SUMMARY
Differences in expression, protein interactions and DNA binding of paralogous transcription factors (“TF parameters”) are thought to be important determinants of regulatory and biological specificity. However, both the extent of TF divergence and the relative contribution of individual TF parameters remain undetermined. We comprehensively identify dimerization partners, spatiotemporal expression patterns and DNA binding specificities for the C. elegans bHLH family of TFs, and model these data into an integrated network. This network displays both specificity and promiscuity, as some bHLH proteins, DNA sequences, and tissues are highly connected, whereas others are not. By comparing all bHLH TFs, we find extensive divergence, and that all three parameters contribute equally to bHLH divergence. Our approach provides a framework for examining divergence for other protein families in C. elegans and in other complex multicellular organisms, including humans. Cross-species comparisons of integrated networks may provide further insights into molecular features underlying protein family evolution.
INTRODUCTION
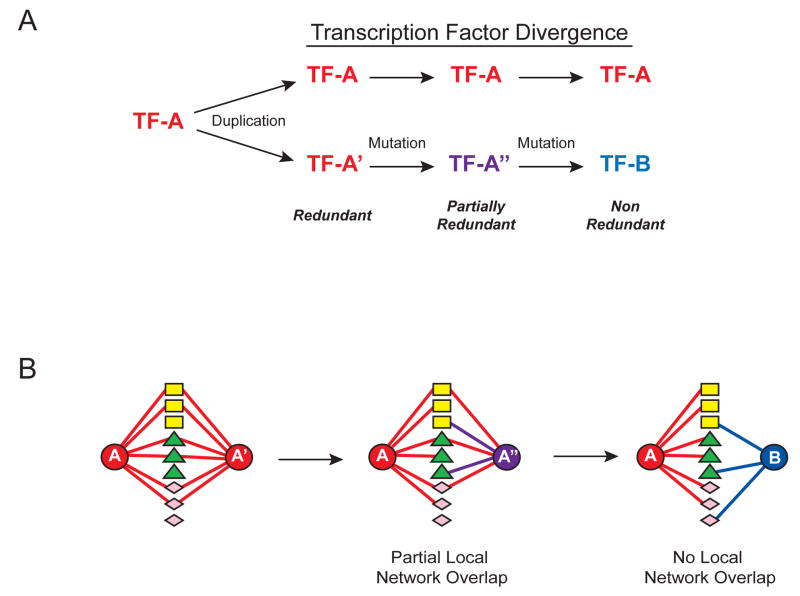
Transcription regulatory networks capture physical and regulatory relationships between sequence-specific transcription factors (TFs), and between TFs and their target genes (Walhout, 2006). Paralogous TFs are grouped into families based on the type of DNA binding domain they possess. Such families grow by gene duplications upon which identical and therefore fully redundant TFs emerge. After acquiring mutations, duplicate TFs diverge and may become partially redundant. Upon further mutation completely non-redundant, yet paralogous TFs may emerge (Figure 1A).
Figure 1. Functional and Molecular Divergence in Paralogous TF Families.
(A) Paralogous TFs arise by gene duplication and mutation.
(B) TF divergence can be achieved by the accumulation of molecular and functional differences. Differently shaped nodes (rectangles, triangles and diamonds) between TFs (circles) represent different TF parameters (e.g. dimerization partners, spatiotemporal expression and DNA binding specificities).
Paralogous TF families often expand with organismal complexity. For instance, whereas the nematode Caenorhabditis elegans has 42 basic helix-loop-helix (bHLH) proteins (Reece-Hoyes et al., 2005), the human genome encodes more than 100 (Simionato et al., 2007). The expansion and divergence of TFs has been proposed to lead to increased regulatory complexity, biological specificity and organismal complexity.
Paralogous TFs often have different biological functions. For example, loss of C. elegans bHLH TFs results in phenotypes ranging from neuronal defects to embryonic lethality (see e.g. Chen et al., 1994; Hallam et al., 2000; Portman and Emmons, 2000). In humans, mutations in paralogous TFs can result in different diseases. For instance, mutations in TWIST and HAND1, both bHLH TFs, can result in Saethre-Chotzen syndrome and heart hypoplasia, respectively (Howard et al., 1997; Reamon-Buettner et al., 2008).
