Abstract
Neurodegenerative diseases are often defined pathologically by the presence of protein aggregates. These aggregates, including amyloid plaques in Alzheimer’s disease (AD), result from the abnormal accumulation and processing of proteins, and may ultimately lead to neuronal dysfunction and cell death. To date, conventional biochemical studies have revealed abundant core components in protein aggregates. However, rapidly improving proteomics technologies offer opportunities to revisit pathologic aggregate composition, and to identify less abundant but potentially important functional molecules that participate in neurodegeneration. The purpose of this study was to establish a proteomic strategy for the profiling of neurodegenerative disease tissues for disease-specific changes in protein abundance. Using high resolution liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) we analyzed detergent-insoluble frontal cortex samples from AD and unaffected control cases. In addition, we analyzed samples from frontotemporal lobar degeneration (FTLD) cases to identify AD-specific changes not present in other neurodegenerative diseases. We used a labeling-free quantification technique to compare the abundance of identified peptides in the samples based on extracted ion current (XIC) of their corresponding ions. Of the 512 identified proteins, quantitation demonstrated significant changes in 81 AD-specific proteins. Following additional manual filtering, 11 proteins were accepted with high confidence as increased in AD compared to control and FTLD brains, including β-amyloid, tau and apolipoprotein E, all well-established AD-linked proteins. In addition, we identified and validated the presence of serine protease 15, ankyrin B, and 14-3-3η in the detergent-insoluble fraction. Our results provide further evidence for the capacity of proteomics applications to identify conserved sets of disease-specific proteins in AD, to enhance our understanding of disease pathogenesis, and to deliver new candidates for the development of effective therapies for this, and other, devastating neurodegenerative disorders.
Keywords: Proteomics, Alzheimer’s disease, Detergent-insoluble fractions
Introduction
Alzheimer’s disease (AD) is a progressive neurological disorder characterized by impairment of memory and cognitive decline1. Accounting for more than 20 million cases worldwide, AD is both the most common neurodegenerative disorder and the most prevalent form of dementia2, 3. Multiple pathways have been implicated in AD pathogenesis, including neuronal cell death, oxidative damage, chronic inflammation, and protein aggregation4–6. Pathologically, AD is characterized by neuronal loss, gliosis, and the accumulation of two major lesions: senile plaques composed primarily of amyloid-β peptide (Aβ)7 and neurofibrillary tangles (NFTs) composed primarily of hyperphosphorylated microtubule-associated protein, tau8. Most lesion components, including Aβ and tau7–9, have been identified using brute-force biochemical or immunological approaches10. However, these methods allow only the identification of a limited number of high-abundance targets. Due to the complex pathogenesis of AD and the dynamic nature of cellular proteins, proteomics methodologies have quickly emerged as invaluable high-throughput strategies for the unbiased identification of disease-specific proteins.
Recent efforts have been directed at the analysis of protein abnormalities in the AD brain using proteomic approaches11. Large-scale proteomics analyses were first conducted using traditional two-dimensional (2D) gel separation, often in combination with mass spectrometry, identifying important proteins in key pathogenic pathways12–16. However, this approach is limited by the narrow sensitivity of 2D gel-based technique, susceptibility to human error, and bias against proteins of extreme pI, molecular weight, and hydrophobicity17. To alleviate these limitations, liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) has emerged as the preferred first-stage strategy for large-scale proteomics18. This approach allows the identification and quantification of thousands of differentially-expressed proteins directly from complex protein mixtures with superb sensitivity in the low femtomole range19. Moreover, both quantitative and descriptive LC-MS/MS strategies have been successfully used in the characterization of AD related proteins. We first combined laser capture microdissection (LCM) with LC-MS/MS to identify a total of 488 proteins that co-isolate with senile plaques20, 21. Further label-free quantitative comparison of plaque and non-plaque tissues based on extracted ion currents of identified peptides revealed at least two-fold enrichment of 26 proteins in the plaque regions. LC-MS/MS analysis of NFTs isolated by LCM quickly followed22, demonstrating an additional 72 NFT-associated proteins identified by multiple unique peptides. Finally, a descriptive profile of detergent-insoluble proteins from temporal cortex of late onset AD post-mortem samples revealed an additional 125 proteins with reduced detergent solubility23. These data highlight the diversity of cellular processes involved in aggregate formation, and emphasize the need for thorough analysis of the complex neuropathological lesions associated with AD.
Although the rapid developments in proteomic technologies offer the opportunity to revisit pathologic aggregate composition, significant optimization of experimental design strategies and data processing is necessary to reduce false positives and improve analysis depth. A variety of factors can affect protein expression, quality, and stability in the post-mortem tissues used to study neurodegenerative disorders24. For example, the interval between death and the processing of the brain (post-mortem interval, PMI) must be matched across cases to control for protein degradation that begins immediately after death25. In addition, clinical and pathophysiological heterogeneity, age, sex, and race are important sources of biological variability26, and may be controlled by implementation of a pooling strategy. Although pooling may complicate subsequent statistical comparisons, combining samples dilutes the between-subject variance27, and helps reduce false-positive rates in proteomic experiments. Additionally, detection of disease-specific proteins can be enhanced by reducing the complexity of the analytes. Despite the exquisite sensitivity of LC-MS/MS, large-scale analyses of complex samples, such as total homogenates, often result in the identification of only the most abundant cellular proteins. Thus, it may be useful to simplify the sample by focusing on the examination of the subproteomes with highest relevance to neurodegeneration.
In this study we establish a strategy for unbiased proteomic characterization of post-mortem human neurodegenerative disease tissues. To validate our approach and further identify new protein targets, we used proteomics to profile disease-specific protein changes in detergent-insoluble extracts of pooled AD frontal cortex cases. Frontal cortex is a selectively vulnerable region in AD28, and has been shown to contain abundant protein aggregates characterized by reduced solubility in multiple neurodegenerative disorders22, 23, 29, 30. Thus, we used a label-free quantitation strategy based on extracted ion currents of identified peptides to compare the detergent-insoluble subproteome with the profiles obtained from pools of both unaffected control and frontotemporal lobar degeneration (FTLD) cases. Inclusion of the FTLD cohort to filter the selection of proteins that are specific to AD rather than non-selectively altered as a result of common processes involved in neurodegeneration resulted in our identification of 11 proteins with disproportionate enrichment in AD extracts. These included several proteins already implicated in AD along with a group of proteins not previously linked to AD. Identification of these proteins validates our proteomic strategy for the examination of human neurodegenerative disease samples, and suggests that our systematic examination of insoluble brain extracts can provide important new clues for understanding the pathways underlying the formation of AD pathological lesions.
Experimental Procedures
Case Material
Post-mortem human brain tissues corresponding to diagnoses of AD, frontotemporal lobar degeneration with ubiquitinated inclusions (FTLD-U), and unaffected control were obtained from the Alzheimer’s Disease Research Center (ADRC) and Center for Neurodegenerative Disease (CND) Brain Bank at Emory University School of Medicine. Neuropathological assessment of AD was made according to the Consortium to Establish a Registry for Alzheimer’s Disease (CERAD) criteria31, as well as the National Institute on Aging (NIA)-Reagan32 criteria. The diagnosis of FTLD-U was based on consensus criteria33, 34, with all cases exhibiting small, ubiquitin-positive, tau- and α-synuclein-negative neuronal cytoplasmic inclusions and immunoreactive dystrophic neurites in the hippocampal dentate gyrus or frontotemporal cortex. Additionally, TDP-43 immunoreactivity was histochemically confirmed in all FTLD-U cases. Finally, control cases had neither a clinical history nor a neuropathologic diagnosis of neurologic disease.
For proteomic analysis, small blocks of fresh frozen frontal cortex were obtained from 10 cases each of AD, FTLD-U, and control cases. Selected cases were matched according to sex, post-mortem interval (PMI), and age at death (see Tables 1 and S1 for detailed demographic information and Table 2 for detailed neuropathologic analysis of included AD cases). Immunohistochemical and biochemical validation of identified proteins was conducted on aldehyde-fixed and frozen brain tissues from additional cases. These included 9 AD, 2 FTLD-U, and 3 control cases for which neocortex and hippocampus was examined.
Table 1.
Demographics of patient pools used for proteomic characterization1
| Diagnosis | n2 | PMI (hr)3 | Age at Death | Duration (years)4 | Sex (#F/#M)5 |
|---|---|---|---|---|---|
| AD | 10 | 11.5 | 66.1 | 9.4 | 6/4 |
| Normal Control | 10 | 8.1 | 64.6 | NA | 6/4 |
| FTLD-U | 10 | 12.1 | 65.7 | 7.1 | 6/4 |
Complete demographic data for individual cases in pool are listed in Table S1
Number of cases in pool
Average post-mortem interval in hours
Average duration of disease from diagnosis to death in years
Sex: M male F female.
