Abstract
Short hairpin RNA (shRNA) libraries are limited by the low efficacy of many shRNAs, giving false negatives, and off-target effects, giving false positives. Here we present a strategy for rapidly creating expanded shRNA pools (∼30 shRNAs/gene) that are analyzed by deep-sequencing (EXPAND). This approach enables identification of multiple effective target-specific shRNAs from a complex pool, allowing a rigorous statistical evaluation of whether a gene is a true hit.
Several viral-based shRNA library methods have been described, and have become valuable tools for conducting RNAi screens (reviewed in 1–3). Microarray synthesis of shRNAs has been used to generate diverse libraries of shRNAs or microRNA-designed shRNAs, which are then cloned, sequence-verified, and arrayed into 96-well plate format 2, 4, 5. To simplify screening, these barcoded shRNA constructs can be used as pools, and the resulting hits identified by recovering and hybridizing the barcodes to a microarray 6–9. These strategies have been widely implemented with many successes6–9, and such libraries are commercially available (e.g. Open Biosystems, Sigma, TRC libraries).
A central shortcoming of existing lentiviral libraries is their low diversity (typically 3–5 shRNAs/gene), which results in high rates of both false negatives and false positives. False negatives occur because currently available algorithms (e.g., 10) cannot ensure the presence of effective hairpins specific to a target gene. For the same reason, false positives become problematic because more than one effective shRNA/target must be present to rule out off-target effects. The use of high diversity libraries would allow for identification of multiple potent shRNAs/target (a critical control in RNAi experiments11), while increasing the sensitivity of RNAi screens.
Existing shRNA libraries have typically utilized extensive cloning, sequencing, and often addition of a vector-specific “barcode” sequence to each shRNA before it is included in the library, a time-consuming and costly process. Recent improvements in microarray-based oligonucleotide synthesis allow the production of long oligos (>100 bp) with an error rate of less than 1/250 bp (data not shown). These oligos should allow a direct clone-and-use strategy that would allow easy adoption of changes in RNAi technology, vector choice, or assay design.
