Abstract
The kinetics and magnitude of cytokine gene expression are tightly regulated to elicit a balanced response to pathogens and result from integrated changes in transcription and mRNA stability. Yet, how a single microbial stimulus induces peak transcription of some genes (TNFα) within minutes whereas others (IP-10) require hours remains unclear. Here, we dissect activation of several lipopolysaccharide (LPS)-inducible genes in macrophages, an essential cell type mediating inflammatory response in mammals. We show that a key difference between the genes is the step of the transcription cycle at which they are regulated. Specifically, at TNFα, RNA Polymerase II initiates transcription in resting macrophages, but stalls near the promoter until LPS triggers rapid and transient release of the negative elongation factor (NELF) complex and productive elongation. In contrast, no NELF or polymerase is detectible near the IP-10 promoter before induction, and LPS-dependent polymerase recruitment is rate limiting for transcription. We further demonstrate that this strategy is shared by other immune mediators and is independent of the inducer and signaling pathway responsible for gene activation. Finally, as a striking example of evolutionary conservation, the Drosophila homolog of the TNFα gene, eiger, displayed all of the hallmarks of NELF-dependent polymerase stalling. We propose that polymerase stalling ensures the coordinated, timely activation the inflammatory gene expression program from Drosophila to mammals.
Keywords: cytokine gene expression, inflammation, TNFα, transcription initiation and elongation, NELF
Innate immune responses are ancient programs that evolved to provide protection against invading pathogens. In addition to mobilizing professional phagocytic cells, e.g., macrophages (MΦ) and neutrophils, which actively prevent the dissemination of a given microbial agent and create a hostile extracellular environment for its replication, the innate immune system in higher organisms triggers much slower-acting adaptive immunity to mount a response should the primary defenses fail to contain the infection.
In recent years our understanding of the key molecular mechanisms underlying innate responses in mammals has greatly advanced. Host cells sense the presence of pathogens through a family of receptors, such as Toll-like receptors (TLRs), which detect conserved microbial components including lipopolysaccharide (LPS), peptidoglycans, single-stranded (ss) and double-stranded (ds) DNA and RNA, and common bacterial surface proteins, collectively known as pathogen-associated molecular patterns (PAMPs) (1, 2). Upon activation, TLRs initiate a signaling cascade through adaptors and protein kinases that converge on transcriptional regulators nuclear factor (NF)κB, activator protein (AP)1, and interferon (IFN) regulatory factors (IRFs) that, in turn, induce transcription of diverse proinflammatory cytokines and chemokines (3). These will ultimately activate MΦ and dendritic cells and stimulate their migration to the site of invasion.
Because the innate response is rapid and relatively nonspecific, it imposes an intrinsic threat to the host if not properly contained. Indeed, uncontrolled production of proinflammatory cytokines, e.g., tumor necrosis factor (TNF)α, IL-1β, and IFNs, is the underlying cause of the toxic shock syndrome. Persistent inflammation is also a hallmark of most autoimmune diseases: TNFα has been causally linked to joint destruction in rheumatoid arthritis, whereas pathogenesis of systemic lupus has been associated with dysregulated production of type I IFN (4–7).
Not surprisingly, numerous mechanisms have evolved to limit the levels of proinflammatory cytokines. In fact, many of them directly trigger negative feedback loops, including synthesis of anti-inflammatory cytokines such as IL-10 (8). Certain cytokine and chemokine proteins are synthesized as proforms (IL-1β) or membrane-bound precursors (TNFα), requiring proteolysis for activation (9, 10). Secreted IL-1β can be sequestered by a decoy receptor (IL-1RII) or a soluble receptor antagonist (IL-1Ra), precluding it from signaling (10). mRNAs encoding many proinflammatory mediators are intrinsically unstable because of the AU-rich elements (AREs) that bind proteins like tristetraprolin (TTP), which target the transcripts for mRNA decay (11); TNFα, IL-6, IL-1β, and IL-10 are all thought to be subject to TTP-mediated degradation (12, 13). In fact, recent genomewide analysis described functionally distinct classes of proinflammatory molecules on the basis of their mRNA stability (14).
Finally, proinflammatory gene transcription is one of the most sophisticated and tightly controlled gene expression programs. Indeed, DNA sequences associated with these genes are rich in binding sites for numerous transcriptional regulators, and the roles of AP1, NFκB/Rel, and IRF families in cytokine gene expression have been studied extensively long before these factors were identified as the effectors of TLR signaling. A transcriptional response of a given cytokine gene may be capped by a sequential recruitment of different regulator family members to the element. For example, upon sustained exposure of MΦ to LPS, cRel/p50 and p65/p50 heterodimers at the TNFα and CXCL2 genes are replaced by p50 homodimers, which correlates with cessation of transcription and LPS tolerance (15, 16). Similarly, cofactor switch because of a signaling event, leading to modification of either the cofactor itself or the DNA-bound regulator responsible for its recruitment, can alter the rate of transcription or even reverse its polarity. For instance, cJun homodimers occupying the AP1 site of several proinflammatory genes in resting MΦ mediate binding of the corepressor NCoR, which imposes a silent transcriptional state; exposure to LPS leads to cJun phosphorylation, heterodimerization with cFos, NCoR dismissal, and derepression (17). Recently, nucleosome positioning was proposed to confer a particular transcriptional profile to genes with common chromatin structure (18). On the basis of the differential requirements for Brg1/BRM and Mi-2b chromatin remodeling complexes, the authors described 3 types of genes encoding proinflammatory mediators that shared specific induction profiles under conditions tested (18).