Table 2.
Detailed neuropathological analysis of Alzheimer’s disease cases
| Diagnosis | Case # | Frontal Cortex Neuritic Plaques | Frontal Cortex Neurofibrillary Tangles | CERAD AD | Braak Stage | Reagan AD | Other Pathology |
|---|---|---|---|---|---|---|---|
| AD | 1 | Frequent | Frequent | Definite | 5 | High | - |
| 2 | Frequent | Frequent | Definite | 5/6 | High | - | |
| 3 | Frequent | Frequent | Definite | 5/6 | High | - | |
| 4 | Frequent | Frequent | Definite | 5 | High | - | |
| 5 | Frequent | Frequent | Definite | 6 | High | 1 LB - amygdala, 2 ECx | |
| 6 | Frequent | Frequent | Definite | 6 | High | Microinfarcts-3 Put, 4 GP | |
| 7 | Frequent | 0 | Definite | 4 | Moderate | Microinfarcts-5 FCx, 6 PCx, amygdala; Lacunes-Put | |
| 8 | Frequent | Frequent | Definite | 6 | High | LB - amygdala, Ecx, 7 TCx, 8 Icx | |
| 9 | Frequent | Frequent | Definite | 6 | High | Lacunes-Hypothalamus, striatum | |
| 10 | Frequent | Frequent | Definite | 6 | High | Microinfarcts-PCx |
Lewy Bodies
Entorhinal Cortex
Putamen
Globus Pallidus
Frontal Cortex
Parietal Cortex
Temporal Cortex
Insular Cortex
Antibodies
Primary antibodies used in these studies were Tau2 (1:5000, mouse monoclonal; Chemicon International, Temecula CA), ApoE (1:500, goat polyclonal; Calbiochem, Darmstadt, Germany), ankyrin B (1:200, mouse monoclonal; Santa Cruz Biotechnology, Santa Cruz CA), serine protease 15 (1:200, rabbit polyclonal; Atlas Antibodies, Stockholm, Sweden), YWHAH (0.5 μg/mL, rabbit polyclonal; Aviva Antibody Corporation; San Diego, CA), and ATP6V0D1 (1:200, mouse monoclonal; Novus Biologicals, Littleton CO). The antibody dilutions noted above reflect prior dilution of each antibody (1:1) with glycerol.
Sequential Biochemical Fractionation
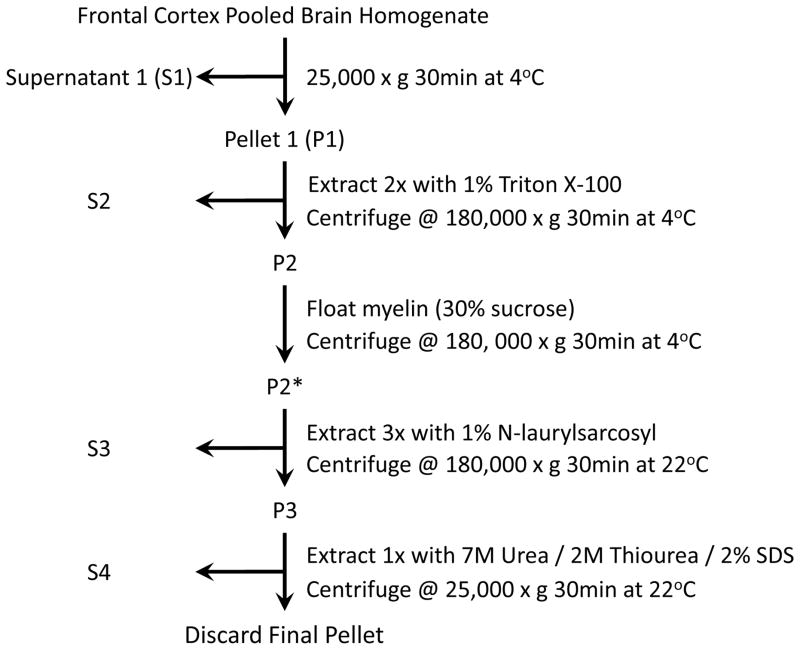
Sequential extractions were performed as described30 with slight modification (Figure 1). Briefly, post-mortem frontal cortex was extracted at 5mL/g (volume/weight) with ice-cold low salt (LS) buffer (10mM Tris, pH 7.5, 5mM EDTA, 1mM DTT, 10% sucrose, 10mM b-glycerophosphate, 10mM sodium orthovanadate, 10mM tetrasodium pyrophosphate, 50mM sodium fluoride, 1 × Roche complete protease inhibitor cocktail). Protein concentration of the resulting homogenate was determined by the bicinchoninic acid (BCA) method (Pierce, Rockford, IL) and used to equally pool ten cases each of AD, FTLD-U, and unaffected controls by diagnosis (3mg total protein per case). Notably, the FTLD-U pool and subsequent extraction was duplicated to discern experimental variance. The four pools were centrifuged at 25,000 × g for 30 minutes at 4°C and then washed with additional LS buffer. The resultant pellets were sequentially extracted with buffers of increasing stringency including Triton X-100 (TX) buffer (LS buffer + 1% Triton X-100, 0.5M NaCl), myelin floatation (MF) buffer (LS buffer with 30% sucrose + 1 % Triton X-100, 0.5M NaCl), and sarkosyl (SK) buffer (LS buffer + 1% N-Lauroyl-sarcosine, 0.5M NaCl). Specifically, each pooled sample was extracted with TX buffer and spun at 180,000 × g for 30 minutes at 4°C to generate a Triton-insoluble fraction. Following two washes with TX buffer, this fraction was incubated with MF buffer and centrifuged at 180,000 × g as noted above. Pellets were then sonicated briefly, incubated in SK buffer for 30 minutes at room temperature, and centrifuged at 180,000 × g for 30 minutes at 22°C. Two additional SK buffer washes were followed by extraction of the sarkosyl-insoluble fraction with urea buffer (30mM Tris, pH 8.5, 7M urea, 2M thiourea, 1% SDS), a brief sonication, and a final 30 minute centrifugation at 25,000 × g at 22°C. Since the BCA protein assay is incompatible with thiol reagents, final protein concentrations of urea-soluble fractions were obtained by estimating Coomassie Blue G-250 staining intensity of extracts (~2%) following electrophoresis in polyacrylamide gels using titrated BSA as a standard (10-fold range). The gel was run ~5mm in order to increase the accuracy of quantification by concentrating the proteins in a narrow region. Signal intensity of proteins in stained gel was quantified by densitometry with an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). Generally, the protein yield in urea samples during sequential extraction was approximately 2% of the starting material (see Table S2 for detailed sequential detergent extraction insoluble protein yields).
Fig. 1.
Diagram of sequential extraction protocol used to generate detergent-insoluble fractions for proteomic analysis.
Proteomic Analysis
Comprehensive shotgun sequencing and label-free quantification of reduced, alkylated, and trypsin-digested proteins from urea fractions was performed. Urea fraction proteins (~50μg per sample) corresponding to pooled AD, control, or FTLD-U (replicates) were reduced with 10mM DTT and alkylated in the dark with 50mM iodoacetamide for 30 minutes. The samples were then separated on a 10% SDS gel (0.75 mm thickness) and stained with Coomassie Blue G-250 to both confirm equal sample loading and visualize proteins for subsequent processing. Each sample lane was then cut into five gel bands and subjected to in-gel digestion (12.5 μg/mL trypsin)35. The resulting peptides from each gel piece were dissolved in buffer A (0.4% acetic acid, 0.005% heptafluorobutyric acid, 5% AcN), loaded onto a C18 column (75 μm i.d., 10 cm long, ~300 nl/min flow rate) as described36, and eluted over 2 hours during a 10–30% gradient of buffer B (0.4% acetic acid, 0.005% heptafluorobutyric acid, 95% AcN). The eluted peptides were detected by Orbitrap (350–1600 m/z, 1,000,000 AGC target, 1,000 ms maximum ion time, resolution 30,000 fwhm) followed by ten data-dependent MS/MS scans in LTQ (3 m/z isolation width, 35% collision energy, 5,000 AGC target, 200 ms maximum ion time) on a hybrid mass spectrometer (Thermo Finnigan, San Jose, CA). Peaklists were generated by Xcalibur 2.0 SR2 software (Thermo Finnigan, San Jose, CA).
The acquired MS/MS spectra were searched against the human reference database (29,575 proteins) of the National Center for Biotechnology Information (January 2007) using the SEQUEST-Sorcerer algorithm version 3.11 r11 (Sage-N-Research, San Jose CA)37. Searching parameters included parent ion mass tolerance (50 ppm), partially tryptic restriction, and mass shifts for modification of carboxyamidomethylated Cys (+57.0215 Da), and for oxidized Met (+15.9949 Da). Only b and y series ions were considered. A target-decoy composite database was used to evaluate the level of false positive matches38. The peptide matches were grouped by charge state (+1, +2, and +3) and trypticity (fully and partially tryptic), and then stringently filtered by (i) mass accuracy of 15 ppm, (ii) minimal peptide length of 7 amino acids, (iii) three maximal modification sites, (iv) two maximal miscleavages, and (v) matching scores (XCorr and ΔCn). In each group, the matching scores were dynamically increased until all decoy matches were discarded, suggesting that estimated false discovery rate was close to zero. To remove redundancy during the assignment of identified peptides to proteins, all accepted proteins sharing peptides were grouped together and represented by the protein with the highest spectral count. Generally, following manual validation of the spectra38, we accepted proteins identified by at least one unique peptide.
Quantification of proteins was based on the comparison of paired peptides from the AD and control/FTLD-U samples. Ion current intensities for identified peptides were extracted in MS survey scans of high-resolution, and a ratio of the peak intensities for the peptide precursor ion was calculated39, 40. For peptides identified in only one sample, corresponding non-sequenced ion peaks were identified for quantification in MS survey scans using the predicted m/z, and adjusted retention time. The resultant ratio is a measure of the relative abundance of the peptide in the two separate samples41. Statistical analysis to evaluate the significance of the protein changes and to correct for technical errors was performed as previously described with modifications42, 43. Briefly, the peptide abundance ratios were logarithmically transformed (log2). The mean and associated variance of the transformed ratios was calculated for peptides quantified multiple times. Abundance ratios for all peptides of a particular protein were then averaged to determine the protein abundance ratio. A histogram of all protein abundance ratios was fitted with a normal distribution on the basis of the central limit theorem (bins of 0.3 = log2(ratio)). Because the majority of proteins likely display similar abundance in disease and control tissues, the abundance ratio for each protein was normalized by subtracting the fitted mean, as the mean should have been zero if sample preparation was ideal. Although most proteins in the data set correlate well with the fitted normal distribution, there are a few proteins demonstrating larger changes that distort the fit of the experimental distribution. These perturbations from the curve of the normal distribution represent proteins that are associated with disease, and presumably deviate from the normal population of unaltered proteins. To be considered significantly altered in AD, we established that a protein must have a consistently elevated log2(ratio) ≥ ±1.0 (i.e. 2-fold change) in both AD/Control and AD/FTLDavg distributions.
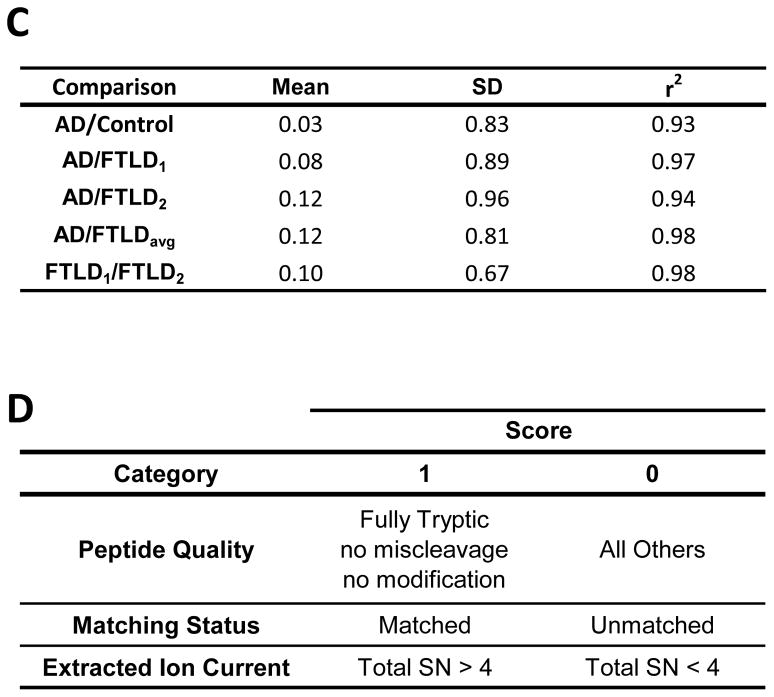
The quantified proteins were further manually examined to verify MS/MS assignment, ion peak matching, and ion intensity (signal-to-noise ratio ≥ 4). Proteins were scored in three categories, with 1 point awarded in each category: peptide quality, matching status, and extracted ion current. Proteins were scored for peptide quality if their quantitation was based only on fully tryptic peptides with no missed trypsin cleavage sites and no modifications. Only proteins whose quantitation was based on correctly matched peaks in each sample were scored for matching status. Finally, a score was awarded if the total extracted ion current for each peptide exceeded 4 times the moving average for noise level (signal to noise ratio ≥ 4).
Immunohistochemistry
50μm-thick brain sections were prepared with a freezing microtome (Microm, Heidelberg, Germany) with AD, FTLD-U, and control brain blocks from frontal cortex, cingulated cortex, and hippocampus. Sections were incubated in 3% hydrogen peroxide (H2O2) to quench endogenous peroxidase activity. Sections were subsequently incubated with normal serum followed by primary antibody overnight at 4°C. After extensive washes with TBS, sections were incubated with biotinylated secondary antibody for 1 hour at 4°C and an additional hour with avidin-biotin-peroxidase complex (Vector Elite ABC Kit, Vector Laboratories, Burlingame CA). Staining was visualized by light microscopy using 3,3′-diaminobenzidine (DAB) as a chromogen.
Western Blotting
Detergent-insoluble urea fractions from pooled homogenates or individual cases were separated by SDS-PAGE and transferred overnight to PVDF Immobilon-P membranes (Millipore, Billercia MA). To ensure both equal loading and complete transfer of proteins from the gel, membranes were reversibly stained with Ponceau S (Diasys Europe Ltd, Workingham, England). Blots were subsequently blocked for 1 hour at room temperature, probed with primary antibody overnight at 4°C, and incubated for 1 hour at room temperature with secondary antibodies (1:20,000) conjugated to fluorophores (Molecular Probes, Eugene OR; Rockland, Gilbertsville PA). Blots were dried, scanned, and quantified with an Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). Statistical analysis was performed using Student’s t-test for independent samples.
Results
Protein identification in urea samples by LC-MS/MS
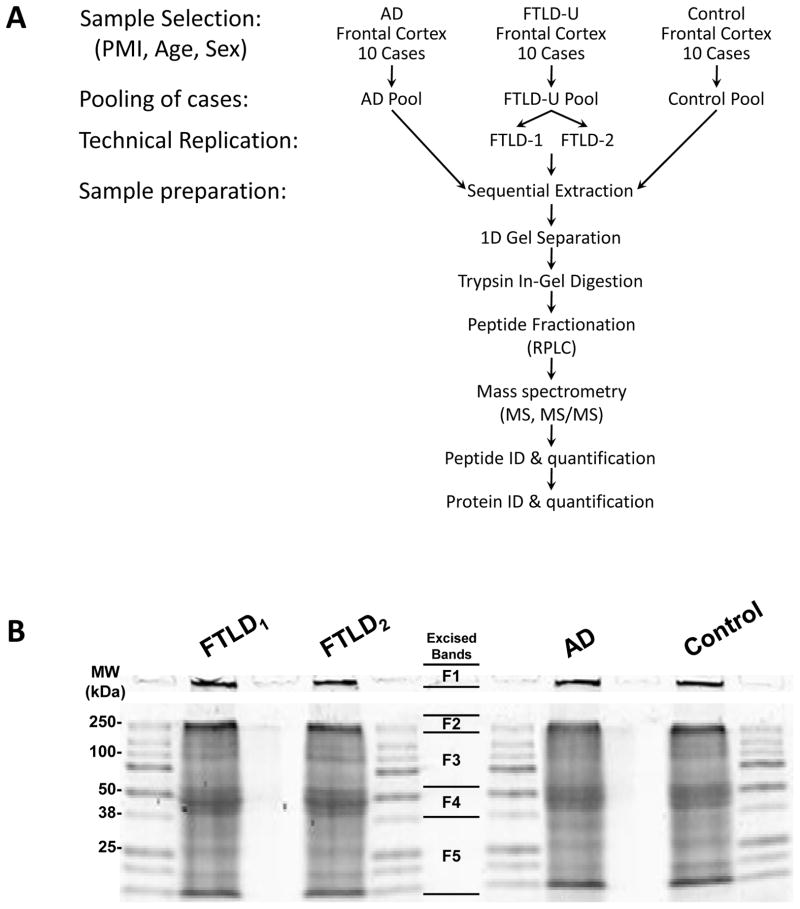
The proteomic profiling of AD detergent-insoluble aggregated protein fractions was initialized with sample optimization and preparation as illustrated in Figure 2A. In our strategy, frontal cortical samples from postmortem AD, FTLD-U and control cases were individually homogenized and examined prior to pooling by SDS-PAGE followed by silver staining (data not shown). Although there were some differences in band intensities, there was little degradation of proteins such that the pattern of major bands was similar between cases. To minimize inter-case variation, homogenates from 10 cases of each diagnosis (supplemental Table S1) were then combined into a single pool with each case contributing 3 mg total protein. The FTLD-U pooled samples were divided into two identical samples that were processed in parallel as “technical replicates” to discern experimental variances and improve capacity to accurately identify disease-specific proteins44, 45. AD, control, and both FTLD-U pools were serially extracted with triton X-100, sarkosyl, and urea buffers. Urea-soluble fractions were then separated by mass via SDS-PAGE (Figure 2B), excised in 5 bands from the gel, and trypsin-digested. To minimize variation associated with HPLC column performance and ionization efficiency, the tryptic peptide mixtures for each sample were analyzed sequentially under identical LC-MS/MS conditions on a high-resolution mass spectrometer. These spectra were searched against a human protein database, and further stringently filtered by mass accuracy and matching scores. A total of 1,045 proteins (3,216 peptides) were identified in all four samples in all gel bands and these were clustered into 512 protein groups based on shared peptides (Supplemental Table S3 and Table S4).
Fig. 2. Sample preparation for proteomic analysis.
(A) Diagram of strategy for sample preparation and “bottom-up” proteomics analysis in which proteins are digested into peptides for improved separation and ionization. (B) SDS-PAGE gel of the isolated urea fractions stained with Coomassie Blue G-250. The gel lanes were excised in 5 pieces as indicated.
Relative quantification of proteins in urea samples by the label-free strategy
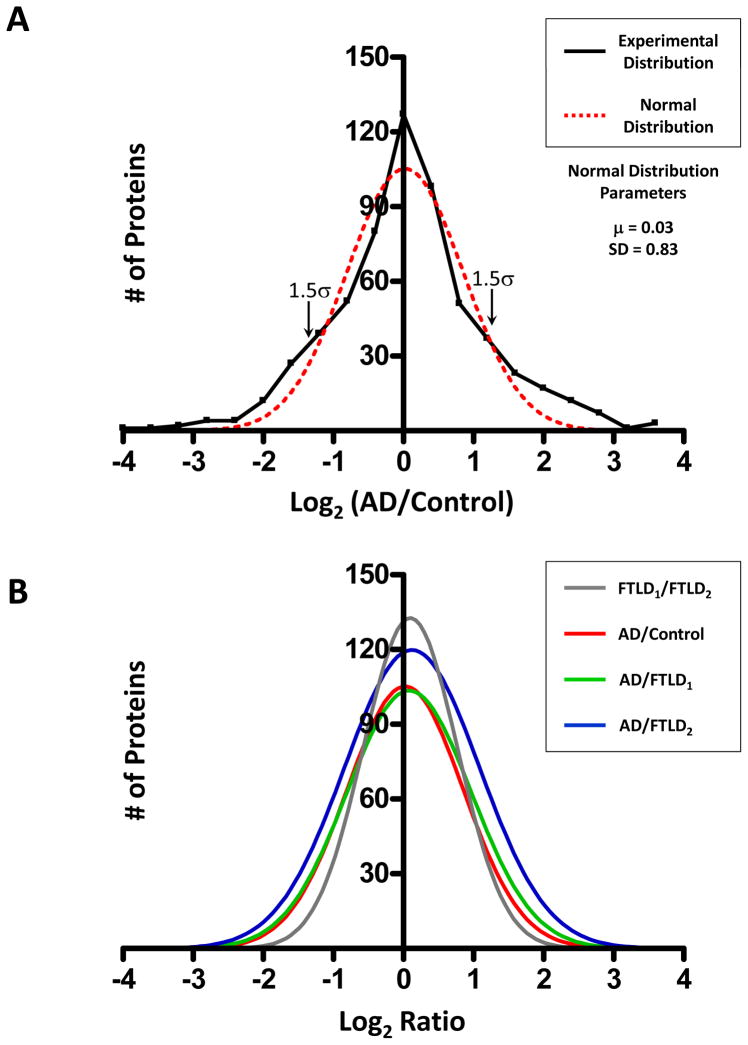
The comprehensive profiling of protein expression in AD detergent-insoluble preparation requires both the identification and the quantitation of proteins. To determine which of the identified proteins were preferentially enriched in AD, a quantitative protein comparison based on the extracted ion current (XIC) of identified peptides was performed. Abundance ratios corresponding to the relative protein abundance between AD and FTLD-U or control samples were calculated for all 512 proteins identified using peptide data from all gel bands. Abundance ratios for each possible comparison (e.g. AD/Control) were converted logarithmically and plotted as a histogram (Figure 3A). Theoretically, only the abundance of disease-related proteins should be altered in each comparison, and each Gaussian distribution fits the data very well (range of r2 = 0.93 to 0.98), confirming the assumption that the majority of the proteins display similar abundance in disease and control urea extracts. Additionally, the fitted normal distribution was used to approximate the common variance of the whole data set, a feature that was used to establish significance thresholds for protein changes42. The means and standard deviations (SD) for all relevant case comparisons are shown in Figure 3C. As an initial filter for significantly changed proteins in AD, we required at least a 2-fold change in protein abundance for both AD/Control and AD/FTLDavg comparisons. Although for a single comparison the probability associated with this 2-fold threshold is approximately 20% according to the average of the SD values (0.82, Figure 3C), when two comparisons were simultaneously considered, the probability of false positives falls to ~4% (20% × 20%). According to the null hypothesis, this probability is equivalent to 21 false positives in a list of 512 proteins. However, using the 2-fold cutoff in our data, 81 proteins were accepted in the filtering, suggesting ~60 (equivalent to 81-21) proteins could theoretically be altered specifically in the AD sample. To remove the false positives in the 81 remaining proteins, we manually examined these proteins and filtered according to additional criteria (Figure 3D, see Experimental Procedures), resulting in a final list of 11 AD-specific proteins (Table 3). Although this stringent manual processing increased our confidence in the quantitative measures, our criteria favor specificity over sensitivity in populating a list of less than the predicted 60 proteins.
Fig. 3. Statistical evaluation and filtering of proteomics data.
(A) Abundance ratios for AD/Control comparison for all proteins across all gel bands were transformed (logarithmic base 2) and plotted with each point corresponding to the number of proteins in 0.3 unit windows (black line). A Gaussian curve was subsequently fitted to the data (red line) and used to determine significance levels for protein change. (B) Fitted normal distributions for all possible case comparisons with (C) statistical means, standard deviations, and regression coefficients. (D) Additional filtering criteria for removal of false positives were based on quality assessment of protein quantification by manual scoring of corresponding peptides. A top scoring protein would receive 1 point in each of the above categories for a total of 3 points. SN, Signal to Noise Ratio.
Table 3.
Proteins specifically altered in AD urea fractions compared with control and FTLD-U
| Protein | GeneBank™ Accession Number | Log2 Ratio AD/Control | mean Log2 Ratio AD/FTLD | Quantified Peptides |
|---|---|---|---|---|
| microtubule-associated protein tau | NP_058518.1 | 5.2 | 5.5 | 16 |
| serum amyloid P component precursor | NP_001630.1 | 4.0 | 3.3 | 2 |
| amyloid beta A4 protein precursor | NP_958817.1 | 3.5 | 3.6 | 5 |
| complement component 4B | NP_001002029.1 | 3.4 | 3.1 | 5 |
| serine protease 15 | NP_004784.2 | 2.3 | 1.2 | 6 |
| apolipoprotein E | NP_000032.1 | 2.1 | 2.5 | 5 |
| 14-3-3, eta polypeptide | NP_003396.1 | 1.9 | 1.4 | 3 |
| 14-3-3, zeta polypeptide | NP_663723.1 | 1.7 | 1.6 | 3 |
| ankyrin B | NP_066187.2 | 1.3 | 1.4 | 1 |
| dynamin 1 | NP_004399.2 | 1.3 | 1.2 | 5 |
| aquaporin 1 | NP_000376.1 | −2.0 | −2.0 | 2 |
The difference between the quantitative information obtained using the XIC method and the qualitative information obtained during peptide identification is further emphasized by the observation that, of the 11 AD-specific proteins in our final list, only the inflammatory complement C4 was identified in the AD sample alone. Since identification of a peptide relies on its fragmentation to produce a unique MS/MS spectrum, complex protein samples can easily saturate the sampling rates of the mass spectrometer46. Therefore, due to inherent limitations in the duty cycle of the instrument, only a fraction of ionized peptides can be selected for sequencing. Hence, many nonsequenced ion peaks remain in the MS survey scans, an issue known as “undersampling” in shotgun LC-MS/MS analyses. Even a comparison of the technical replicates, FTLD1 and FTLD2, which are identical samples, reveals only a 66% overlap in identified proteins. Certainly, those remaining proteins not identified in both FTLD1 and FTLD2 do not represent true differences between these replicates, but instead reflect the capacity for the instrument to be overwhelmed by particularly complex samples. In contrast, quantification using the XIC method is derived from the MS survey scan and is, thus, not entirely dependent on the acquisition of MS/MS scans. Once a peptide has been detected in one of the samples, its corresponding non-sequenced ion peak in all of the other samples can be localized for quantitation using the predicted high resolution m/z value and adjusted retention time21. Using this strategy, we quantified peptides for all 35 proteins detected in the AD sample alone, and only complement C4 showed significant quantitative change.
Validation of selected AD-specific, detergent-insoluble proteins
To independently verify the accuracy of the LC-MS/MS generated data, we analyzed several of the proteins identified as altered in AD by immunoblotting and immunohistochemical methods. Among these, we selected several proteins that have been previously implicated in AD or have been associated with known pathogenic pathways. Notably, microtubule-associated protein tau and apolipoprotein E (ApoE) were among the most altered proteins, demonstrating enrichment in AD tissues of greater than 30-fold and 5-fold respectively by quantitative LC-MS/MS. Quantitative immunoblot analysis demonstrated significant enrichment of both tau and ApoE in urea fractions from pooled AD postmortem frontal cortex samples (Figure 4A & 4B). As insoluble proteins typically aggregate focally in disease pathology, immunohistochemistry provides a complementary method to visualize the distribution of altered proteins and their relationship to neurodegenerative changes. AD frontal cortex showed extensive neuropathology with striking enrichment of immunoreactivity for ApoE (Figure 5A & 5B) and tau (data not shown) as has been well-established previously8, 47, 48. Although apolipoprotein E was primarily localized to amyloid plaques, there was also a striking association in multiple cases with neurofibrillary tangles in frontal cortex.
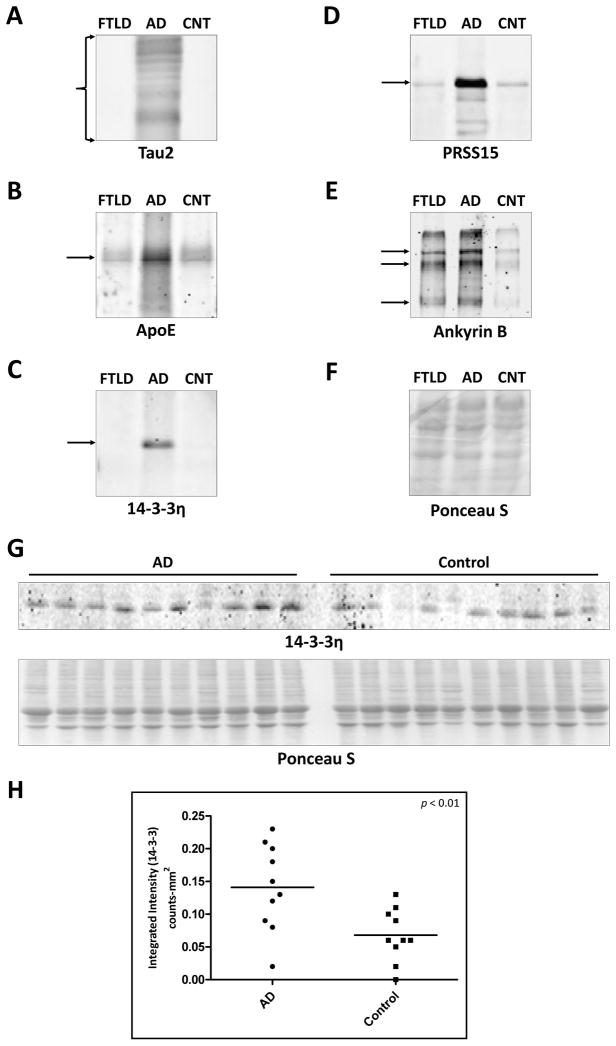
Fig. 4. Confirmation of proteomic candidates by immunoblot analysis.
Frontal cortex from FTLD-U, AD, and unaffected control cases were pooled by diagnosis and sequentially extracted with buffers containing triton X-100, sarkosyl, and urea. Sarkosyl insoluble (urea) samples were immunoblotted with antibodies to (A) Tau2 which recognizes both non-phosphorylated and abnormal phosphorylated tau (region shown ~30kD–60kD) (B) apolipoprotein E (36kD denoted by arrow) (C) 14-3-3η (28kD denoted by arrow) (D) Serine protease 15 (106kD denoted by arrow) and (E) Ankyrin B (multiple isoforms denoted by arrows at 205kD, 163kD, and 100kD) (F) Representative Ponceau S reversible membrane stain was performed to ensure both equal loading and complete transfer of proteins from the gel. (G) Unpooled AD (n = 10) and control (n = 10) cases were individually detergent-extracted and immunoblotted with an antibody to 14-3-3η. The corresponding Ponceau S reversible membrane stain is shown. (H) Quantification of immunoreactive 14-3-3η bands revealed a 2.1-fold enrichment of this protein in AD urea extract (p < 0.01).
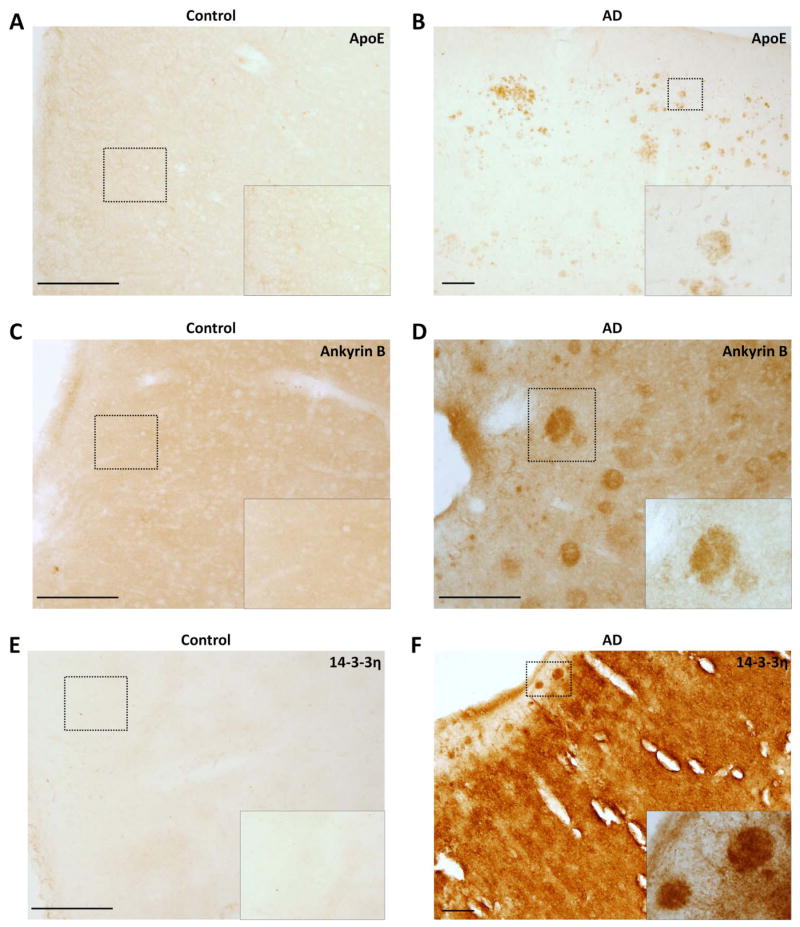
Fig. 5. Confirmation of proteomic candidates by immunohistochemistry in frontal cortex floating sections (50μm).
(A) Apolipoprotein E staining in control tissue (200μm) and (B) AD (100μm). (B) Ankyrin B staining in control tissue (200μm) and (B) AD (200μm). (C) 14-3-3η staining in control tissue (200μm) and (B) AD (100μm). (Scale Bar in μm)
Having validated well-known AD-linked proteins by proteomics and immunological methods, we used similar methods to independently validate and characterize several novel proteins identified as increased in AD from our LC-MS/MS-based approach. Compared with control and FTLD-U cases, pooled urea samples from AD frontal cortex demonstrated up-regulation of ankyrin B, serine protease 15 (PRSS15), and 14-3-3η (Figure 4C–F) in Western blot analysis. Moreover, quantification of immunoblot band intensities in frontal cortex urea fractions from individual, non-pooled AD and control cases used in the proteomics analysis (n = 10 for both AD and control) showed a significant 2.1-fold enrichment (p = 0.008) of 14-3-3η in AD samples (Figure 4G–H). Immunohistochemical examination of both ankyrin B and 14-3-3η revealed thread-like structures with striking localization in plaque regions, confirming the enrichment suggested by the proteomic findings (Figure 5). In addition to being a plaque component, 14-3-3η also appeared to be significantly enriched in the surrounding neuropil in AD tissues. These broad changes in 14-3-3η expression may suggest a more general role in AD pathogenesis. Despite extensive overexpression in AD urea fractions, serine protease 15 (PRSS15) was not associated with classical AD pathology, and its localization remained unaltered in both hippocampus and frontal cortex (data not shown). Finally, although we attempted to validate changes in expression of dynamin 1 and aquaporin 1, the lack of specific immunoreactivity of the purchased commercially available reagents precluded confirmation of the proteomic results in these studies.
Discussion
Protein accumulation and aggregation in the brain is one of the pathologic hallmarks of neurodegenerative diseases. Interestingly, an important characteristic of these aggregates is their spontaneous conversion from functional soluble proteins to pathologic, detergent-insoluble fibrillar forms identified in diseased tissues49–52. The altered insoluble structure of aggregated proteins can result in both functional deficits and, more importantly, in a toxic gain of function associated with the aggregate itself or its transient intermediates53, 54. Thorough characterization of neurodegenerative disease aggregates and the aggregation process is, therefore, essential for our understanding of the molecular pathways underlying disease pathogenesis. Here, we describe an unbiased proteomics strategy for the large-scale identification of novel disease-specific proteins that may serve as biomarkers and targets for therapeutic intervention. Our study revealed a total of 512 proteins present in pathologic detergent-insoluble fractions prepared from AD frontal cortex. Using strict filtering criteria to remove proteins that were either unchanged in disease extracts compared to control or were non-specifically altered in both FTLD-U and AD, we identified 11 proteins with high specificity for AD. These included established proteins that are classically associated with AD pathologic lesions, as well as several novel associated proteins encompassing a broad range of cellular activities. We subsequently verified five targets by immunoblotting and immunohistochemistry, highlighting the power of our proteomics approach for identifying new proteins as well as a more complete understanding of the complement of proteomic changes that are involved in neurodegeneration.
The widespread transition of proteins to detergent insolubility and the complexity of aggregate composition are well-established in neurodegeneration10, 20, 22, 23, 30. As is reflected in our data, development of LC-MS/MS based proteomics has allowed for examination of complex disease samples with increasing sensitivity. Our data provide evidence for reduced detergent solubility in greater than 500 proteins in AD. Yet, when quantitatively compared with control and FTLD-U samples, only 11 proteins show significant changes that are specific to AD. Many of the unchanged proteins may be contaminants acquired during the extraction procedure or proteins that are altered generally in either neurodegeneration or during normal aging. Nevertheless, in the context of AD, the unchanged proteins contribute to the false-positive rate and suggest that, due to improved sensitivity, the identification of proteins without quantitation is insufficient for establishing a specific association with disease.
Despite improved sensitivity of LC-MS/MS analyses, limitations of “undersampling” in LC-MS/MS restrict the number of peptides that can be identified in a specific sample. This technical limitation can be assessed by the inclusion of technical replicates, or identical samples that are processed in parallel. Our comparison of the replicates FTLD1 and FTLD2 revealed that only two-thirds of the proteins were identified in the identical samples. Thus, since during a shotgun proteomics experiment, only a fraction of possible peptides from a complex mixture can be selected and sequenced by MS/MS36, the failure to identify a peptide in a complex sample in shotgun mode does not imply the absence of that peptide in the mixture. By using the XIC method for quantitation, we quantified many peptides selected for MS/MS analysis in only one of the samples by matching their corresponding non-sequenced ion peak in all other samples. Our data, therefore, demonstrate the importance of the application of a quantitative strategy in the profiling of complex neurodegenerative disease samples.
In addition to the variability inherent in the process of mass spectrometry, identification of important disease-specific changes is complicated by the variation resulting from the use of patient samples in neuroproteomics. Theoretically, by pooling equal volumes of different patient samples with a well-characterized common pathologic phenotype, we reduce the potential for aberrations in a single sample to sway the mean55. This reduction in inter-sample, or biological, variation can improve the capacity to identify the most significant and consistent changes between diseases. The potential loss of statistical power is thereby compensated by the increased confidence in the measurement of each protein concentration. However, as is typical with analysis of human tissues, each sample comparison in this study was associated with a relatively large standard deviation, including the comparison of technical replicates (FTLD1/FTLD2). Since FTLD1 and FTLD2 are identical pools separated prior to sequential extraction, the large variations in this comparison may be primarily attributed to the technical error during sample preparation, SDS gel electrophoresis, in-gel digestion and the following LC-MS/MS runs. This implies that the larger standard deviations measured in the other comparisons (e.g. AD/Control) are only partially affected by true biological differences. Accordingly, our proteomic strategy could be modified to include multiple pools for each diagnosis to allow an assessment of this biological variability.
Analysis of the detergent-insoluble proteome revealed robust differences in 11 proteins with high specificity to AD. Among these, we corroborate the extensive enrichment of key AD-associated proteins, including the microtubule-associated protein, tau, Aβ, apolipoprotein E56, serum amyloid P57, and complement component 458. The known association of these proteins with the pathologic features of AD, and their disproportionate enrichment in our AD detergent-insoluble extracts, substantiate the specificity of our proteomic strategy and validate the quality of our urea extract preparations. Interestingly, our strict filtering criteria removed multiple proteins known to colocalize with AD pathologic lesions, including apolipoprotein D59, αB-crystallin60, and cathepsin-D61. While it is probable that in the effort to reduce the false discovery rate we also eliminated certain molecules with relevance to AD pathogenesis, it is possible that some of these molecules play a more general role in neurodegeneration, and were removed by the inclusion of FTLD-U as a disease control. It is also possible that some of these molecules may be associated with AD pathology without a corresponding increase in abundance in insoluble fractions prepared from frontal cortex. Alternatively, these molecules may non-specifically associate with AD lesions by immunohistochemistry, and may have limited involvement in the pathogenesis of the disease23. Their exclusion, coupled with the presence of proteins such as tau and Aβ among the candidates, may help validate the specificity of the remaining targets.
In addition to well-established disease proteins, we demonstrated and immunologically validated the enrichment of several novel proteins in AD detergent-insoluble fractions. Although their specific association with AD pathogenesis is not well characterized, these proteins, including ankyrin B and PRSS15, play important roles in functional pathways of particular relevance to AD pathophysiology. For example, the disruption of cytoskeletal integrity is a fundamental feature of AD and can impact microtubule stability, alter essential axonal or synaptic signaling, and influence vesicular trafficking and biogenesis62. Our recently published phosphoproteomic analysis revealed that, in addition to tau, there are other abundant phosphorylated proteins the AD brain63. One of these proteins, ankyrin B, is a cytoskeletal component that plays a important role in synaptogenesis, stabilization of the synapse, and organization of the plasma membrane64. These actions depend on the ability of ankyrin B to establish specialized protein complexes by coordinating the interactions of another skeletal protein, spectrin, with various integral membrane proteins65. In the current study, ankyrin B was preferentially enriched in AD detergent-insoluble preparations, and was localized in plaque regions by immunohistochemistry. It is possible that the aggregation of ankyrin B results in a loss of function phenotype with pervasive implications for AD pathogenesis. The absence of ankyrin-binding activity, as is seen in ankyrin B-knockout mice64, can trigger a loss of interneuron synapses that may contribute to cognitive decline in AD.
Another protein with altered abundance in AD urea fractions was PRSS15, an ATP-dependent mitochondrial matrix protein that is responsible for the degradation of oxidized proteins66, 67. The involvement of mitochondrial dysfunction and oxidative stress in neurodegeneration is well-established68. In AD, multiple studies link oxidative stress and the modulation of both Aβ levels and hyperphosphorylation of tau69. Additionally, under conditions of cellular stress, many proteins become highly susceptible to oxidative modification and must be selectively removed by proteolytic digestion67, 70. Although PRSS15 is normally present in relatively low abundance66, in our study its expression was significantly upregulated in the pathologic insoluble preparation. Despite this upregulation, PRSS15 demonstrated no association with characteristic AD neuropathology by immunohistochemistry. We speculate that, during AD, the capacity for PRSS15 to remove damaged proteins is overwhelmed, as evidenced by the accumulation of aconitase71, a mitochondrial matrix protein that is preferentially degraded by PRSS1567. The subsequent accrual and aggregation of damaged or misfolded proteins result in the exacerbation of mitochondrial dysfunction, the increased production of reactive oxygen species (ROS), and the triggering of programmed cell death.
In summary, this study illustrates the potential of proteomic applications to identify sets of disease-specific proteins linked to individual neurodegenerative diseases, as well as conserved sets of proteins involved in processes common to many neurodegenerative diseases. We were able to use a labeling-free strategy to assign quantitative information to thousands of peptides identified directly from clinical samples. As a result, we generated a highly specific profile of altered AD proteins. This profile included both well-characterized components of AD pathology and novel potential therapeutic targets with roles in various important functional pathways. Since pathologic protein aggregation is ubiquitous to all neurodegenerative diseases, the approach detailed herein may be uniformly applied to generate disease-specific profiles that may be customized to improve diagnosis or to shed light on the pathogenesis of these complex disorders.
Supplementary Material
Acknowledgments
We thank Dr. Howard Rees for technical assistance and Dr. Nicholas Seyfried for valuable comments regarding the manuscript. This work was supported by the National Institutes of Health through the Emory Alzheimer’s Disease Center grant (P50AG025688), the Emory Neuroscience NINDS Core Facilities (P30NS055077), and NIH training grant (F30NS057902 to Y.G.).
Footnotes
Supporting Information Available
Supplementary tables S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Blennow K, de Leon MJ, Zetterberg H. Alzheimer’s disease. Lancet. 2006;368(9533):387–403. doi: 10.1016/S0140-6736(06)69113-7. [DOI] [PubMed] [Google Scholar]
- 2.Bergamaschini L, Canziani S, Bottasso B, Cugno M, Braidotti P, Agostoni A. Alzheimer’s beta-amyloid peptides can activate the early components of complement classical pathway in a C1q-independent manner. Clin Exp Immunol. 1999;115(3):526–33. doi: 10.1046/j.1365-2249.1999.00835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goedert M, Spillantini MG. A century of Alzheimer’s disease. Science. 2006;314(5800):777–81. doi: 10.1126/science.1132814. [DOI] [PubMed] [Google Scholar]
- 4.Nunomura A, Perry G, Aliev G, Hirai K, Takeda A, Balraj EK, Jones PK, Ghanbari H, Wataya T, Shimohama S, Chiba S, Atwood CS, Petersen RB, Smith MA. Oxidative damage is the earliest event in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60(8):759–67. doi: 10.1093/jnen/60.8.759. [DOI] [PubMed] [Google Scholar]
- 5.Tuppo EE, Arias HR. The role of inflammation in Alzheimer’s disease. Int J Biochem Cell Biol. 2005;37(2):289–305. doi: 10.1016/j.biocel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10 (Suppl):S10–7. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 7.Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120(3):885–90. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 8.Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science. 1991;251(4994):675–8. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 9.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82(12):4245–9. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atwood CS, Martins RN, Smith MA, Perry G. Senile plaque composition and posttranslational modification of amyloid-beta peptide and associated proteins. Peptides. 2002;23(7):1343–50. doi: 10.1016/s0196-9781(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 11.Butterfield DA, Castegna A. Proteomics for the identification of specifically oxidized proteins in brain: technology and application to the study of neurodegenerative disorders. Amino Acids. 2003;25(3–4):419–25. doi: 10.1007/s00726-003-0027-7. [DOI] [PubMed] [Google Scholar]
- 12.Tsuji T, Shimohama S. Analysis of the proteomic profiling of brain tissue in Alzheimer’s disease. Dis Markers. 2001;17(4):247–57. doi: 10.1155/2001/386284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattila KM, Frey H. Alzheimer brain proteins investigated by two-dimensional gel electrophoresis with immobilized pH gradients in the first dimension. Electrophoresis. 1994;15(5):721–5. doi: 10.1002/elps.1150150199. [DOI] [PubMed] [Google Scholar]
- 14.Tsuji T, Shimohama S, Kamiya S, Sazuka T, Ohara O. Analysis of brain proteins in Alzheimer’s disease using high-resolution two-dimensional gel electrophoresis. J Neurol Sci. 1999;166(2):100–6. doi: 10.1016/s0022-510x(99)00120-3. [DOI] [PubMed] [Google Scholar]
- 15.Tsuji T, Shiozaki A, Kohno R, Yoshizato K, Shimohama S. Proteomic profiling and neurodegeneration in Alzheimer’s disease. Neurochem Res. 2002;27(10):1245–53. doi: 10.1023/a:1020941929414. [DOI] [PubMed] [Google Scholar]
- 16.Cottrell BA, Galvan V, Banwait S, Gorostiza O, Lombardo CR, Williams T, Schilling B, Peel A, Gibson B, Koo EH, Link CD, Bredesen DE. A pilot proteomic study of amyloid precursor interactors in Alzheimer’s disease. Ann Neurol. 2005;58(2):277–89. doi: 10.1002/ana.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palzkill T. Proteomics. Kluwer Academic Publishers; Boston: 2002. p. 127. [Google Scholar]
- 18.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 19.McCormack AL, Schieltz DM, Goode B, Yang S, Barnes G, Drubin D, Yates JR., 3rd Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal Chem. 1997;69(4):767–76. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 20.Liao L, Cheng D, Wang J, Duong DM, Losik TG, Gearing M, Rees HD, Lah JJ, Levey AI, Peng J. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279(35):37061–8. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- 21.Gozal YM, Cheng D, Duong DM, Lah JJ, Levey AI, Peng J. Merger of laser capture microdissection and mass spectrometry: a window into the amyloid plaque proteome. Methods Enzymol. 2006;412:77–93. doi: 10.1016/S0076-6879(06)12006-6. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Woltjer RL, Cimino PJ, Pan C, Montine KS, Zhang J, Montine TJ. Proteomic analysis of neurofibrillary tangles in Alzheimer disease identifies GAPDH as a detergent-insoluble paired helical filament tau binding protein. Faseb J. 2005;19(7):869–71. doi: 10.1096/fj.04-3210fje. [DOI] [PubMed] [Google Scholar]
- 23.Woltjer RL, Cimino PJ, Boutte AM, Schantz AM, Montine KS, Larson EB, Bird T, Quinn JF, Zhang J, Montine TJ. Proteomic determination of widespread detergent-insolubility including Abeta but not tau early in the pathogenesis of Alzheimer’s disease. Faseb J. 2005;19(13):1923–5. doi: 10.1096/fj.05-4263fje. [DOI] [PubMed] [Google Scholar]
- 24.Hynd MR, Lewohl JM, Scott HL, Dodd PR. Biochemical and molecular studies using human autopsy brain tissue. J Neurochem. 2003;85(3):543–62. doi: 10.1046/j.1471-4159.2003.01747.x. [DOI] [PubMed] [Google Scholar]
- 25.Fountoulakis M, Hardmeier R, Hoger H, Lubec G. Postmortem changes in the level of brain proteins. Exp Neurol. 2001;167(1):86–94. doi: 10.1006/exnr.2000.7529. [DOI] [PubMed] [Google Scholar]
- 26.Kim SI, Voshol H, van Oostrum J, Hastings TG, Cascio M, Glucksman MJ. Neuroproteomics: expression profiling of the brain’s proteomes in health and disease. Neurochem Res. 2004;29(6):1317–31. doi: 10.1023/b:nere.0000023618.35579.7c. [DOI] [PubMed] [Google Scholar]
- 27.Leger DW, Didrichsons I. An assessment of data pooling and some alternatives. Anim Behav. 1994;48:823–832. [Google Scholar]
- 28.Terry RD, Katzman R. Senile dementia of the Alzheimer type. Ann Neurol. 1983;14(5):497–506. doi: 10.1002/ana.410140502. [DOI] [PubMed] [Google Scholar]
- 29.Mitsui K, Doi H, Nukina N. Proteomics of polyglutamine aggregates. Methods in enzymology. 2006;412:63–76. doi: 10.1016/S0076-6879(06)12005-4. [DOI] [PubMed] [Google Scholar]