TFs engage in numerous molecular interactions; they bind DNA and often dimerize with each other (Grove and Walhout, 2008). In addition, they exhibit specific spatiotemporal expression patterns (Reece-Hoyes et al., 2007). Together we refer to such interactions and expression patterns as “TF parameters”. A main challenge in regulatory and genome biology is to understand the mechanisms of TF divergence and to disentangle the contribution of each of the parameters to this process. Specific questions are to what extent members of a TF family differ in each of these parameters, and if differences in any one parameter are more prevalent than differences in another (Figure 1B).
Assessment of metazoan TF divergence requires the comprehensive and standardized measurement of multiple TF parameters and the incorporation of these parameters into a single, integrated network. Initial studies in the yeast revealed a large degree of redundancy for the eight Yap TFs, as well as functional divergence through DNA binding specificities and interactions with chromatin proteins (Fernandes et al., 1997; Tan et al., 2008). However, the mechanisms of divergence in large metazoan TF families remain unexplored (Figure 1B). Numerous metazoan TFs have been studied individually, but the resulting data are sparse due to assay incompleteness and heterogeneity. Therefore, such data could not be used to determine the extent and mechanisms of divergence of complete TF families.
Here, we comprehensively determined the dimerization, spatiotemporal expression and DNA binding specificities for nearly all members of the C. elegans bHLH family, and modeled these data into an integrated network (Supplemental Figure S1). We systematically compared all the nodes in this network and asked whether they have a high connectivity, i.e. are “promiscuous”, or if they display low connectivity, i.e. are “specific”. Together, these analyses reveal the overall extent of divergence within the C. elegans bHLH family, as well as the relative contribution of each parameter to TF divergence.
RESULTS
A C. elegans bHLH Dimerization Network
We first grouped the C. elegans bHLH proteins according to the classes outlined previously, and supplemented by our own data described below (Massari and Murre, 2000)(Supplemental Figure S2).
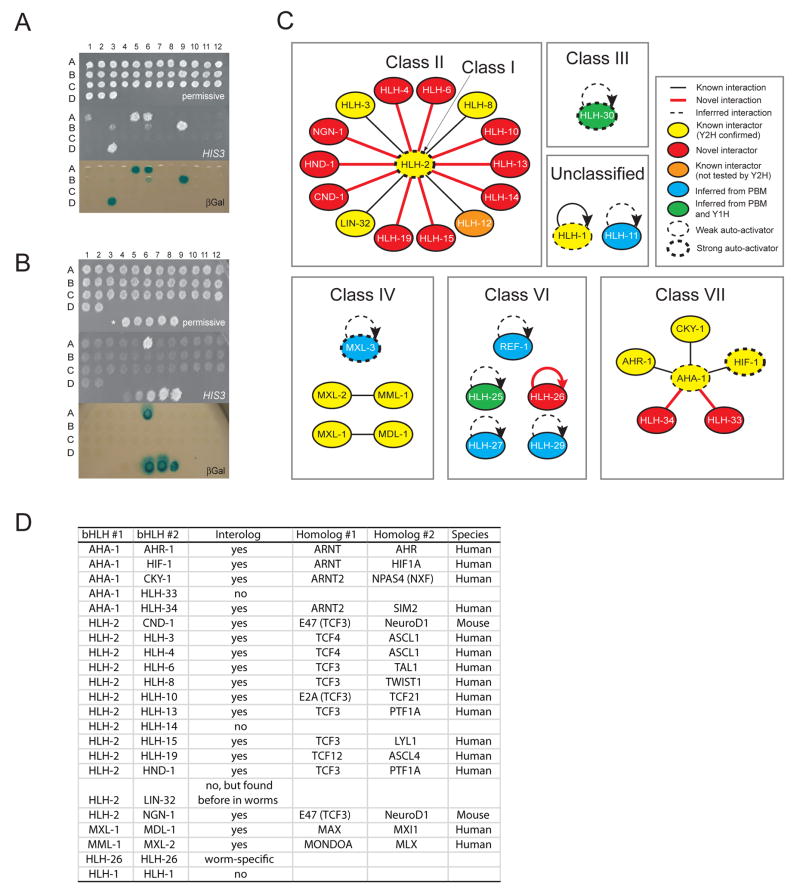
Previous studies in C. elegans have identified ten bHLH homo- and heterodimers involving 14 TFs (Harfe et al., 1998; Jiang et al., 2001; Krause et al., 1997; Ooe et al., 2007; Pickett et al., 2007; Portman and Emmons, 2000; Powell-Coffman et al., 1998; Tamai and Nishiwaki, 2007; Yuan et al., 1998). However, the dimerization partners of the majority of C. elegans bHLH TFs remained unidentified. We performed pair-wise yeast two-hybrid (Y2H) assays to identify bHLH-bHLH dimers (Walhout et al., 2000). In total, we examined 765 bHLH-bHLH combinations involving 39 