Although the studies above explain expression patterns of individual molecules, coordinate and sharp waves of expression of large sets of proinflammatory genes in response to a common stimulus imply that a broader regulatory mechanism may be in place. Notably, events in the transcription cycle subsequent to sequence-specific factor binding, cofactor recruitment, chromatin remodeling, and preinitiation complex (PIC) assembly may represent important points of regulation. In fact, recent work in Drosophila and human cells estimates that ≈20% of genes are regulated after RNA Polymerase (Pol) II recruitment to the gene promoter, by controlling the efficiency of early transcription elongation through the promoter-proximal region (19–21). Importantly, this strategy for gene regulation was found to be enriched at stimulus-responsive genes in Drosophila (19, 22). How widespread this regulatory checkpoint is in the mammalian innate immune system is unknown.
To understand the patterns of transcriptional responses to TLR signaling in MΦ, we examined transcription complex assembly at TNFα, the prototypic proinflammatory cytokine, and several other mediators of inflammation. Our data indicate that these genes fall into distinct classes, depending on whether their transcription is controlled at the level of PIC recruitment vs. during transcription elongation, and that their induction profile correlates with the rate-limiting step in a transcription cycle rather than signaling events leading to their activation.
Results
RNA Polymerase II Occupies the TNFα Proximal Promoter in MΦ Before Gene Activation.
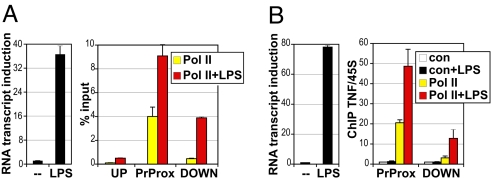
TNFα gene expression is strongly induced in response to microbial products: LPS treatment of the murine RAW264.7 MΦ-like cell line and of primary bone marrow-derived MΦ (BMMΦ) led to a dramatic increase in TNFα transcript (Fig. 1 A and B, Left). Unexpectedly, chromatin immunoprecipitation (ChIP) analysis revealed a significant RNA Pol II occupancy near the TNFα promoter under basal conditions in both cell contexts (Fig. 1 A and B, Right). This high “basal” Pol II occupancy was focused in the promoter-proximal region; far less Pol II was detected upstream or farther downstream of the transcription start site (TSS). LPS treatment triggered a significant increase in Pol II signal particularly within the downstream region of TNFα, suggesting that LPS-mediated gene activation facilitates the release of Pol II from the promoter into the gene.
Fig. 1.
Pol II occupies the promoter of the TNFα gene before activation. RAW264.7 cells (A) and day 7 BMMΦ (B) were treated with LPS (1 μg/mL for 0.5 h and 10 ng/mL for 1 h, respectively) and the induction of TNFα transcript was measured by real-time qPCR (Left) or cells were subjected to ChIP (Right). For RAW264.7 cells (A) ChIP material precipitated with antibodies to Pol II was quantified by real-time qPCR using primers centered around the upstream (UP), promoter-proximal (PrProx), and downstream (DOWN) regions of the TNFα gene and expressed as percentage of input DNA obtained in each sample. n = 3, error is SEM. For BMMΦ (B) ChIP material precipitated with normal rabbit IgG (con) or equivalent amount of antibodies to Pol II was qPCR quantified with primers to PrProx and DOWN regions of the TNFα gene and normalized to corresponding signals at the unrelated 45S rRNA gene as an internal control; the value of IgG control in untreated cells was set to 1 for both locations. n = 3, error is SEM.
This Pol II binding pattern resembled that described for genes whose promoters are regulated at a step subsequent to Pol II recruitment and are constitutively occupied by “stalled” or “paused” Pol II. Pol II stalling, increasingly appreciated as a widespread regulatory checkpoint in transcription (19–21), occurs when Pol II initiates and produces a short RNA transcript but stalls during elongation through the promoter-proximal region.
LPS Target Genes in BMMΦ Display Different Patterns of Pol II Occupancy and Phosphorylation.
The C-terminal domain (CTD) of the Pol II largest subunit contains a series of heptapeptide repeats (YS2PTS5PS) that can be differentially phosphorylated during the transcription cycle. Interestingly, the stalled Poll II displays a characteristic phosphorylation pattern, whereby levels of Serine5 phosphorylation (P-S5), a marker for initiated Pol II, are high, whereas phosphorylation levels at Serine2 (P-S2), a marker for the transition to productive elongation, are low (23, 24). Hence, we examined CTD phosphorylation across the TNFα gene in resting and LPS-stimulated MΦ.