Accordingly, we generated pooled shRNA libraries that (1) have highly expanded per-gene coverage (∼30 shRNAs/gene), (2) are easy to construct and inexpensive to screen, (3) can be used directly as shRNA pools, and (4) can be readily quantitated by microarray and deep sequencing following a screen (Fig. 1). To this end, we designed a pilot library encoding 22,000 shRNAs to target ∼600 genes, including nearly all known human CD antigens (CD antigen shRNA library). To maintain the high fidelity and diversity of the input oligo library, we carefully optimized conditions for PCR amplification, cloning, and propagation of the shRNA library (see methods, and Supplementary Note 1).
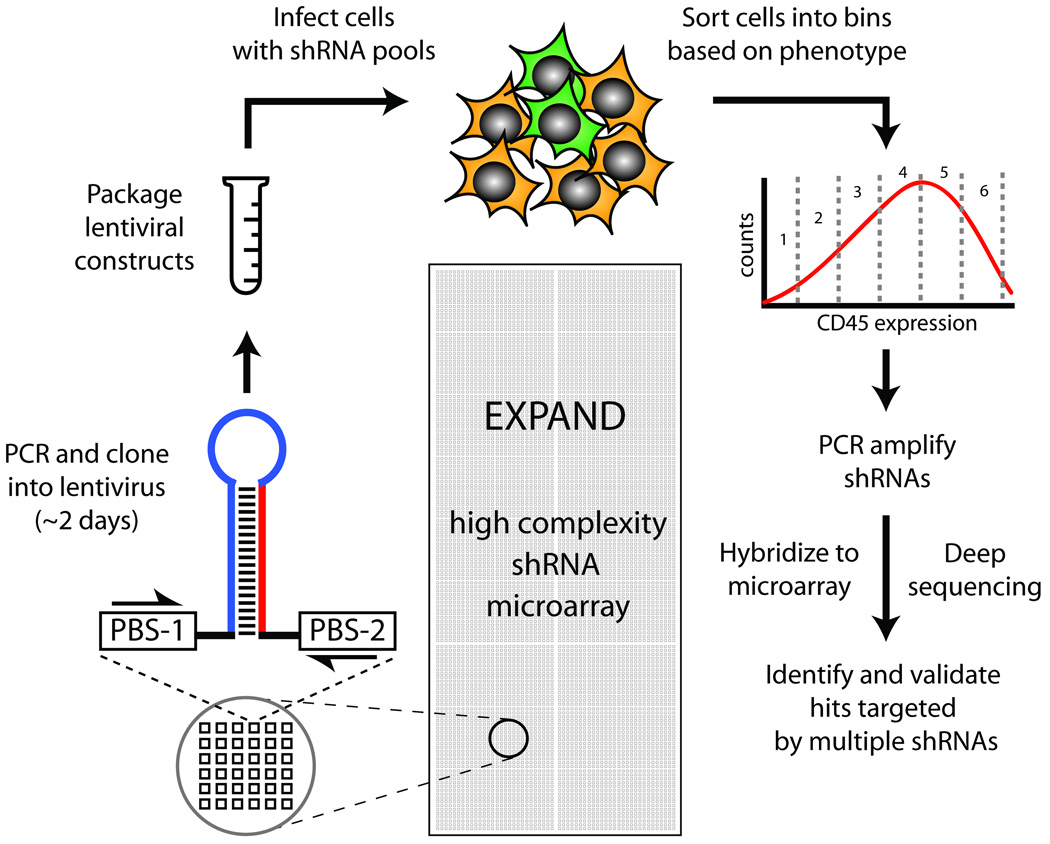
Figure 1. Schematic for Microarray Synthesis, Cloning, and Enrichment.
Microarrays containing 22,000 shRNAs with ∼30 shRNAs per gene are synthesized, and released from the array into a single pool. shRNAs are amplified, cloned, and packaged into lentivirus. Cells infected with the lentiviral shRNA library are sorted by FACS according to phenotype. shRNAs from different fractions are PCR amplified and quantitated either by microarray hybridization or deep sequencing.
To determine the mutation frequency in the shRNA library, we sequenced 122 random shRNA inserts. 64% of the clones were correct (Supplementary Table 1). The errors in the remainder generally consisted of 1–2 nucleotide mutations or deletions. While it is likely that many of the imperfect shRNA sequences retain effectiveness in gene knockdown12–14, these mutants can be identified by deep sequencing and removed from downstream analysis. We repeatedly created CD antigen shRNA libraries, with 60–80% correct shRNA sequences (Supplementary Table 1).
PCR amplification can lead to a reduction in complexity of an shRNA mixture during multiple cycling steps. To assess the library complexity, we deep sequenced PCR-amplified shRNAs and identified ∼95% of the expected shRNAs (Supplementary Fig. 1), with error rates consistent with our previous single clone measurements (Supplementary Table 2). It is also possible to monitor these libraries using microarrays and improved half-hairpin probes7–9. Either approach would allow the direct identification of shRNAs, and eliminate the need to independently barcode each vector.
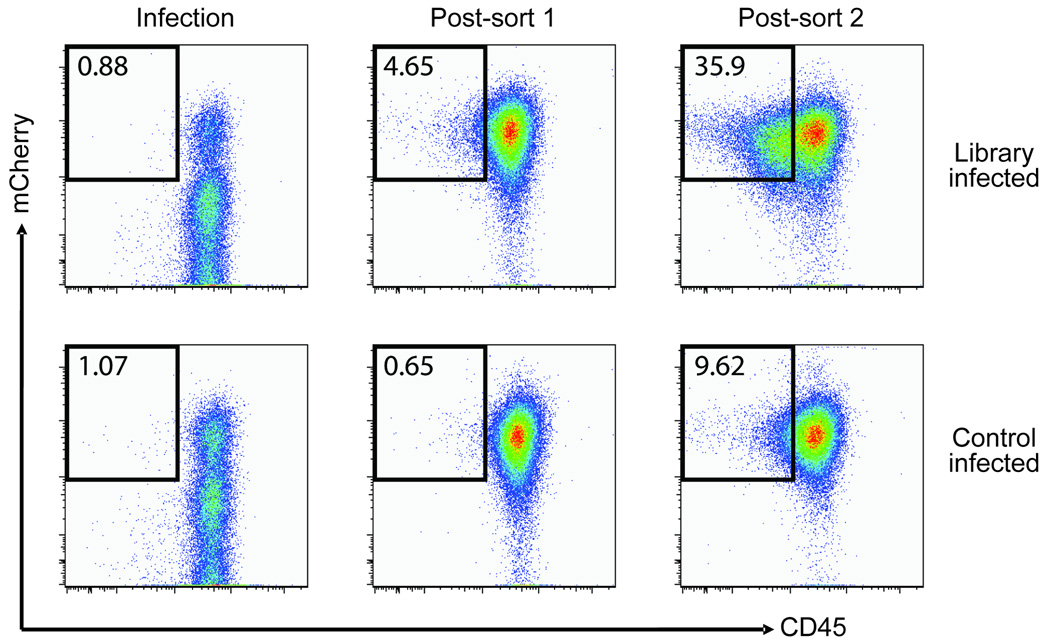
Since we obtained a reasonably low error rate and nearly complete shRNA coverage, it seemed likely that the libraries could be used directly in RNAi experiments. As a functional test, we infected human Raji B cells with the CD antigen shRNA library, and sorted infected cells (expressing mCherry as a marker for infection) that also displayed reduced expression of CD45. The initial comparison of cells seven days after infection with either the control virus or the shRNA library showed no notable difference during the first sort. However, after two rounds of sorting with cells cultured for seven days between sorts, we observed a substantial enrichment for mCherry+ CD45low cells as compared to cells infected with vector alone (35.9% vs. 9.62%) (Fig. 2). To identify active anti-CD45 shRNAs, we PCR-amplified and cloned the lentiviral shRNA inserts from genomic DNA isolated from the CD45low sorted cells. Of 83 sequenced shRNA clones, 39 targeted CD45 (46%). Given that the starting population of the library contained 0.15% CD45-targeting shRNAs (33 out of 22,000), this represents an enrichment of > 300 fold accomplished in a two step sort procedure.
Figure 2. Raji cells sorted for low CD45 expression are enriched for anti-CD45 shRNAs.
Raji B cells were infected with the CD antigen shRNA library (top panels) or control virus (no insert, lower panels), allowed to grow for 7 days, and sorted by FACS for reduced expression of CD45 and expression of mCherry in two rounds (7 days between sorts). Boxes indicate the gates used to sort CD45-downregulated cells. The percentage of mCherry+ cells displaying reduced CD45 expression is indicated in the figure.
Although other shRNAs were detected in the enriched fraction, we only detected multiple distinct shRNAs for CD45, highlighting the power of the expanded library to unambiguously detect ‘hits’ in a single screen. In this initial experiment we recovered 6 unique CD45-targeting shRNAs in the sorted CD45low fraction from the total of 33 CD45-targeting shRNAs present in the library. These active shRNAs differed markedly in abundance within the sorted fraction (Supplementary Table 3). In general, the shRNAs that were recovered in larger numbers were also more potent when they were individually re-tested for CD45 knockdown (Supplementary Table 3, Fig. 2). Therefore, the expanded library yielded a diverse population of target-specific shRNAs in a single experiment, and allowed us to obtain information about the potency of each individual shRNA from the relative number of each shRNA recovered.