bHLH proteins (Supplemental Table S1). Five bHLH proteins exhibited medium to strong levels of auto-activation (HLH-2, HLH-30, SBP-1, MXL-3 and HIF-1) and could only be tested as preys (Figure 2A). In total, we detected 22 dimers (2 homodimers and 20 heterodimers) involving 26 bHLH proteins (Figure 2B, Supplemental Figure S3). The complete dimerization network is shown in Figure 2C. We supplemented this network with homodimeric interactions for HLH-25, HLH-27, HLH-29, REF-1, HLH-11, MXL-3, and HLH-30, because we detected their specific DNA binding in protein binding microarray (PBM) and/or yeast one-hybrid assays (Deplancke et al., 2006)(see below). Together, the resulting bHLH network contains 9 homodimers and 21 heterodimers involving 34 proteins.
Figure 2. The C. elegans bHLH Dimerization Network.
(A) Auto-activation of DB-bHLH Y2H baits. Top - DB-bHLH strains were plated in spots on permissive media; middle - activation of the HIS3 reporter gene ; bottom - activation of the lacZ reporter gene (βGal). Auto-activators are: A1 - DB-AHA-1; A5 - DB-HLH-30; A6 - DB-HLH-2; B3 - DB-HLH-1; B6 - DB-MXL-3; B9 - DB-SBP-1; D3 - DB-HIF-1.
(B) Example of Y2H matrix assay using DB-HLH-15 as bait. Top – permissive media; middle - activation of the HIS3 reporter gene; bottom - activation of the lacZ reporter gene (βGal). Bottom spots in each panel - Y2H controls (Walhout and Vidal, 2001).
(C) The bHLH dimerization network. Y1H – yeast one-hybrid.
(D) Several bHLH dimers identified are evolutionarily conserved interologs.
The majority of bHLH proteins exhibit highly specific dimerization as they interact with only a single other bHLH protein (Figure 2C). However, there are two bHLH proteins that interact with multiple other bHLH proteins. The first is AHA-1, the C. elegans Arnt ortholog that dimerizes with all known class VII members. Members of this class contain a PAS domain that mediates protein-protein interactions (Crews, 1998). The second is HLH-2, which binds to 14 other bHLH proteins, many orthologs of which are known to interact with the HLH-2 ortholog in other organisms (interologs, Figure 2D). Taken together, the dimerization network displays both specificity and promiscuous as most bHLH proteins interact with one, but some interact with many other bHLH proteins.
Spatiotemporal Activity of bHLH Promoters
To analyze the spatiotemporal expression pattern of bHLH genes, we generated transgenic animals that express the green fluorescent protein (GFP) under the control of bHLH gene promoters. The earliest GFP expression that we observed was at the ~24-cell stage with Pcnd-1 and Pngn-1. For five bHLH promoters we did not detect any GFP expression (Supplemental Tables S2–S4). This may be because they are missing distal activating elements, because they are incorrectly annotated, or because they are active under conditions that we did not examine (e.g., in dauers or males).
We found that some hlh promoters are active broadly, whereas others drive GFP expression in a more restricted fashion (Supplemental Table S4). The promoters corresponding to both bHLH proteins that dimerize with multiple partners, AHA-1 and HLH-2, confer broad GFP expression, whereas their partners are generally expressed in a more restricted manner. Conversely, some tissues express few bHLH TFs, whereas other tissues express many. For instance, numerous hlh promoters drive expression in the vulva, but only the ref-1 promoter is active in the pharyngeal-intestinal valve.
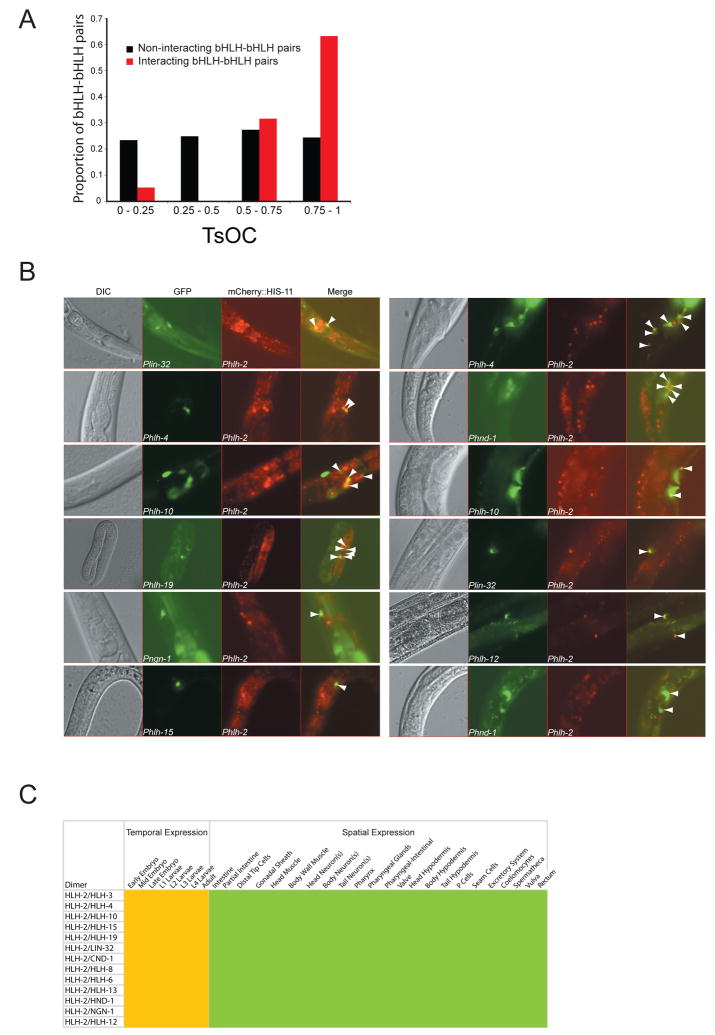
If spatiotemporal expression plays an important role in functional TF divergence, one could expect that proteins that dimerize exhibit greater co-expression than proteins that do not dimerize. To test this, we annotated the spatiotemporal expression of the bHLH gene promoters using a controlled vocabulary and calculated the tissue overlap coefficient (TsOC)(Martinez et al., 2008) between all bHLH-bHLH pairs. As expected, dimerization partners are more likely to be co-expressed than bHLH proteins that do not dimerize with each other (Figure 3A, Fisher’s exact test p < 0.001).
Figure 3. Post-Embryonic Co-Expression of HLH-2 and Its Partners.
(A) Tissue overlap coefficient (TsOC) analysis was done as described (Martinez et al., 2008). , where HLH-X is the number of tissues where HLH-X is expressed, and HLH-Y is the number of tissues where HLH-Y is expressed. HLH-N is the smallest total number of tissues for either HLH-X or HLH-Y.
(B) