting protein for Western blotting.
Western Blotting
Western blot assays were carried out as described previously (24). Proteins were resolved on 8–12% SDS-polyacrylamide gels, as required and transferred to Hybond ECL (Amersham Biosciences). The membranes were probed with appropriate primary antibodies and detected using peroxidase-conjugated anti-mouse or anti-rabbit antibodies (1:5000) and visualized by ECL (Pierce). The primary antibodies used were: mouse monoclonal anti-Tbx2 62-2 antibody (1:2000), rabbit polyclonal anti-phospho-H3 (1:2000, Upstate Biotechnology), anti-phospho-p38 (1:1000), anti-p38 (1:5000), anti-phospho- MSK1 (1:1000), anti-phospho-ERK1/2 (1:1000), anti-phospho-JNK (1:1000) (Cell Signaling Technology Inc., Beverly, MA), goat polyclonal anti-MSK1 (1:1000), rabbit polyclonal anti-c-Jun, JunB, ERK2, Sp1 (1:500), and mouse monoclonal α-tubulin (1:500) from Santa Cruz Biotechnology. Densitometric analysis of Western blots was done with a Syngene scanner and GeneTool and GeneSnap software (Syngene).
Real-time RT-PCR
Total RNA was extracted from cultured cells and quantitative RT-PCR assays were carried out using the LightCycler as previously described (24). Primers used for RT-PCR were human TBX2: (forward) 5′-ATGGGCATGGGTCACCTACT-3′; (reverse) 5′-GGTGTAGGGGTATTTTAAGA-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH): forward 5′-GAAGGCTGGGGCTCATTT-3′; reverse 5′-CAGGAGGCATTGCTGATGAT-3′ 3 PCR conditions were as follows: 95 °C for 10 min; and 45 cycles of 95 °C for 45 s (denaturation), 59 °C for 20 s (annealing), and 72 °C for 20 s (extension). The PCR products of TBX2 were cloned into the pGEM-T easy vector (Promega) and verified by sequencing.
DNA Affinity Immunoblotting (DAI)
Nuclear extracts were prepared from WI-38 cells as previously described (25). For DAI assays, biotinylated DNA probes were generated by PCR using primer pairs that were synthesized and 5′-end-labeled with biotin (Forward: 5′-TGGCCTGAGCTGTCAAAAC-3′; Reverse: 5′-GCGCGACTGGTTAGATCTTG-3′), and immobilized on Dynabeads Streptavidin (Dynal Invitrogen) according to the manufacturer's instructions. For each reaction, 40 μg of nuclear extract was incubated with 1 μg of DNA probes in binding buffer (20 mm Tris-HCl (pH 7.6), 50 mm NaCl, 1 mm MgCl2, 0.2 mm EDTA, 0.5 mm dithiothreitol, 5% glycerol, and 2 μg of poly(dI-dC)) in a final volume of 400 μl. The beads were extensively washed with binding buffer and then boiled in 25 μl of 2× loading buffer before SDS-polyacrylamide gel electrophoresis followed by immunoblotting.
Chromatin Immunoprecipitation Assays (ChIP)
ChIP assays were carried out as previously described (13). Briefly, WI-38 cells (60–70% confluence) were serum-starved 24 h prior to treatment with TPA or vehicle. Cells were fixed in 1% formaldehyde after 8-h treatment. The chromatin was extracted, sonicated, and immunoprecipitated using antibodies against Sp1 (Santa Cruz Biotechnology) or phospho-histone H3 (Upstate Biotechnology). DNA precipitation was analyzed by PCR using human TBX2-specific primer pairs (Forward: 5′-TGGCCTGAGCTGTCAAAAC-3′; Reverse: 5′-GCGCGACTGGTTAGATCTTG-3′).
RESULTS
Induction of TBX2 Gene Expression by TPA in Human Fibroblasts
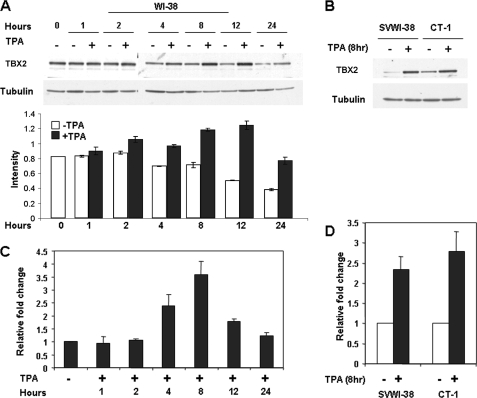
We have previously shown that TBX2 is expressed at high levels in the normal human lung fibroblast cell line, WI-38, but that it is expressed at very low levels in WI-38-transformed cell lines, CT-1 and SVWI-38 (24). To identify signaling pathways that regulate TBX2 expression, WI-38 cells were treated with TPA and TBX2 levels were examined by Western blot analysis. Our results showed that TPA significantly induced TBX2 expression in a time-dependent manner with levels increasing from 2 h and peaking at 8 h (Fig. 1A). Interestingly, TBX2 expression was strongly induced in the SVWI-38 and CT-1 cells treated with TPA for 8 h (Fig. 1B) suggesting that the TPA signaling pathway plays an important role in the regulation of TBX2 expression.