Recent developments in deep sequencing technology make it possible to simultaneously measure the presence of > 80 million distinct sequences at a typical read length of ∼50–70 nucleotides, which is ideally suited to monitor the shRNA sequences in the library described here. A digital readout allows the clear resolution of even very similar shRNA species, which is important for the determination of efficacy of individual shRNAs in high complexity libraries. In addition, mutant shRNAs can be directly detected and discarded from further analysis, which is not possible using microarray hybridization assays. This should be particularly useful when measuring the loss of shRNAs that cause cell death or slow growth (dropout screens), where the presence of inactive mutant shRNAs would complicate the interpretation of results. Finally, the large capacity of deep sequencing allows the detection of subtle changes in abundance within genome-scale populations of shRNAs.
To evaluate the capacity of deep sequencing to accurately measure sequence abundance over a broad range of concentrations, we performed a dilution series with a known set of 32 oligonucleotides with unique 28-mer sequence tags. We could detect a highly linear distribution of oligonucleotide counts over an ∼ 1 × 106 concentration range (Supplementary Fig. 2). Sequences which were read less than ∼10 times were less reliably measured, but can be accurately measured by simply increasing the sequencing coverage.
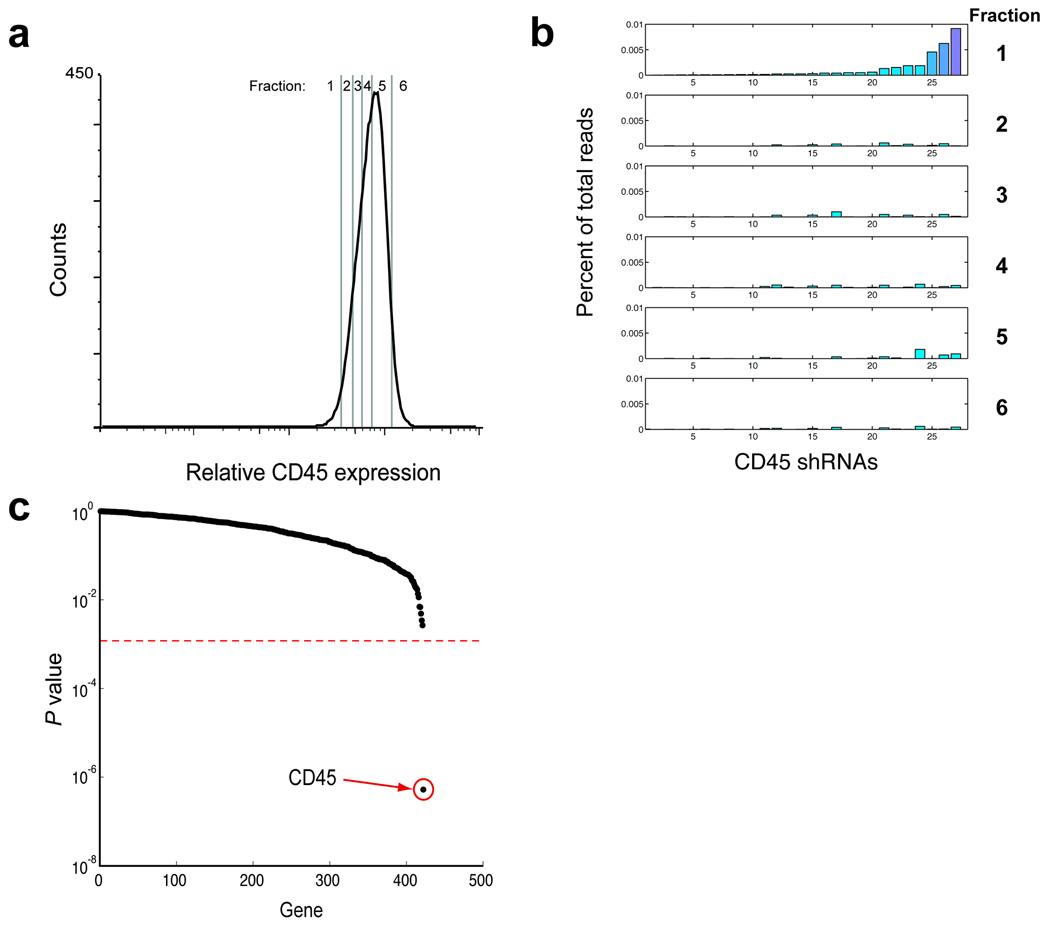
The large dynamic range and linearity of this counting approach suggested that deep sequencing could be used to measure the change in abundance of shRNA species at early time points in our test screen. To this end, we used deep sequencing coupled with binned flow cytometry based-sorting to search for shRNAs targeting CD45. Human Raji B cells were infected with the CD antigen shRNA library and grown for 1 week, after which they were sorted into 6 fractions representing different levels of CD45 expression (Fig. 3a, Supplementary Fig. 3). Genomic DNA was prepared from the sorted fractions, shRNAs were amplified by PCR, and the abundance of each shRNA in the various fractions was assessed by deep sequencing. Remarkably, even though a population of CD45low cells was undetectable by flow cytometry at this early time point (Fig. 2), we could readily measure substantial enrichment of multiple active anti-CD45 shRNAs in the CD45low fractions (Fig. 3b). This included all anti-CD45 shRNAs identified in the previous experiment, which involved two rounds of highly selective sorting performed over several weeks. Importantly, although the shRNAs were present at unequal levels at the beginning of the experiment, normalization of their abundance in CD45low fractions to a fraction with high CD45 expression could clearly identify enrichment for multiple active anti-CD45 shRNAs.