- 30.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 31.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology. 1991;41(4):479–86. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 32.Consensus recommendations for the postmortem diagnosis of Alzheimer’s disease. The National Institute on Aging, and Reagan Institute Working Group on Diagnostic Criteria for the Neuropathological Assessment of Alzheimer’s Disease. Neurobiol Aging. 1997;18(4 Suppl):S1–2. [PubMed] [Google Scholar]
- 33.McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick’s Disease. Arch Neurol. 2001;58(11):1803–9. doi: 10.1001/archneur.58.11.1803. [DOI] [PubMed] [Google Scholar]
- 34.Trojanowski JQ, Dickson D. Update on the neuropathological diagnosis of frontotemporal dementias. J Neuropathol Exp Neurol. 2001;60(12):1123–6. doi: 10.1093/jnen/60.12.1123. [DOI] [PubMed] [Google Scholar]
- 35.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 36.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom. 2001;36(10):1083–91. doi: 10.1002/jms.229. [DOI] [PubMed] [Google Scholar]
- 37.Eng J, McCormack AL, Yates JR. 3rd, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 38.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. Journal of proteome research. 2003;2(1):43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 39.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426(6966):570–4. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 40.Bondarenko PV, Chelius D, Shaler TA. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem. 2002;74(18):4741–9. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 41.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal Chem. 2003;75(18):4818–26. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 42.Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol Cell Proteomics. 2006;5(6):1158–70. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Li XJ, Zhang H, Ranish JA, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal Chem. 2003;75(23):6648–57. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 44.Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature reviews. 2006;7(1):55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- 45.Kendziorski CM, Zhang Y, Lan H, Attie AD. The efficiency of pooling mRNA in microarray experiments. Biostatistics (Oxford, England) 2003;4(3):465–77. doi: 10.1093/biostatistics/4.3.465. [DOI] [PubMed] [Google Scholar]
- 46.Roxas BA, Li Q. Significance analysis of microarray for relative quantitation of LC/MS data in proteomics. BMC Bioinformatics. 2008;9:187. doi: 10.1186/1471-2105-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu F, Vitek MP, Colton CA, Previti ML, Gharkholonarehe N, Davis J, Van Nostrand WE. Human apolipoprotein E redistributes fibrillar amyloid deposition in Tg-SwDI mice. J Neurosci. 2008;28(20):5312–20. doi: 10.1523/JNEUROSCI.1042-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fryer JD, Simmons K, Parsadanian M, Bales KR, Paul SM, Sullivan PM, Holtzman DM. Human apolipoprotein E4 alters the amyloid-beta 40:42 ratio and promotes the formation of cerebral amyloid angiopathy in an amyloid precursor protein transgenic model. J Neurosci. 2005;25(11):2803–10. doi: 10.1523/JNEUROSCI.5170-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pallitto MM, Murphy RM. A mathematical model of the kinetics of beta-amyloid fibril growth from the denatured state. Biophys J. 2001;81(3):1805–22. doi: 10.1016/S0006-3495(01)75831-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahara N, Lewis J, DeTure M, McGowan E, Dickson DW, Hutton M, Yen SH. Assembly of tau in transgenic animals expressing P301L tau: alteration of phosphorylation and solubility. J Neurochem. 2002;83(6):1498–508. doi: 10.1046/j.1471-4159.2002.01241.x. [DOI] [PubMed] [Google Scholar]
- 51.Morishima-Kawashima M, Ihara Y. Alzheimer’s disease: beta-Amyloid protein and tau. J Neurosci Res. 2002;70(3):392–401. doi: 10.1002/jnr.10355. [DOI] [PubMed] [Google Scholar]
- 52.Kuret J, Congdon EE, Li G, Yin H, Yu X, Zhong Q. Evaluating triggers and enhancers of tau fibrillization. Microsc Res Tech. 2005;67(3–4):141–55. doi: 10.1002/jemt.20187. [DOI] [PubMed] [Google Scholar]
- 53.Lansbury PT., Jr Evolution of amyloid: what normal protein folding may tell us about fibrillogenesis and disease. Proc Natl Acad Sci U S A. 1999;96(7):3342–4. doi: 10.1073/pnas.96.7.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JR, Muresan A, Lee KY, Murphy RM. Urea modulation of beta-amyloid fibril growth: experimental studies and kinetic models. Protein Sci. 2004;13(11):2888–98. doi: 10.1110/ps.04847404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinkauf M, Hiddemann W, Dreyling M. Sample pooling in 2-D gel electrophoresis: a new approach to reduce nonspecific expression background. Electrophoresis. 2006;27(22):4555–8. doi: 10.1002/elps.200600207. [DOI] [PubMed] [Google Scholar]
- 56.Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer disease. Proc Natl Acad Sci U S A. 1995;92(11):4725–7. doi: 10.1073/pnas.92.11.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rostagno A, Lashley T, Ng D, Meyerson J, Braendgaard H, Plant G, Bojsen-Moller M, Holton J, Frangione B, Revesz T, Ghiso J. Preferential association of serum amyloid P component with fibrillar deposits in familial British and Danish dementias: similarities with Alzheimer’s disease. J Neurol Sci. 2007;257(1–2):88–96. doi: 10.1016/j.jns.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 58.Yasojima K, Schwab C, McGeer EG, McGeer PL. Up-regulated production and activation of the complement system in Alzheimer’s disease brain. Am J Pathol. 1999;154(3):927–36. doi: 10.1016/S0002-9440(10)65340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Navarro A, Del Valle E, Astudillo A, Gonzalez del Rey C, Tolivia J. Immunohistochemical study of distribution of apolipoproteins E and D in human cerebral beta amyloid deposits. Exp Neurol. 2003;184(2):697–704. doi: 10.1016/S0014-4886(03)00315-7. [DOI] [PubMed] [Google Scholar]
- 60.Wilhelmus MM, Otte-Holler I, Wesseling P, de Waal RM, Boelens WC, Verbeek MM. Specific association of small heat shock proteins with the pathological hallmarks of Alzheimer’s disease brains. Neuropathol Appl Neurobiol. 2006;32(2):119–30. doi: 10.1111/j.1365-2990.2006.00689.x. [DOI] [PubMed] [Google Scholar]
- 61.Nakamura Y, Takeda M, Suzuki H, Hattori H, Tada K, Hariguchi S, Hashimoto S, Nishimura T. Abnormal distribution of cathepsins in the brain of patients with Alzheimer’s disease. Neurosci Lett. 1991;130(2):195–8. doi: 10.1016/0304-3940(91)90395-a. [DOI] [PubMed] [Google Scholar]
- 62.McMurray CT. Neurodegeneration: diseases of the cytoskeleton? . Cell Death Differ. 2000;7(10):861–5. doi: 10.1038/sj.cdd.4400764. [DOI] [PubMed] [Google Scholar]
- 63.Xia Q, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J. Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. J Proteome Res. 2008;7(7):2845–51. doi: 10.1021/pr8000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bennett V, Healy J. Organizing the fluid membrane bilayer: diseases linked to spectrin and ankyrin. Trends Mol Med. 2008;14(1):28–36. doi: 10.1016/j.molmed.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Bennett V, Lambert S. Physiological roles of axonal ankyrins in survival of premyelinated axons and localization of voltage-gated sodium channels. J Neurocytol. 1999;28(4–5):303–18. doi: 10.1023/a:1007005528505. [DOI] [PubMed] [Google Scholar]
- 66.von Janowsky B, Knapp K, Major T, Krayl M, Guiard B, Voos W. Structural properties of substrate proteins determine their proteolysis by the mitochondrial AAA+ protease Pim1. Biol Chem. 2005;386(12):1307–17. doi: 10.1515/BC.2005.149. [DOI] [PubMed] [Google Scholar]
- 67.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4(9):674–80. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 68.Atamna H, Frey WH., 2nd Mechanisms of mitochondrial dysfunction and energy deficiency in Alzheimer’s disease. Mitochondrion. 2007;7(5):297–310. doi: 10.1016/j.mito.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 69.Fukui H, Moraes CT. The mitochondrial impairment, oxidative stress and neurodegeneration connection: reality or just an attractive hypothesis? . Trends Neurosci. 2008;31(5):251–6. doi: 10.1016/j.tins.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Marcillat O, Giulivi C, Ernster L, Davies KJ. The oxidative inactivation of mitochondrial electron transport chain components and ATPase. J Biol Chem. 1990;265(27):16330–6. [PubMed] [Google Scholar]
- 71.Shin SJ, Lee SE, Boo JH, Kim M, Yoon YD, Kim SI, Mook-Jung I. Profiling proteins related to amyloid deposited brain of Tg2576 mice. Proteomics. 2004;4(11):3359–68. doi: 10.1002/pmic.200400961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.