FIGURE 1.
Induction of TBX2 expression by TPA in human fibroblasts. WI-38 (A and C) and SVWI-38 and CT-1 (B and D) cells were treated with either vehicle (control) or TPA (100 nm) for the indicated times. Whole cell lysates and total RNA were harvested and subjected to Western blot (A and B) or real-time PCR (C and D) analyses, respectively. The bar graph in A compares the intensity of the TBX2 band in each sample normalized to the tubulin loading control.
To determine whether the above induction of TBX2 expression by TPA is due to an increase in transcription, cells were treated with or without TPA for a period spanning 1 h to 24 h and TBX2 mRNA levels assessed by real-time PCR. Our results showed that TBX2 mRNA levels were up-regulated from 2 h with levels peaking at 8 h (Fig. 1C) in WI-38 cells. Similarly, TBX2 mRNA levels increased when SVWI-38 and CT-1 cells were treated with TPA for 8 h (Fig. 1D). TBX2 mRNA levels therefore closely correlated with levels of TBX2 protein, which suggested that TPA induced TBX2 expression at a transcriptional level.
Induction of TBX2 Gene Expression by TPA in WI-38 Cells Is PKC-dependent but MAPK-independent
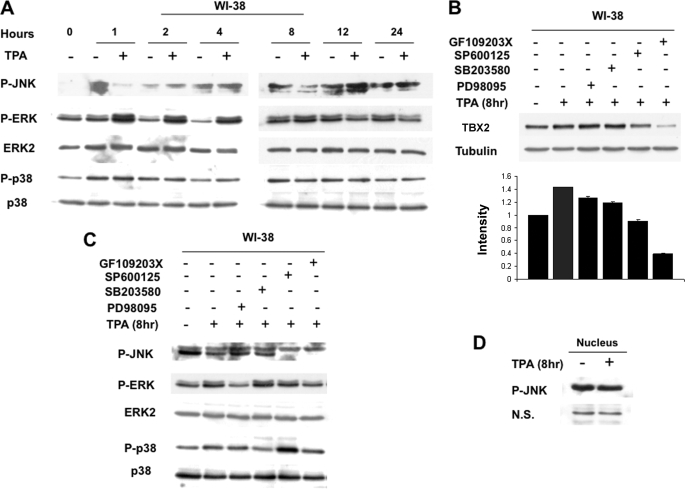
It is well established that in response to TPA, PKCs can activate the Ras/Raf/MEK/ERK MAPK pathway. Thus to investigate the pathway(s) by which TPA up-regulates TBX2 gene expression in WI-38 cells, we first examined the effect of TPA on the phosphorylation status of MAPKs by Western blot analyses. Fig. 2A shows that while TPA treatment had no effect on JNK phosphorylation, it led to an increase in phosphorylation of ERK1/2 up to 8 h and to a marginal increase in p38 phosphorylation at the 4-h time point.
FIGURE 2.
Induction of TBX2 gene expression by TPA in WI-38 cells is PKC-dependent but MAPK-independent. A, cells were treated with either vehicle (control) or TPA (100 nm) for the indicated times and whole cell lysates harvested and subjected to Western blot analysis with the indicated antibodies. B, cells were pretreated with vehicle or inhibitors to PKC (20 μm GF109203X), ERK1/2 (25 μm PD98095), JNK (20 μm SP600125), and p38 (10 μm SB203580) for 1 h and then treated with TPA (100 nm) for 8 h. Western blots were carried out to detect levels of TBX2, and the bar graph compares the intensity of the TBX2 band in each sample normalized to the tubulin loading control. C, cells were treated as for B and Western blotting carried out with the indicated antibodies. D, WI-38 cells were treated with vehicle or TPA for 8 h, and nuclear proteins were prepared and subjected to Western blotting to detect phosphorylated JNK. N.S. represents a nonspecific band, which is included to show equal loading.
To assess the involvement of the above signaling molecules in TPA-induced expression of TBX2, we next pretreated WI-38 cells with inhibitors to PKC, ERK1/2, p38, and JNK before TPA treatment and then analyzed TBX2 expression by Western blotting. As expected, the PKC inhibitor (GF109203X) repressed TPA-induced TBX2 expression confirming that PKC is an important mediator of TBX2 regulation by the TPA pathway (Fig. 2B). It is important to note, that inhibiting PKC also resulted in reduced basal TBX2 expression, which implies that PKC may in fact be essential for TBX2 expression in WI-38 cells. Moreover, as shown in Fig. 2B, at 8 h of TPA treatment both ERK1/2 (PD98059) and p38 (SB203580) inhibitors had no effect on the induction of TBX2 expression but the JNK inhibitor (SP600125) blocked TPA-induced TBX2 expression. Similar results were also obtained at 4 h of TPA treatment (data not shown) which implicated the JNK pathway in the TPA-mediated induction of TBX2.
To determine the blocking efficiency and specificity of the above inhibitors, their effect on phosphorylation of their respective MAPKs were examined. Our results showed that PD98059 and SB203580 can effectively reduce phosphorylation of ERK1/2 and p38, respectively (Fig. 2C). Surprisingly, while TPA-mediated TBX2 expression was repressed by SP600125 (see Fig. 2B), TPA had no effect on JNK phosphorylation (Fig. 2, C and D). These results suggested that the effect of the SP600125 inhibitor on TBX2 expression was not due to the JNK pathway.
TPA and AP-1 Does Not Activate a TBX2 Gene Reporter Construct
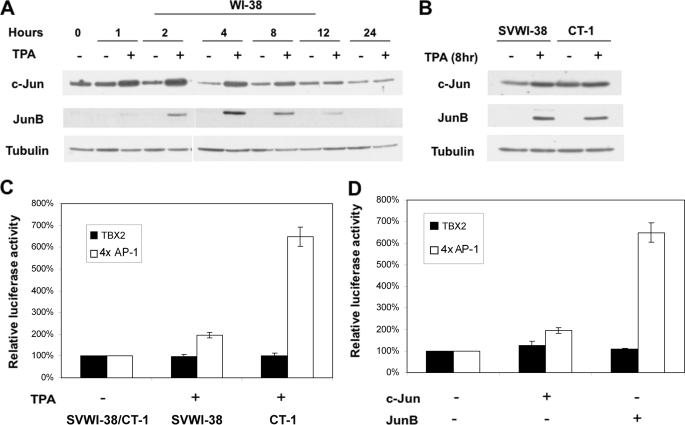
It is well documented that TPA induces the expression of numerous genes through up-regulation of AP-1, and we therefore examined the expression of c-Jun and JunB in WI-38, SVWI-38, and CT-1 cells treated with or without TPA by Western blot analyses. Results showed that TPA increased both c-Jun and JunB protein levels in a time-dependent manner, which correlated with an up-regulation of TBX2 in all three cell lines tested (Fig. 3, A and B and see Fig. 1, A and B).
FIGURE 3.
TPA, c-Jun, and JunB do not affect human TBX2 promoter activity. A and B, TPA-stimulated expression of c-Jun and JunB in human fibroblasts. WI-38, SVWI-38, and CT-1 cells were treated with either vehicle (control) or TPA (100 nm) for the indicated times. Whole cell lysates (the same used in Fig. 1, A and B) were subjected to Western blot analysis with the indicated antibodies. C, TBX2 promoter-luciferase reporter (500 ng) or a 4×AP-1-luciferase construct (50 ng) containing 4 AP-1 binding sites was co-transfected into SVWI-38 and CT-1 cells with the internal control pRL-TK (50 ng) in the presence or absence of TPA (100 nm for 12 h). Luciferase activity was measured 30 h post-transfection and normalized to Renilla luciferase activity. Relative luciferase activity was calculated by setting untreated promoter activity to 100%. Mean values (±) were calculated from three independent experiments. D, TBX2 promoter-luciferase reporter (500 ng) or the 4×AP-1-luciferase construct (50 ng) was co-transfected into HT1080 cells either with the empty pCMV (200 ng) vector or c-Jun or JunB (200 ng) expression vectors, and luciferase activity determined as in C above. Relative luciferase activity was calculated by setting the effect of empty pCMV vector on promoter activity to 100%. Mean values (±) were calculated from three independent experiments.
To determine whether AP-1 can indeed regulate the TBX2 promoter we searched the 3000 bp 5′-flanking regulatory sequence of the human TBX2 gene, which was obtained from the human genome sequence data base, for potential cis-acting elements using the TFSEARCH program. A putative AP-1 site was found at −1069 relative to the transcriptional start site (data not shown), and we thus performed luciferase assays using a reporter construct driven by a 1638-bp sequence of the TBX2 gene containing this putative site (26). TPA failed to stimulate transcriptional activity of the human TBX2 promoter in SVWI-38 and CT-1 cells (Fig. 3C), and overexpression of c-Jun or JunB did not significantly increase TBX2 promoter activity in HT1080 cells (Fig. 3D). As can been seen in Fig. 3, C and D, a reporter plasmid containing four AP-1 DNA binding sites fused to luciferase (4×AP-1-luciferase), which served as a positive control, was responsive to TPA, c-Jun, and JunB. These findings led us to hypothesize that either distal elements not present in the 1638 bp TBX2 regulatory fragment may be mediating the effects of TPA or that a chromatin remodeling mechanism may be involved in transcriptional regulation of TBX2 gene expression by TPA.
Phosphorylation of Histone H3-Ser-10 Is Required for Binding of Sp1 to the TBX2 Gene in WI-38 Cells
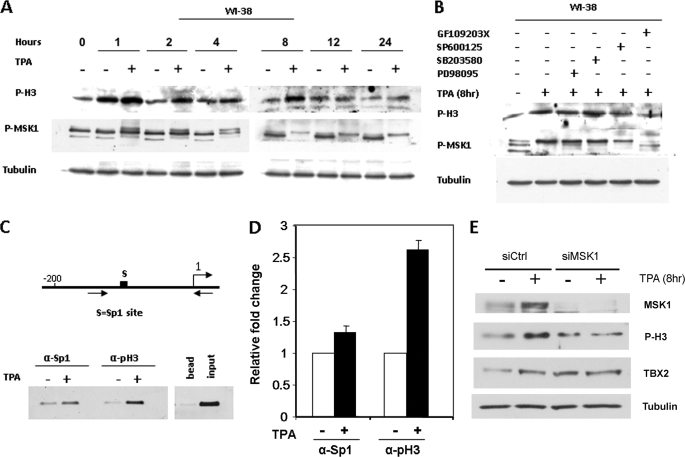
Several lines of evidence suggest that translational modification of histone H3 leads to the activation of genes in a Sp1 site-dependent manner (24, 27–29). We have previously demonstrated that the basal transcription of the human TBX2 gene is dependent on a Sp1 site in its proximal promoter (26). This led us to ask the question as to whether TPA induces TBX2 expression through phosphorylating histone H3 and consequently recruiting Sp1 to the Sp1 site identified above. Western blot analyses were therefore performed to examine the effect of TPA on the phosphorylation status of histone H3 in WI-38 cells. Our results show that TPA treatment resulted in an increase in the levels of phosphorylated histone H3 at Ser-10 for up to 8 h (Fig. 4A), which was blocked by the PKC (GF109203X) and JNK (SP600125) inhibitors (Fig. 4B). Phosphorylation of histone H3-Ser-10 by TPA, therefore, correlated well with the regulation of TBX2 expression by TPA.
FIGURE 4.
Induction of TBX2 expression by TPA in WI-38 cells is associated with histone H3-Ser-10 phosphorylation and recruitment of Sp1 to the TBX2 promoter. A, TPA-stimulated increase of global histone H3-Ser-10 phosphorylation and activation of MSK1. Cells were treated with either vehicle (control) or TPA (100 nm) for the indicated times. Whole cell lysates were harvested and subjected to Western blot analysis to detect phosphorylated histone H3-Ser-10 and MSK1 and tubulin was included as a loading control. For observing phosphorylated MSK1, proteins were separated on 6% SDS-PAGE and the patterns of electrophoretic retardation were observed. B, cells were pretreated with vehicle or inhibitors to PKC (20 μm GF109203X), ERK1/2 (25 μm PD98095), JNK (20 μm SP600125), and p38 (10 μm SB203580) for 1 h and then treated with TPA (100 nm) for 8 h. Western blots were carried out as described for A above. C, top schematic illustrates the region of TBX2 proximal promoter showing the putative Sp1 site (boxed). Arrows represent primer pairs used for PCR. Cells were treated with either vehicle (control) or TPA (100 nm) for 8 h, and ChIP assays were performed with antibodies against Sp1 or phospho-H3. Co-immunoprecipitated DNA was assayed by PCR with primer pairs indicated. Inputs and no antibody are positive and negative controls, respectively. D, co-immunoprecipitated DNA was assayed by real-time PCR and was normalized to input. Fold change values were calculated by setting untreated samples to 1. E, siRNA-mediated down-regulation of MSK1 abrogates the TPA-induced increase in phosphorylated histone H3-Ser-10 and activated TBX2 expression. WI-38 cells were treated with siRNA specific for MSK1 or a control siRNA for 72 h. Cells were treated with TPA (100 nm) for 8 h prior to harvesting protein, which were analyzed by Western blotting using antibodies specific for MSK1, phosphorylated histone H3-Ser-10, TBX2, and tubulin.
We next examined whether TPA induced histone H3-Ser-10 phosphorylation at the TBX2 promoter using ChIP assays. Primers spanning the proximal promoter of TBX2 (Fig. 4C) were used for PCR amplification of immunoprecipitated DNA fragments and quantitative analysis was performed by real-time PCR. Compared with untreated cells, a ∼2-fold increase in histone H3-Ser-10 phosphorylation was obtained at the TBX2 promoter in WI-38 cells treated with TPA for 8 h (Fig. 4, C and D). To explore the possibility that TPA enhanced Sp1 binding to the TBX2 promoter, the same primers shown in Fig. 4C were used to amplify DNA fragments immunoprecipitated with an antibody specific to Sp1. A significant increase of Sp1 was observed at the TBX2 promoter in WI-38 cells treated with TPA (Fig. 4, C and D). Our results clearly show that TPA increased both histone H3-Ser-10 phosphorylation and Sp1 recruitment to a Sp1 site in the TBX2 proximal promoter, which correlated with the induction of TBX2 expression. These findings suggested that TPA-activated transcription of TBX2 through a mechanism involving the phosphorylation of histone H3 and consequent recruitment of Sp1 to the TBX2 proximal promoter.
PKC-dependent Phosphorylation of Histone H3-Ser-10 Correlated with Activation of MSK1
Whereas several kinases, including MSK1/2 (30, 31), ribosomal S6 kinase 2 (RSK2) (32), PKA (33), PKC (34), and IκB kinase-α (IKK-α) (35) have been proposed to phosphorylate histone H3 in response to various stimuli, current evidence suggests that MSK1 and 2 are the primary kinases involved (22). We, therefore, investigated whether MSK1 was activated in WI-38 cells treated with TPA by reprobing the Western blots shown in Fig. 4, A and B with an antibody to phosphorylated MSK1. An increase in MSK1 phosphorylation was evident from 1 h and persisted over a period of 24 h of TPA treatment (Fig. 4A) and as seen for histone H3-Ser-10, phosphorylation was abolished by the PKC (GF109203X) and JNK (SP600125) inhibitors. The result obtained with the JNK inhibitor is worth commenting on at this stage because while results shown in Fig. 2, A, C, and D indicated that TPA did not significantly activate JNK in our system, the JNK inhibitor, SP600125, blocked TPA-induced expression of TBX2 (Fig. 2B), and the phosphorylation of histone H3-Ser-10 and MSK1 (Fig. 4B). Interestingly, SP600125 has recently been identified to act as an inhibitor of histone H3-Ser-10 phosphorylation through an unknown mechanism (36). These findings suggested that SP600125 may repress phosphorylation of histone H3-Ser-10 through blocking MSK1 activation. Furthermore, these results raised the possibility that the TPA-induced TBX2 expression resulted from the phosphorylation of histone H3 by MSK1. To test this possibility, we knocked down MSK1 in WI-38 cells treated with or without TPA and examined the effect on phosphorylated histone H3-Ser-10 and TBX2 levels. Western blot analyses show that MSK1-specific siRNA results in substantial reduction in MSK1 levels since it was undetectable (Fig. 4E). As expected, cells transfected with control siRNA had increased levels of MSK1, phosphorylated histone H3-Ser-10, and TBX2. In contrast, these changes were abrogated in cells transfected with siMSK1. Our results thus confirm that in the absence of MSK1, TPA has no effect on levels of both phosphorylated histone H3-Ser-10 and TBX2. In summary, the above findings revealed a potential functional link between PKC and MSK1 activation during TPA-mediated phosphorylation of histone H3-Ser-10 and TBX2 expression.
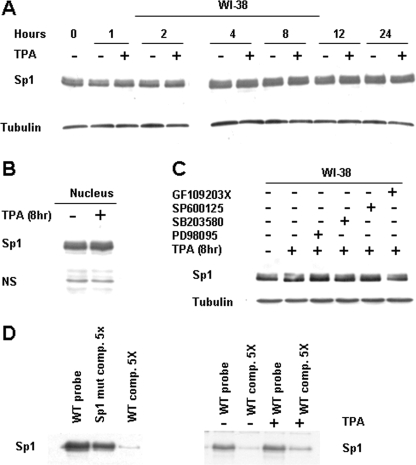
Effect of TPA on Sp1 Expression and DNA Binding Activity
We have clearly shown here that TPA-induced phosphorylation of histone H3-Ser-10 was associated with recruitment of Sp1 to the human TBX2 gene. However, several lines of evidence have also shown that TPA-mediated expression of certain genes is associated with increased levels of total and/or phosphorylated Sp1 (37). To test whether the TPA-mediated increase in TBX2 gene expression is also due to the latter mechanism involving increased levels of Sp1 we next examined the expression pattern of Sp1 in WI-38 cells treated with or without TPA. Western blotting, however, revealed that TPA treatment had no significant effect on total (Fig. 5A) and nuclear (Fig. 5B) Sp1 protein levels at time points tested. In addition, Sp1 protein levels remained unchanged when cells were pretreated with the PKC, JNK, p38, and ERK inhibitors (Fig. 5C).
FIGURE 5.
TPA has no effect on Sp1 expression but stabilizes its binding to the TBX2 promoter. A, WI-38 cells were treated with either vehicle (control) or TPA (100 nm) for the indicated times and cell lysates subjected to Western blot analysis to detect Sp1. B, as for A above but nuclei were prepared from WI-38 cells treated with either vehicle (control) or TPA (100 nm) for 8 h. N.S. represents a nonspecific band that is included to show equal loading. C, as for A above but cells were pretreated with vehicle or inhibitors to PKC (20 μm GF109203X), ERK1/2 (25 μm PD98095), JNK (20 μm SP600125), and p38 (10 μm SB203580) for 1 h prior to TPA treatment (8 h). D, for DAI analysis, biotinylated DNA fragments were generated by PCR using indicated primer pairs and immobilized on streptavidin beads. After incubation with nuclear extracts (40 μg), the DNA-bound Sp1 complexes were analyzed by gel electrophoresis followed by immunoblotting using an antibody to Sp1. Competition assays were performed in the presence of a 5-fold excess of unlabeled wild-type or mutant CCAAT-box DNA fragments.
We finally explored the possibility that TPA treatment may enhance the affinity of Sp1 to bind to the proximal TBX2 promoter. Results from DNA affinity immunoblotting experiments showed that while the Sp1 site in the proximal TBX2 promoter was specifically bound by Sp1 (Fig. 5D, left), TPA treatment did not enhance this binding (Fig. 5D, right). Interestingly, an unlabeled probe was less able to compete with the labeled probe in nuclear extracts from TPA-treated cells, which suggests that TPA treatment may increase the stability of Sp1 binding (Fig. 5D, right).
DISCUSSION
The mechanisms involved in the transcriptional regulation of the TBX2 gene are still poorly understood. In view of the detrimental consequences resulting from altered levels of T-box proteins, as seen in both developmental disorders and in certain cancers, the need to identify such mechanisms is important. Here we demonstrate for the first time that TPA treatment of normal human fibroblasts (WI-38) leads to a time-dependent increase in TBX2 mRNA, and protein levels and we provide compelling evidence that this results from TPA-regulating TBX2 transcription through a process involving phosphorylation of histone H3.
While it is well established that phosphorylation of histone H3 plays a critical role during transcriptional activation of IE genes in response to various stimuli, it has recently also been linked to transcriptional activation of several relative late response genes (22, 38). Here we show that TPA treatment leads to increases in histone H3-Ser-10 phosphorylation, which interestingly is associated with the recruitment of Sp1 to its cognate site in the TBX2 proximal promoter and the induction of TBX2 expression. This is in keeping with our previously published data, which identified this Sp1 site to play an important role in the regulation of TBX2 gene expression as it was shown to be essential in maintaining basal activity of a human TBX2 promoter (26). Moreover, when the phosphorylation of histone H3-Ser-10 was blocked, the TPA-mediated induction of TBX2 expression was abolished. These findings suggest that the mechanism by which TPA activates TBX2 gene expression involves phosphorylation of histone H3 and recruitment of Sp1 to the Sp1 site in the TBX2 proximal promoter.
This study provides compelling data to show that MSK1 is the kinase responsible for phosphorylating histone H3 in response to TPA treatment. While MSK1 has been well characterized as a histone H3 kinase (30, 31), the ERK and/or p38 MAPK cascades have been demonstrated to phosphorylate histone H3 in response to mitogen- and stress-induced pathways in many different systems (22). Interestingly, PKC has also been proposed to be responsible for histone H3-Ser-10 phosphorylation at the LDL receptor promoter in response to TPA in the human hepatocellular carcinoma HepG2 cells (34). Although we cannot rule out the possibility that PKC may also directly phosphorylate histone H3-Ser-10, our findings clearly suggest that MSK1 is a potential downstream target of PKC in normal human fibroblasts and is linked to phosphorylation of histone H3. Interestingly, we found that MSK1 activation appears to occur via the Erk and/or p38 signaling pathways in our transformed CT-1 and SVWI38 fibroblasts and we failed to detect alterations in global phosphorylation of histone H3-Ser-10 in these cells (data not shown). These results points to a different mechanism of Tbx2 regulation by TPA in normal and transformed fibroblasts. This is not surprising considering that signal transduction pathways are frequently altered during transformation. In summary, we show that TPA activates MSK1, which is associated with phosphorylation of histone H3 in a PKC-dependent and MAPK-independent manner in normal human fibroblasts.
In the current study, phosphorylated MSK1 levels increased at all the time points of TPA treatment tested while levels of global phosphorylated histone H3 remained unchanged for the later time points. This is consistent with the literature, which suggests that while global phosphorylation of histone H3 is required for chromosome condensation and segregation during mitosis (39, 40), in stimulus-mediated gene expression only a small fraction of histone H3 is transiently phosphorylated (22, 41). Stimulus-mediated phosphorylation of histone H3 may therefore not be observed at a global level, and this may explain why we did not find an increase in global histone H3-Ser-10 phosphorylation at later time points of TPA treatment.
Although we did not detect activation of JNK by TPA in our system, it is intriguing to note that the JNK inhibitor, SP600125, resulted in an apparent repression of TBX2 expression, which was associated with a reduction of global phosphorylation of histone H3. Consistent with our finding, SP600125 has been shown to reduce global histone H3-Ser-10 phosphorylation in a JNK MAPK-independent manner in the human hepatocellular carcinoma HepG2 cells (36). Importantly, SP600125 has also been shown to be capable of inhibiting a variety of kinases (42), and our results indicated that SP600125 inhibits MSK1 activity. It is thus possible that in our system, SP600125 acts as an inhibitor of MSK1 and thus blocks phosphorylation of histone H3 leading to a reduction in TPA-stimulated TBX2 expression. This possibility is worthy of further investigation.
Although TPA was reported to lead to an increase in Sp1 levels in several systems (43, 44, 45), our data show that TPA does not affect Sp1 protein levels. However, TPA was shown to significantly enhance stability of nuclear factor binding to the Sp1 site in the TBX2 proximal promoter. Interestingly, TPA-induced phosphorylation of Sp1 has been shown to enhance Sp1 binding and transcription activity in the human HepG2 cells (46). Thus, whether Sp1 phosphorylation is also implicated in TPA-mediated transcription activation of the TBX2 gene remains to be elucidated.
Finally, we show that while TPA up-regulates c-Jun/AP1 and JunB/AP1 expression, which correlates well with up-regulation of Tbx2, TPA, and c-Jun/JunB failed to activate a reporter gene driven by a 5′-regulatory region of the TBX2 gene. These findings provide indirect support for the possibility that TPA-mediated phosphorylation of histone H3-Ser-10 plays a critical role in transcriptional activation of the TBX2 gene.
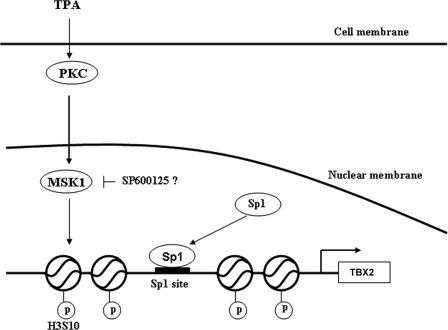
In conclusion, we show that in response to TPA, phosphorylation of histone H3 leads to the recruitment of Sp1 to a site in the proximal TBX2 gene promoter, which is required for TPA-mediated induction of TBX2 gene expression. Importantly, we show that this process involves the activation of MSK1 in a PKC-dependent manner (Fig. 6). Our findings give insight into the molecular mechanism(s) that regulate TBX2 gene expression.
FIGURE 6.
Proposed model for TBX2 regulation by TPA. In response to TPA treatment, PKC activates MSK1 which in turn phosphorylates histone H3 leading to chromatin remodeling at the TBX2 promoter. This results in the recruitment of Sp1 to the TBX2 promoter and subsequent activation of TBX2 gene expression. Indicated in the model is the possibility that the SP600125 inhibitor may block MSK1 acitivity.
Acknowledgments
We thank Professors Iqbal Parker and Colin Goding, Dr. Amaal Abrahams, and Emily Davis for helpful discussions and comments on the manuscript.
This work was supported by grants from the SA Medical Research Council, the National Research Foundation, and the University of Cape Town.
- PKC
- protein kinase C
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- JNK
- c-Jun N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- DAI
- DNA affinity immunoblotting
- siRNA
- small interfering RNA
- ChIP
- chromatin immunoprecipitation assay
- IE
- immediate-early gene
- ERK
- extracellular signal-regulated kinase
- siRNA
- small interfering RNA.
REFERENCES
- 1.Bollag R. J., Siegfried Z., Cebra-Thomas J. A., Garvey N., Davison E. M., Silver L. M. (1994) Nat. Genet. 7, 383–389 [DOI] [PubMed] [Google Scholar]
- 2.Campbell C., Goodrich K., Casey G., Beatty B. (1995) Genomics 28, 255–260 [DOI] [PubMed] [Google Scholar]
- 3.Carreira S., Dexter T. J., Yavuzer U., Easty D. J., Goding C. R. (1998) Mol. Cell. Biol. 18, 5099–5108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Law D. J., Gebuhr T., Garvey N., Agulnik S. I., Silver L. M. (1995) Mamm. Genome 6, 793–797 [DOI] [PubMed] [Google Scholar]
- 5.Habets P. E., Moorman A. F., Clout D. E., van , Roon M. A., Lingbeek M., van , Lohuizen M., Campione M., Christoffels V. M. (2002) Genes Dev. 16, 1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nissim S., Allard P., Bandyopadhyay A., Harfe B. D., Tabin C. J. (2007) Dev. Biol. 304, 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki T., Takeuchi J., Koshiba-Takeuchi K., Ogura T. (2004) Dev. Cell 6, 43–53 [DOI] [PubMed] [Google Scholar]
- 8.Mahlamäki E. H., Bärlund M., Tanner M., Gorunova L., Höglund M., Karhu R., Kallioniemi A. (2002) Genes Chromosomes Cancer 35, 353–358 [DOI] [PubMed] [Google Scholar]
- 9.Sinclair C. S., Adem C., Naderi A., Soderberg C. L., Johnson M., Wu K., Wadum L., Couch V. L., Sellers T. A., Schaid D., Slezak J., Fredericksen Z., Ingle J. N., Hartmann L., Jenkins R. B., Couch F. J. (2002) Cancer Res. 62, 3587–3591 [PubMed] [Google Scholar]
- 10.Vance K. W., Carreira S., Brosch G., Goding C. R. (2005) Cancer Res. 65, 2260–2268 [DOI] [PubMed] [Google Scholar]
- 11.Davis E., Teng H., Bilican B., Parker M. I., Liu B., Carriera S., Goding C. R., Prince S. (2008) Oncogene 27, 976–984 [DOI] [PubMed] [Google Scholar]
- 12.Jacobs J. J. L., Keblusek P., Robanus-Maandag E., Kristel P., Lingbeek M., Nederlof P. M., van Welsem T., van de Vijver M. J., Koh E. Y., Daley G. Q., van Lohuizen M. (2000) Nat. Genet. 26, 291–299 [DOI] [PubMed] [Google Scholar]
- 13.Prince S., Carreira S., Vance K. W., Abrahams A., Goding C. R. (2004) Cancer Res. 64, 1669–1674 [DOI] [PubMed] [Google Scholar]
- 14.Abrahams A., Mowla S., Parker M. I., Goding C.R., Prince S. (2008) J. Biol. Chem. 283, 2223–2230 [DOI] [PubMed] [Google Scholar]
- 15.Griner E. M., Kazanietz M. G. (2007) Nat. Rev. Cancer 7, 281–294 [DOI] [PubMed] [Google Scholar]
- 16.Newton A. C. (1997) Curr. Opin. Cell Biol. 9, 161–167 [DOI] [PubMed] [Google Scholar]