Figure 3. Binned flow-sorting coupled with deep sequencing can quantitatively resolve active shRNAs after a single sort.
(a) Raji B cells infected with the CD antigen shRNA library were grown for 7 days and sorted into the indicated fractions. (b) Genomic DNA was prepared from cells collected from the fractions in A, and the shRNAs were PCR amplified and deep sequenced. The percentage of total reads for each shRNA in each fraction for CD45 is depicted; shRNAs are sorted in order of abundance in fraction 1. (c) The abundance of each shRNA in fraction 1 was normalized to its abundance in fraction 5. Only CD45 gave a significant P value (< 2 × 10−7).
The ability to identify multiple active shRNAs specific for each gene is one of the most critical improvements of our approach over existing methodologies, and is a direct consequence of including 33 shRNAs per gene in the library. Taking into account the full range of shRNAs for each gene, a rigorous statistical test can be performed to differentiate true hits from genes that by chance have one or two off-target shRNAs. This allows the assignment of a P value to every screened gene (rather than to single shRNAs). Using this method, we could readily resolve anti-CD45 shRNAs from the rest of the library (Fig. 3c); (P < 2 × 10−7, see Supplementary Note 2). In contrast, the large majority of shRNAs were not significantly enriched in any fraction. This result was not unique to a particular CD antigen or cell type, as we obtained similar results when sorting for different CD antigens (LAIR1/CD305 in U937 cells or CD3 in Jurkat cells). LAIR1 and CD3-specific shRNAs could be clearly resolved from the rest of the library (P = 2.6 × 10−5 and P = 1.1 × 10−7, respectively)(Supplementary Fig. 4).
To further test the ability of our method to enrich for active target-specific shRNAs, we individually cloned and analyzed the potency of 33 anti-CD45 shRNAs predicted by an algorithm15 (Supplementary Fig. 5). Only about 50% of these had minimal activity (> 25% knockdown), and only 6 had > 60% knockdown. In general, while we could see substantial enrichment for active shRNAs after a single sort, there was little enrichment for shRNAs with low activity. Nonetheless, the correlation between activity and enrichment was not perfect, possibly because of off-target effects of some shRNAs.
Interestingly, the highly active shRNAs were not restricted to those predicted to be most active by the algorithm; indeed, the most active species were often quite low on the list. We observed similar results when testing shRNAs directed against LAIR1 (Supplementary Fig. 6). This analysis illustrates a key advantage of our expanded-coverage library: without including 30 shRNAs per gene it would have been impossible to predict enough functional shRNAs to corroborate hits. As data accumulates on which hairpins are most active, shRNA prediction algorithms can be improved and library sizes reduced.