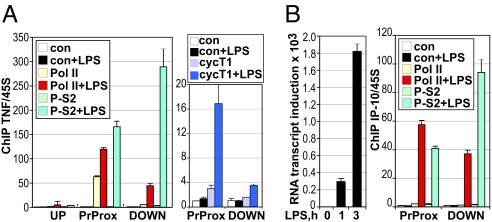
Consistent with polymerase stalling, Pol II adjacent to the TNFα TSS was phosphorylated on S5 in untreated BMMΦ [supporting information (SI) Fig. S1A], whereas P-S2 Pol II was undetectable (Fig. 2A Left). Upon LPS treatment, Pol II was strongly phosphorylated on S2 and associated with the downstream regions of TNFα. Hence CTD phosphorylation at the TNFα gene suggested that Pol II initiated transcription in resting cells but required an additional signal (e.g., LPS-induced transcription factor binding) to undergo S2 phosphorylation, be released from the promoter, and enter productive elongation.
Fig. 2.
TNFα but not IP-10 display the hallmarks of Pol II stalling. BMMΦ derived and treated as in Fig. 1B were subjected to ChIP with IgG control, antibodies to Pol II, P-S2, or cyclin T1, as indicated. Occupancy at the UP, PrProx, and DOWN regions of the TNFα gene (A) or PrProx and DOWN regions of the IP-10 gene (B Right) was assessed as in Fig. 1B. IP-10 RNA induction (B Left) in BMMΦ treated with LPS for 0, 1, or 3 h was assessed as in Fig. 1B.
Indeed, cyclin T1, the regulatory subunit of the positive transcription elongation factor (P-TEFb) kinase complex responsible for S2 phosphorylation and release of stalled Pol II into the gene, was recruited to the TNFα proximal promoter specifically in response to LPS (Fig. 2A Right). A modest LPS-induced increase in cyclin T1 occupancy was also seen downstream within the TNFα gene. ChIP analysis revealed a similar LPS-dependent increase in occupancy by Cdk9, the catalytic subunit of P-TEFb, at the TNFα promoter-proximal region (Fig. S1B).
In stark contrast to TNFα, a gene encoding an LPS-inducible chemokine IP-10 (Fig. 2B Right) was devoid of detectible Pol II in resting BMMΦ; Pol II recruitment to the IP-10 promoter-proximal region and CTD phosphorylation on S5 and S2 were strictly LPS dependent (Fig. 2B Right and Fig. S1). Consistent with the data at TNFα (Fig. 2A) and previous studies (24), levels of S2 phosphorylation were greater on the fully elongation-competent Pol II within the IP-10 gene than for the Pol II beginning productive elongation near the IP-10 promoter. Thus, IP-10 transcription appears to be controlled at the level of LPS-induced Pol II recruitment and preinitiation complex assembly.
The TNFα Promoter Is Occupied by the Negative Elongation Factor (NELF) Complex in MΦ.
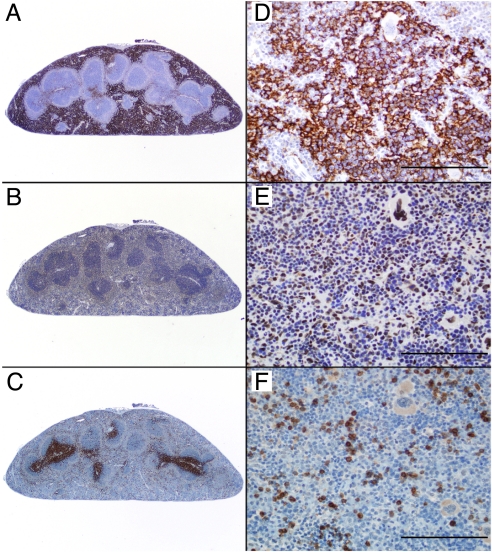
It has been previously shown that the 4-subunit NELF complex plays a key role in Pol II stalling by inhibiting early transcription elongation (25–27). The negative effects of NELF are counteracted by phosphorylation of the CTD, and potentially NELF itself, by the P-TEFb kinase, whose recruitment is concomitant with release of stalled Pol II from the promoter region. To evaluate a possible role for NELF in regulating TNFα transcription in vivo, we first confirmed that NELF was expressed in MΦ (Fig. S2). Furthermore, immunohistochemical analysis revealed abundant NELF-E staining (Fig. 3 B and E) in the nuclei of the resident MΦ occupying the majority of the red pulp of the mouse spleen (Fig. 3 A and D; F4/80 staining); NELF-E expression appeared to be particularly high in MΦ around and immediately adjacent to the white pulp of the follicles (Fig. 3B; compare to Fig. 3 C and F; CD3 staining for T cells).
Fig. 3.
NELF complex is present in mouse splenic MΦ. F4/80, NELF-E, and CD3 immunohistochemistry in a C57BL/6 mouse spleen is shown. Serial 5-μm sections are shown with 2× (Left) and 40× (Right) magnification. (Scale bar, 100 μm.) F4/80 positive cells consistent with MΦ represent the predominant cell population throughout the splenic red pulp (A and D). Note the strong and specific immunostaining for NELF-E within cell nuclei in these MΦ-rich areas (B and E). CD3-positive T cells are largely confined to the periarteriolar lymphoid sheath region of the white pulp, but small numbers of T cells are also scattered throughout the red pulp (C and F).
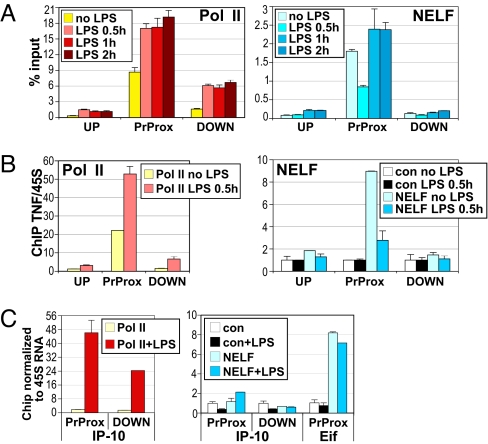
We next assessed NELF occupancy at several LPS target genes under basal and inducing conditions. Consistent with the idea that NELF inhibits early elongation, both NELF-E and NELF-A were present specifically in the promoter-proximal region of TNFα in resting RAW264.7 cells (Fig. 4A Right and Fig. S3A, respectively). Similarly, NELF-A associated with the TNFα promoter in untreated BMMΦ (Fig. 4B Right). Interestingly, in both cell contexts, NELF occupancy rapidly and transiently decreased upon addition of LPS, which inversely correlated with TNFα RNA induction. These data suggested that gene activation involves the temporary dissociation of NELF from the TNFα promoter, allowing Pol II to efficiently elongate into the gene. Strikingly, by 1–2 h of LPS treatment, NELF resumed occupancy at the TNFα promoter (Fig. 4A and Fig. S3).
Fig. 4.
The NELF complex dissociates from the TNFα promoter during the peak of gene activation. LPS-dependent dismissal of NELF from the TNFα promoter in RAW264.7 cells (A) and primary BMMΦ (B) is shown. Cells were derived and treated and ChIP assays performed as in Fig. 1. Pol II and NELF occupancy at the UP, PrProx, and DOWN regions of the TNFα gene is shown. (C) The IP-10 promoter is not occupied by the NELF complex in primary BMMΦ. Cells were treated with LPS for 1 h, as indicated, and Pol II and NELF-A occupancy at the PrProx and DOWN regions of IP-10 and the PrProx region of the Eif4a1 gene was assessed as in Fig. 1B.
In contrast, we observed no appreciable NELF-A occupancy above the background of control IgG at the IP-10 gene (promoter proximally or downstream) in either resting or LPS-treated BMMΦ whereas Pol II was, as expected, recruited in response to LPS (Fig. 4C). In the same experiment NELF occupied the promoter of Eif4a1, a control gene whose transcription is LPS unresponsive, and importantly this occupancy was not altered in LPS-stimulated BMMΦ (Fig. 4C).
Permanganate Footprinting in BMMΦ Reveals an Open Transcription Bubble in the TNFα Promoter-Proximal Region.
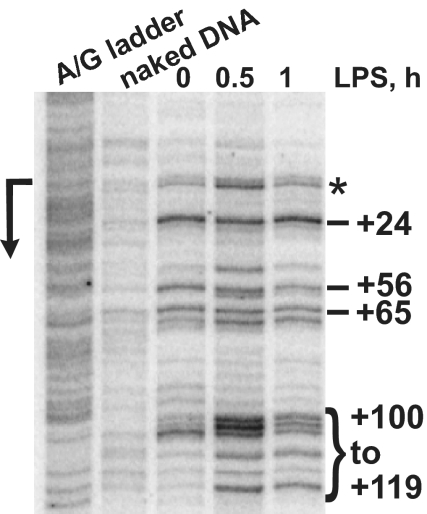
The above experiments suggested that Pol II recruited to the TNFα promoter in resting MΦ initiated transcription but was halted within the promoter-proximal region by NELF. To confirm the presence of an engaged but stalled Pol II at TNFα, we used in vivo permanganate footprinting, a technique that capitalizes on the ability of KMnO4 to specifically interact with thymine bases in ssDNA, such as those in the open transcription bubble. When Pol II stalls near the promoter for an extended period, ss regions associated with an engaged Pol II can be readily detected by their KMnO4 hyperreactivity. Fig. 5(lane “0”) illustrates strong permanganate reactivity in resting BMMΦ from near the TNFα TSS through the promoter-proximal region (dashes). This finding is consistent with the presence of an open transcription bubble at the uninduced TNFα gene generated by Pol II, which has produced a short transcript and stalled during early transcription elongation. LPS treatment both enhanced (particularly at 0.5 h) and extended KMnO4 reactivity further downstream into the gene (bracket), suggesting that Pol II has been released from the promoter to enter productive transcription elongation. This downstream expansion of the KMnO4-reactive region is reminiscent of the patterns seen at the induced heat-shock genes (28), as is the appearance of detectable reactivity at the TSS (asterisk), which indicates a very high level of transcription initiation upon gene activation.
Fig. 5.
In vivo permanganate probing of the TNFα gene in BMMΦ reveals a promoter-proximal open transcription bubble consistent with Pol II stalling. Day 7 BMMΦ were treated with LPS for the indicated times, harvested in ice-cold PBS, and subjected to permanganate footprinting (see Methods). Dashes indicate the unpaired thymine bases within the area from the TSS (arrow on the left) to ≈+100 in untreated cells (“0”). A bracket marks additional KMnO4 reactivity appearing from +100 to +119 in response to LPS. Note several reactive bands including that near the promoter (asterisk) that are specifically enhanced at 0.5 h of LPS treatment and decay thereafter. A + G sequencing ladder and KMnO4 probing of purified “naked” DNA are shown.