- 17.López-Bergami P., Habelhah H., Bhoumik A., Zhang W., Wang L. H., Ronai Z. (2005) Mol. Cell 19, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mauro A., Ciccarelli C., De Cesaris P. D., Scoglio A., Bouché M., Molinaro M., Aquino A., Zani B. M. (2002) J. Cell Sci. 115, 3587–3599 [DOI] [PubMed] [Google Scholar]
- 19.Berger S. L. (2002) Curr. Opin. Genet. Dev. 12, 142–148 [DOI] [PubMed] [Google Scholar]
- 20.Jenuwein T., Allis C. D. (2001) Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 21.Mahadevan L. C., Willis A. C., Barratt M. J. (1991) Cell 65, 775–783 [DOI] [PubMed] [Google Scholar]
- 22.Clayton A. L., Mahadevan L. C. (2003) FEBS Lett. 546, 51–58 [DOI] [PubMed] [Google Scholar]
- 23.Namba M., Nishitani K., Kimoto T. (1980) Gann 71, 300–307 [PubMed] [Google Scholar]
- 24.Teng H., Davis E., Abrahams A., Mowla S., Parker M. I., Prince S. (2007) J. Cell. Biochem. 102, 618–625 [DOI] [PubMed] [Google Scholar]
- 25.Lee K. A., Green M. R. (1990) Methods Enzymol. 181, 20–30 [DOI] [PubMed] [Google Scholar]
- 26.Teng H., Parker M. I., Prince S. (2008) Gene 423, 8–13 [DOI] [PubMed] [Google Scholar]
- 27.Han J. W., Ahn S. H., Kim Y. K., Bae G. U., Yoon J. W., Hong S., Lee H. Y., Lee Y. W., Lee H. W. (2001) J. Biol. Chem. 276, 42084–42090 [DOI] [PubMed] [Google Scholar]
- 28.Schuettengruber B., Simboeck E., Khier H., Seiser C. (2003) Mol. Cell. Biol. 23, 6993–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tone Y., Kojima Y., Furuuchi K., Brady M., Yashiro-Ohtani Y., Tykocinski M. L., Tone M. (2007) J. Immunol. 179, 1760–1767 [DOI] [PubMed] [Google Scholar]
- 30.Soloaga A., Thomson S., Wiggin G. R., Rampersaud N., Dyson M. H., Hazzalin C. A., Mahadevan L. C., Arthur J. S. (2003) EMBO J. 22, 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomson S., Clayton A. L., Hazzalin C. A., Rose S., Barratt M. J., Mahadevan L. C. (1999) EMBO J. 18, 4779–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sassone-Corsi P., Mizzen C. A., Cheung P., Crosio C., Monaco L., Jacquot S., Hanauer A., Allis C. D. (1999) Science 285, 886–891 [DOI] [PubMed] [Google Scholar]
- 33.Salvador L. M., Park Y., Cottom J., Maizels E. T., Jones J. C., Schillace R. V., Carr D. W., Cheung P., Allis C. D., Jameson J. L., Hunzicker-Dunn M. (2001) J. Biol. Chem. 276, 40146–40155 [DOI] [PubMed] [Google Scholar]
- 34.Huang W., Mishra V., Batra S., Dillon I., Mehta K. D. (2004) J. Lipid Res. 45, 1519–1527 [DOI] [PubMed] [Google Scholar]
- 35.Park G. Y., Wang X., Hu N., Pedchenko T. V., Blackwell T. S., Christman J. W. (2006) J. Biol. Chem. 281, 18684–18690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W., Batra S., Korrapati S., Mishra V., Mehta K. D. (2006) Mol. Cell. Biol. 26, 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samson S. L., Wong N. C. (2002) J. Mol. Endocrinol. 29, 265–279 [DOI] [PubMed] [Google Scholar]
- 38.Ge Z., Liu C., Björkholm M., Gruber A., Xu D. (2006) Mol. Cell. Biol. 26, 230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurley L. R., D'Anna J. A., Barham S. S., Deaven L. L., Tobey R. A. (1978) Eur. J. Biochem. 84, 1–15 [DOI] [PubMed] [Google Scholar]
- 40.Hendzel M. J., Wei Y., Mancini M. A., Van Hooser A., Ranalli T., Brinkley B. R., Bazett-Jones D. P., Allis C. D. (1997) Chromosoma 106, 348–360 [DOI] [PubMed] [Google Scholar]
- 41.Johansen K. M., Johansen J. (2006) Chromosome Res. 14, 393–404 [DOI] [PubMed] [Google Scholar]
- 42.Bain J., McLauchlan H., Elliott M., Cohen P. (2003) Biochem. J. 371, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D'Angelo D. D., Oliver B. G., Davis M. G., Mccluskey T. S., Dorn G. W. (1996) J. Biol. Chem. 271, 19696–19704 [DOI] [PubMed] [Google Scholar]
- 44.Noé V., Alemany C., Nicolás M., Ciudad C. J. (2001) Eur. J. Biochem. 268, 3163–3173 [DOI] [PubMed] [Google Scholar]
- 45.Porntadavity S., Xu Y., Kiningham K., Rangnekar V. M., Prachayasitikul V., St., Clair D. K. (2001) DNA Cell Biol. 20, 473–481 [DOI] [PubMed] [Google Scholar]
- 46.Zheng X. L., Matsubara S., Diao C., Hollenberg M. D., Wong N. C. W. (2000) J. Biol. Chem. 275, 31747–31754 [DOI] [PubMed] [Google Scholar]