In summary, our approach provides an efficient method for rapidly creating and screening shRNA libraries, which addresses both false negative and false positive problems that commonly plague RNAi screens. With only ∼20–30% of predicted hairpins giving > 50% knockdown (Supplementary Fig. 5–6), low complexity libraries will often not have enough shRNAs to corroborate genuine hits in a screen. We show here that high-coverage shRNA libraries can identify many shRNAs targeting a single gene, which increases confidence in hits obtained in RNAi screens. The increased complexity of these libraries can be deconvoluted through deep sequencing. We further show that this method allows detection of active shRNA hits in a model screen without extensive selection, sorting, and cell proliferation, which will greatly facilitate efforts to identify essential genes whose absence may slow growth or cause cell death. Finally, the direct clone-and-use method provides the flexibility to easily remake libraries to immediately incorporate continual advances in RNAi technology, such as various microRNA contexts for expression and improved algorithms for shRNA prediction.
Methods
Amplification, digestion, cloning, and preparation of shRNA library
To design shRNAs against CD antigens, we used the shRNA prediction algorithm freely available through the Hannon lab website: http://katahdin.cshl.org:9331/homepage/portal/scripts/main2.pl15. The output shRNA sequences were then modified to have a 9-bp loop (TTCAAGAGA) and common primer binding sites compatible with cloning into the lentiviral vector pSicoR-mCherry which drives stable expression of the hairpin via a mouse U6 promoter16: 5': CGCCTGCGAGTCTGGTATg, 3’: GGAATTCGCCAGCTCGAG (reverse complement). A 22,000-element, 96-bp oligonucleotide pilot library was designed using these sequences, and was provided by Agilent Technologies as a pool in a single tube (4 pmol) and dissolved in 200 µL water. Improvements to fidelity of long oligonucleotides were made by minimizing the synthetic cycle loss during coupling and deblock, and by minimizing the depurination side reaction.
We identified optimal conditions for amplification of full-length product (96 bp oligos) by using Phusion DNA polymerase (Finnzymes), varying the amount of DMSO, annealing temperature, and extension times for the PCR on a Bio-Rad iCycler. Final conditions were for 50 µl reaction: 30 µl water, 10 µl 5x Phusion GC buffer, 1 µl 10 mM dNTPs, 5 µl 5 µM primer mix, 0.5 µl template, 1 µl DMSO, 0.5 µl Hot Start Phusion polymerase (Finnzymes). Cycling parameters were 98 °C for 30 s; 15 cycles of (98 °C for 10 s and 72 °C for 30 s); 72 °C for 10 min. Primers: 5': CGCCTGCGAGTCTGGTAT, 3’: GGAATTCGCCAGCTCGAG. Note that all primers used in this study are listed in Supplementary Table 4.
The PCR-amplified oligos were purified using a nucleotide removal kit (Qiagen) according to manufacturer’s recommendations, and subjected to restriction digest in a 70 µl reaction containing 1 µg DNA, 7 µl 10x NEB buffer 4, 0.7 µl 100x BSA, 40 U of XhoI and MlyI (2 µl XhoI, 4 µl MlyI), and incubated in 37 °C waterbath for 6 H. Digested fragments were verified by electrophoresis on 20% PAGE-TBE with 0.5% TBE running buffer, and purified over a second Qiagen nucleotide removal column before cloning into pSicoR-mCherry.
To prepare the vector for cloning, pSicoRmCherry was subject to restriction digest in a 150 µl reaction containing 15 µl NEB buffer 4, 1.5 µl BSA, 5 µg pSicoR-mCherry, 50 U each of XhoI and HpaI (NEB), and incubated overnight at 37 °C. Vector was then treated (without purification) with Antarctic Phosphatase (NEB) in a 150 µl reaction containing 16.9 µl Antarctic Phosphatase buffer and 12.5 U enzyme for 4 H at 37 °C. Vector was then purified on an 0.8% agarose gel, cut out, and subject to gel-extraction using the QIAquick gel purification it (Qiagen). Vector was then phenol-chloroform extracted, ethanol precipitated, and resuspended in 20 µl water. A second library type was cloned using primers for mir30-context shRNAs as previously described4, 5, using the above parameters, except cloning was performed by digestion with XhoI and EcoRI.
Ligations were performed in a 20 µl reaction containing 500 ng vector, 30 ng insert, 2 µl 10X ligase buffer (NEB), 2000 U T4 DNA ligase (NEB), and were incubated at 16 °C for 16 H.
To preserve the diversity of the library, colonies were scraped from twelve 15 cm plates directly after transformation of the ligation mixture (six 100 µl transformations of max-efficiency DH5a cells (Invitrogen)). Plates contained about 50,000 colonies each. The collected cell pellet was used for maxiprep (Qiagen) directly, and enough DNA was recovered from the plates for numerous viral preps.
Isolation of CD45, LAIR1, or CD3-targeting shRNAs
To select shRNAs that specifically target the human CD45 receptor, LAIR1/CD305, or CD3, we prepared concentrated virus from the CD-antigen library and control (no insert) pSicoR-mCherry vectors as described previously16. 2 × 106 human Raji B cells, U937 cells, or Jurkat cells (ATCC) were infected with virus at an MOI of 0.1 to ensure single integration sites of the viruses. After 7 days of culture, ∼ 5 × 107 cells were incubated on ice for 15 min with 500 µl of PBS containing 20% normal mouse serum, 5% BSA, and 10% FCS to block non-specific interactions. Subsequently, PE-conjugated anti-CD45 (BD Biosciences), APC-conjugated anti-LAIR1 (R&D), or APC-conjugated anti-CD3ϵ (BD Biosciences) was added and allowed to interact with the cells for 30 min, followed by two washes with Hanks’ BSS medium supplemented with 2% FCS. mCherry positive (as a marker for infected cells) and CD45 or LAIR-1-reduced cells (to select potential active anti-CD45 or anti-LAIR1 shRNAs) were sorted by using a MoFlow cell sorter (Dako Cytomation). After a week of culture the same procedure was repeated, and the cells were cultured for an additional 7 days. Note that for the binning/deep sequencing experiment, cells were infected and sorted after 7 days. For further details of binning/sorting, see Supplementary Figure 1. All of the sequences for the CD antigen shRNA library as well as the effective shRNAs targeting CD45 and LAIR1 are included as Supplementary Data 1–3 online.
To re-test anti-CD45 shRNAs following the 2-sort experiment, genomic DNA was isolated from the sorted cell fractions, and lentiviral shRNA integrations were amplified by PCR using the primers: 5’: TGCAGGGGAAAGAATAGTAGAC, and 3’: AGTTATGTAACGCGGAACTCC, cloned into the pCR2.1-TOPO vector (Invitrogen), and sequenced. Identified CD45-targeting shRNAs were subcloned into pSicoR-mCherry, packaged into lentivirus and individually validated for their efficiency in CD45 knock-down, essentially as described above. Analysis of cells after viral infection and surface antigen staining was performed on an LSR II flow cytometer (BD Biosciences) and the FACS data was analyzed by FlowJo software (Tree Star, Inc.). Percent knockdown is calculated by ((Geo-mean of uninfected cells minus Geo-mean of infected cells)/Geo-mean of uninfected cells)*100.