Pol II Stalling at the Genes in the TNF Family Is Highly Evolutionarily Conserved.
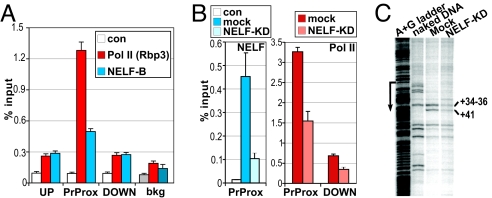
Interestingly, recent studies identified eiger, the sole Drosophila member of the TNF family, as a gene whose expression was attenuated in NELF-depleted cells (19, 22). Eiger, like TNFα, is an essential, stress-inducible regulator of cell death pathways (29, 30), critical for protection against extracellular pathogens. We therefore examined whether eiger is also regulated by Pol II stalling. Our ChIP analysis in Drosophila S2 cells revealed a striking resemblance of the Pol II and NELF-E binding profiles at eiger to those of Pol II and the NELF complex at the mammalian TNFα gene in MΦ. Indeed, both Pol II and NELF-E were strongly enriched at the eiger promoter-proximal region, but not in the upstream or downstream regions (Fig. 6A). Furthermore, RNAi-mediated depletion of NELF significantly decreased Pol II occupancy at eiger (Fig. 6B), consistent with the role of the NELF complex in maintaining stalled Pol II at this promoter.
Fig. 6.
Eiger, the Drosophila homolog of TNFα, is a target of NELF-dependent Pol II stalling. (A) Pol II and NELF occupy the promoter-proximal region of the uninduced eiger gene. ChIP was performed on Drosophila S2 cells with antibodies to the Rpb3 subunit of Pol II or NELF-B or with no antibody (con). Occupancy at the UP, PrProx, and DOWN regions of eiger (or an intergenic region to assess the background of the assay, bkg) was assessed by qPCR and expressed as percentage of input; n = 4, error is SEM. (B) Depletion of NELF reduces Pol II occupancy at the eiger promoter. S2 cells were mock treated or NELF-B depleted using RNAi (NELF KD). ChIP with no antibody (con) or antibodies to NELF-B (Left) or Pol II (Right) was performed as in A. (C) The permanganate footprint in the eiger promoter-proximal region is dependent on NELF. Lanes depict, left to right: the A + G ladder used to determine the position of the promoter (arrow), the KMnO4 reactivity of naked DNA as a control, and KMnO4 reactivity of eiger in S2 cells (mock-treated or NELF KD).
To obtain direct evidence for the transcriptional status of the Pol II bound near the eiger promoter, we performed permanganate footprinting of the gene. As shown in Fig. 6C, Pol II detected by ChIP is associated with a stalled, open transcription bubble in the eiger promoter-proximal region, as evidenced by KMnO4-sensitive sites at positions between +34 and +41 relative to the TSS. Further corroborating the idea that Pol II stalling at the eiger promoter is mediated by the NELF complex (Fig. 6B), NELF-B knockdown by RNAi decreases KMnO4 reactivity (Fig. 6C). Combined, these data demonstrate a remarkable level of evolutionary conservation of the transcriptional regulatory mechanisms governing TNF gene expression from Drosophila to mammals.
Pol II Stalling and the RNA Induction Profile in LPS-Inducible Genes in MΦ.
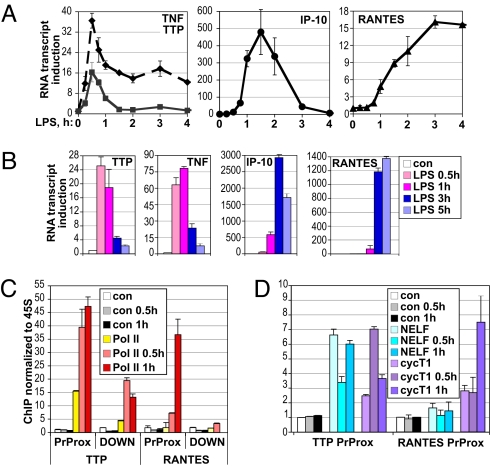
In mammals, MΦ represent an early line of defense against infection that respond to invading pathogens by producing a vast number of proinflammatory cytokines, chemokines, and factors that provide negative feedback loops limiting excessive inflammation. We quantified the expression of several such factors in LPS-treated MΦ over time. As shown in Fig. 7 A and B, TNFα nascent unprocessed transcript, as measured by the amplification of the intronic region, was induced with strikingly rapid kinetics, reaching a maximum in both RAW264.7 cells and BMMΦ by 0.5–1 h and dropping off precipitously thereafter. Interestingly, this down-regulation of TNFα transcription coincides with NELF reloading onto the gene (Fig. 4). Consistent with earlier observations (11), the TTP gene displayed a fast and transient induction profile similar to that of TNFα itself (Fig. 7 A and B). In contrast, IP-10 induction occurred much more gradually and was sustained for several hours; an even slower induction profile was characteristic of RANTES, another well-established LPS-inducible chemokine. Importantly, similarly “delayed” induction of IP-10 or RANTES was seen whether mRNA or nascent transcript was analyzed.
Fig. 7.
Pol II stalling correlates with rapid and transient RNA induction of LPS-inducible genes in MΦ. RAW264.7 cells (A) or BMMΦ (B) were treated for the indicated times with LPS, total RNA was isolated, and the expression of the indicated genes was assessed by real-time qPCR, normalized to β-actin and defined as fold induction over that in untreated cells (set as 1). Shown is a representative of 2–3 independent experiments performed in duplicate. (C and D) TTP but not RANTES promoter is occupied by stalled Pol II in MΦ. BMMΦ were treated with LPS for the indicated times and processed for ChIP as in Fig. 1B. Occupancy of Pol II at the PrProx and DOWN (C) and of NELF-A and cycT1 at the PrProx (D) regions of TTP and RANTES was assessed. Shown is 1 of 2 independent experiments done in duplicate.
To begin to address the potential mechanisms underlying such dramatic differences in the transcriptional response of these 2 pairs of genes to a common stimulus, we assessed Pol II and NELF occupancy at TTP and RANTES. Pol II was significantly enriched at the TTP promoter-proximal region in untreated BMMΦ (Fig. 7C); LPS treatment promoted additional Pol II loading onto the promoter and, particularly, the downstream region of the TTP gene. In addition, NELF complex occupied the TTP promoter-proximal region in resting MΦ, was dismissed upon 0.5 h LPS exposure, coinciding with cycT1 recruitment, but resumed promoter occupancy by 1 h, as cycT1 occupancy began to decline (Fig. 7D). These data were consistent with promoter-proximal Pol II stalling and LPS-dependent release and elongation at TTP, similar to that occurring at TNFα. In contrast, RANTES was not occupied by Pol II or NELF in untreated BMMΦ (Fig. 7 C and D); Pol II loading onto the RANTES promoter was LPS dependent, indicating that, as with IP-10, Pol II recruitment is a rate-limiting step in RANTES transcription. Thus, in a limited subset of genes tested, Pol II stalling correlated with a rapid and transient expression profile.
Pol II Stalling and Not the Nature of an Inducer Determines the Induction Profile of LPS Targets.
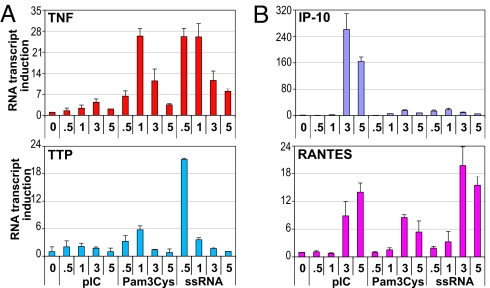
Most cytokines and chemokines are induced by multiple PAMPs, which signal through specific TLRs. For example, bacterial lipopeptide Pam3Cys activates TLR2, dsRNA [and its synthetic analog poly(IC)] activates TLR3, and ssRNA signals through TLR7 (2). Signal transduction cascades initiated by these TLRs are partially overlapping yet distinct, creating a sophisticated regulatory matrix whereby, in principle, each PAMP may trigger a different transcriptional response of a given gene. We therefore examined the induction profile of TNFα, TTP, IP-10, and RANTES in BMMΦ treated with poly(IC), Pam3Cys, and ssRNA. We found that TNFα was induced by Pam3Cys and ssRNA and that TTP was highly ssRNA responsive, but, in all cases, their expression peaked by 0.5–1 h and subsided thereafter (Fig. 8A). In contrast, activation of IP-10 by poly(IC) or RANTES by all 3 PAMPs did not peak until 3–5 h irrespective of the specific TLR engaged (Fig. 8B). We conclude that for a given gene, the kinetics of induction are specific to the gene itself regardless of a signaling pathway responsible for activation and reflect a particular rate-limiting step in transcription (Pol II recruitment vs. release from promoter-proximal stalling).
Fig. 8.
The induction profile of TLR-responsive genes in BMMΦ reflects the rate-limiting step in transcription rather than the nature of an inducer. Day 7 BMMΦ were treated for the indicated times with 5 μg/mL poly(IC), 100 ng/mL Pam3Cys, and 10 ng/mL ssRNA, and total RNA was isolated and reverse transcribed. The expression of (A) TNFα and TTP and (B) IP-10 and RANTES was assessed as in Fig. 7B.
Discussion
A key component of the innate immune system, myeloid cells including monocytes, macrophages, and dentritic cells respond to pathogens by mounting a surge of proinflammatory mediators. Among the numerous factors that contribute to the local or systemic elevation of cytokine and chemokines levels, gene expression has been established as a fundamental mechanism that dictates their balance. Indeed, cytokine-encoding genes are rich in cis-acting elements for numerous transcriptional regulators. The combinatorial dynamics of many factors at a given gene are exemplified by the IFNβ enhanceosome (31), where transcription initiation requires the ordered assembly of the IRF, ATF/AP1, and NFκB family members, followed by histone acetylation, nucleosome remodeling, and Pol II recruitment. Consequently, Pol II recruitment to a promoter has been traditionally considered the critical step in getting transcription underway. Yet, recent studies have begun to paint a very different picture of transcriptional regulation, whereby a sizable fraction of promoters are constitutively occupied by Pol II, and it is the polymerase release into productive elongation that is rate limiting.
The notion that the rate-limiting step in transcription does not need to be Pol II recruitment is not in itself novel: Years ago, elegant studies from J. Lis's group demonstrated that the Drosophila heat-shock genes were regulated during early elongation (32). A surprise, however, came from the recent genomewide studies in Drosophila revealing that up to 20% of genes may be controlled in a similar manner (19, 20). Interestingly, many genes that harbor stalled Pol II encode inducible factors that respond to environmental or developmental triggers (19). If this trend holds true in mammals, genes with promoter-proximally stalled Pol II may be enriched in the innate immune system, which specifically evolved to sense and respond efficiently to pathogens.
Here, we demonstrate that TNFα, a broad spectrum inducer of inflammatory responses and a key molecule in the pathogenesis of many autoimmune and inflammatory diseases, is encoded by a stalled gene. Indeed, in resting cells, the TNFα promoter was co-occupied by NELF and S5- (but not S2-) phosphorylated Pol II that was engaged in early transcription elongation. LPS treatment triggered P-TEFb loading, a dramatic increase in S2 phosphorylation, a rapid yet transient dismissal of NELF from the promoter, and a release of Pol II into the gene.
Pol II stalling has been described for several mammalian immediate early genes including JunB, cFos, cMyc, and a number of estrogen receptor-regulated genes (33–35). However, the regulated dissociation of NELF from the promoter region during gene induction, as shown for Drosophila heat-shock genes (36), has never before been reported in a mammalian system. Furthermore, the fact that Drosophila eiger is a NELF target gene, similarly controlled at the level of promoter proximal stalling, illustrates that the rate-limiting step in the transcription cycle for a given gene could be an ancient evolutionarily conserved mechanism of regulation.
On the basis of these data, we propose the following model for the role of Pol II stalling in the immune response: Before induction, Pol II and the transcription machinery are preloaded onto the promoters of many genes, but are held within the promoter-proximal region by the NELF complex. This stalled elongation complex establishes a “poised” promoter structure, which may include (but is likely not limited to) the active histone marks recently found at such genes before induction (37). Upon immune challenge, P-TEFb recruitment triggers the rapid transition of stalled Pol II to productive elongation and NELF release. During the early phase of induction, NELF binding and Pol II pausing within the promoter-proximal region would be antagonized by the continued presence of high levels of P-TEFb. Over time, P-TEFb levels decline, allowing NELF to reassociate with Pol II and gene transcription to subside. Notably, the decline of the TNFα and TTP transcript levels is a compound of both a decrease in transcription levels and, as previously shown (14), RNA turnover. Future studies, including genetic disruption of mammalian NELF in the immune system, will address the functional role of this complex in transcriptional regulation of the stalled genes.
The global role of Pol II stalling in the innate immune response is currently unknown. Conceivably, genes occupied by the stalled Pol II encode acutely inducible factors in the front line of host defenses, while those regulated via Pol II recruitment represent chemokines and mediators of tissue repair that are required later in the infectious process. It is tempting to speculate that the intrinsic danger imposed by potent proinflammatory cytokines such as TNFα also necessitates multiple mechanisms for a rapid shutoff, including the return of NELF to the promoter and arrest of elongation, prompt degradation of existing mRNA, and inactivation of the signaling pathway via negative feedback. Genomewide ChIP-chip or ChIP-seq approaches should reveal whether Pol II stalling indeed occurs preferentially in a specific functional group of immune mediators.
TNFα is a prototypic inflammatory cytokine whose critical role in the pathogenesis of several autoimmune diseases made it a prime therapeutic target in rheumatoid arthritis, psoriasis, inflammatory bowel disease, ankylosing spondylitis, and other disorders (38). Given the extraordinary importance of this cytokine in human disease, much work has focused on dissecting the pathways responsible for its production. This study uncovered a fundamental, evolutionarily conserved mechanism that controls TNFα transcription in MΦ, which generate a bulk of TNFα during inflammatory processes. While many questions regarding the molecular determinants of Pol II stalling remain, this early elongation checkpoint may be subverted in disease, as suggested by recent data linking NELF dysregulation to enhanced cellular proliferation in breast cancer and gastrointestinal adenocarcinomas (39, 40). Conceivably, altered NELF levels in MΦ may lead to an exaggerated inflammatory response or autoimmunity. A possible correlation of an aberrant Pol II stalling profile with quantitatively or qualitatively different transcriptional response of TNFα would be of great interest to pursue in mouse models of systemic inflammation or autoimmune conditions.
Methods
Cell Culture and BMMΦ Preparation.
Mouse RAW264.7 cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS (HyClone), treated with sterile PBS (vehicle) or LPS (1 μg/mL) as described in the figure legends, and harvested. BMMΦ were prepared from 8-wk-old C57BL/6 mice as described in ref. 41 except L-cell conditioned media were used for the 6-day MΦ expansion; cells were then scraped, incubated in RPMI-20% FBS overnight, and treated as described in the figure legends.
Measurement of RNA Levels.
Total RNA was isolated from RAW264.7 cells and BMMΦ and analyzed (41) for the expression of each gene, using β-actin as an internal control. Primer pairs are listed in Table S1.
Permanganate Footprinting.
In vivo permanganate footprinting in Drosophila was performed as in ref. 19. Day 7 BMMΦ (≈100 × 106) were harvested in ice-cold PBS (400 × g; 5 min), washed in PBS, collected as above, and subjected to KMnO4 probing as in ref. 19 using 1 μg DNA per reaction and 25 cycles of ligation-mediated PCR (see primer sequences in Table S1).
ChIP Experiments and Immunohistochemistry.
See SI Methods.
Supplementary Material
Acknowledgments.
We thank the members of the immunohistochemistry core at the National Institute of Environmental Health Sciences and Drs. Lionel Ivashkiv and Perry Blackshear for critical comments on the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences Grant Z01 ES101987 (to K.A.); by the National Institutes of Health Grant T32 AR07517 (to Y.C.); and by grants (to I.R.) from the National Institutes of Health National Institute of Allergy and Infectious Diseases (R01 AI068820), the Lupus Research Institute, and the Mary Kirkland Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910177106/DCSupplemental.
References
- 1.Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17:338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 3.Foster SL, Medzhitov R. Gene-specific control of the TLR-induced inflammatory response. Clin Immunol. 2009;130:7–15. doi: 10.1016/j.clim.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett L, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu CQ, Field M, Feldmann M, Maini RN. Localization of tumor necrosis factor alpha in synovial tissues and at the cartilage-pannus junction in patients with rheumatoid arthritis. Arthritis Rheum. 1991;34:1125–1132. doi: 10.1002/art.1780340908. [DOI] [PubMed] [Google Scholar]
- 6.Rothe J, et al. Mice lacking the tumour necrosis factor receptor 1 are resistant to TNF-mediated toxicity but highly susceptible to infection by Listeria monocytogenes. Nature. 1993;364:798–802. doi: 10.1038/364798a0. [DOI] [PubMed] [Google Scholar]
- 7.Pfeffer K, et al. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell. 1993;73:457–467. doi: 10.1016/0092-8674(93)90134-c. [DOI] [PubMed] [Google Scholar]
- 8.Hu X, Chakravarty SD, Ivashkiv LB. Regulation of interferon and Toll-like receptor signaling during macrophage activation by opposing feedforward and feedback inhibition mechanisms. Immunol Rev. 2008;226:41–56. doi: 10.1111/j.1600-065X.2008.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horiuchi K, et al. Cutting edge: TNF-alpha-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J Immunol. 2007;179:2686–2689. doi: 10.4049/jimmunol.179.5.2686. [DOI] [PubMed] [Google Scholar]
- 10.Dayer JM. The pivotal role of interleukin-1 in the clinical manifestations of rheumatoid arthritis. Rheumatology. 2003;42(Suppl 2):ii3–ii10. doi: 10.1093/rheumatology/keg326. [DOI] [PubMed] [Google Scholar]
- 11.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 12.Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 13.Baseggio L, Charlot C, Bienvenu J, Felman P, Salles G. Tumor necrosis factor-alpha mRNA stability in human peripheral blood cells after lipopolysaccharide stimulation. Eur Cytokine Netw. 2002;13:92–98. [PubMed] [Google Scholar]
- 14.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Negative regulation of toll-like receptor signaling by NF-kappaB p50 ubiquitination blockade. Science. 2007;317:675–678. doi: 10.1126/science.1142953. [DOI] [PubMed] [Google Scholar]
- 16.Chan C, Li L, McCall CE, Yoza BK. Endotoxin tolerance disrupts chromatin remodeling and NF-kappaB transactivation at the IL-1beta promoter. J Immunol. 2005;175:461–468. doi: 10.4049/jimmunol.175.1.461. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa S, et al. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci USA. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramirez-Carrozzi VR, et al. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeitlinger J, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilchrist DA, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 24.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Yamaguchi Y, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 26.Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH. A highly purified RNA polymerase II elongation control system. J Biol Chem. 2001;276:42601–42609. doi: 10.1074/jbc.M104967200. [DOI] [PubMed] [Google Scholar]
- 27.Cheng B, Price DH. Properties of RNA polymerase II elongation complexes before and after the P-TEFb-mediated transition into productive elongation. J Biol Chem. 2007;282:21901–21912. doi: 10.1074/jbc.M702936200. [DOI] [PubMed] [Google Scholar]
- 28.Giardina C, Perez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- 29.Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol. 2002;12:1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- 30.Igaki T, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002;21:3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 33.Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 34.Aida M, et al. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol. 2009;29:1123–1133. doi: 10.1128/MCB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu CH, et al. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 2005;33:1269–1279. doi: 10.1093/nar/gki274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feldmann M, Maini RN. Lasker Clinical Medical Research Award. TNF defined as a therapeutic target for rheumatoid arthritis and other autoimmune diseases. Nat Med. 2003;9:1245–1250. doi: 10.1038/nm939. [DOI] [PubMed] [Google Scholar]
- 39.Aiyar SE, et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McChesney PA, et al. Cofactor of BRCA1: A novel transcription factor regulator in upper gastrointestinal adenocarcinomas. Cancer Res. 2006;66:1346–1353. doi: 10.1158/0008-5472.CAN-05-3593. [DOI] [PubMed] [Google Scholar]
- 41.Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. EMBO J. 2006;25:108–117. doi: 10.1038/sj.emboj.7600919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogatsky I, Zarember KA, Yamamoto KR. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 2001;20:6071–6083. doi: 10.1093/emboj/20.21.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.