Amplification of shRNA pools from genomic DNA
shRNAs were amplified from genomic DNA in a 50 µl PCR reaction consisting of 30 µl water, 10 µl 5x Phusion GC buffer (Finnzymes), 5 µl 5 µM primer mix, 1 µl 10 mM dNTPs, 1.5 µl DMSO, 750 ng genomic DNA, and 1U (0.5 µl) Phusion polymerase (Finnzymes). Cycling parameters were 98 °C for 30 s, then 25 cycles of (98 °C for 30 s, 56 °C for 15 s, 72 °C for 15 s), then 72 °C for 10 min. Primers for amplification of shRNAs from genomic DNA were 5' ATAAATATCCCTTGGAGAAAAGC, 3' GGCGGTAATACGGTTATCCA. In some cases many PCR reactions were pooled on a minelute column (Qiagen) before electrophoresis on 20% PAGE with 0.5% TBE running buffer, electroelution, and concentration on a second column.
Deep Sequencing, and Quantitation of Read Accuracy
Genomic DNA was prepared as before, and shRNAs were amplified as before, except sequences compatible with annealing to the Illumina flow cell were added. Final primer sequences were: 5’: AATGATACGGCGACCACCGACACTCTTTCCCTCCCTTGGAGAAAAGCCTTGTTtG and 3’: CAAGCAGAAGACGGCATACGA ATGGATCCTA GTACTCGAG. Note that parts of these oligonucleotide sequences are copyrighted by Illumina, Inc. The sequencing primer used was CACTCTTTCCCTCCCTTGGAGAAAAGCCTTGTTTG. Sequencing was performed according to manufacturer’s protocols (Illumina).
For assessing linearity of sequence counting over a dilution series, thirty-one 68mer oligos consisting of a unique 28 nt tag flanked by 21 nt and 19 nt common Illumina primer binding sites were individually amplified by PCR and purified from an acrylamide gel. The dsDNA products were quantified using a BioAnalyzer (Agilent Technologies), and pooled with a dilution strategy designed to give a broad range of expected sequence tag concentrations. This pool was subjected to deep sequencing, and reads were aligned with the tag library allowing up to 3 mismatches against the tag.
Supplementary Material
Acknowledgements
We would like to thank Ali Brincat from the Sandler Lentiviral Core and Cliff McArthur from the Sandler Asthma Basic Research Center (SABRE) for excellent technical assistance. We would also like to thank Quinn Mitrovich and Noel Goddard for technical advice, and David Hirschberg and Tim Baxter of Agilent Technologies. This work was supported by a Rubicon grant from The Netherlands Organization for Scientific Research (NWO, RJL), and by a Career Development Fellowship from the Leukemia and Lymphoma Society (MCB). M.S. Was supported by a post-doctoral fellowship from the Sandler Program in Basic Sciences and is currently supported by an National Institutes of Health K99/R00 (Pathway to Independence) award. N.T.I. is supported by the National Institutes of Health under a Ruth L. Kirschstein National Research Service Award (GM080853) from the National Institute of General Medical Sciences. This work was supported by a Sandler New Technologies grant to J.S.W. and M.T.M., National Institutes of Health grant RO1 GM80783 to M.T.M., and a grant from the Fight For Mike foundation to J.S.W.
Footnotes
Competing Interests Statement:
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemethods/
E.M.L. is employed by Agilent Technologies, Inc., and Agilent reagents are used in the research presented in this article.
References
- 1.Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat. Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- 2.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat. Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 3.Root DE, Hacohen N, Hahn WC, Lander ES, Sabatini DM. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- 4.Cleary MA, et al. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat. Methods. 2004;1:241–248. doi: 10.1038/nmeth724. [DOI] [PubMed] [Google Scholar]
- 5.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 6.Brummelkamp TR, et al. An shRNA barcode screen provides insight into cancer cell vulnerability to MDM2 inhibitors. Nat. Chem. Biol. 2006;2:202–206. doi: 10.1038/nchembio774. [DOI] [PubMed] [Google Scholar]
- 7.Schlabach MR, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silva JM, et al. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. U S A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynolds A, et al. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 11.Echeverri CJ, et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- 12.Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aleman LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paddison PJ, et al. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Methods. 2004;1:163–167. doi: 10.1038/nmeth1104-163. [DOI] [PubMed] [Google Scholar]
- 16.Ventura A, et al. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. U S A. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.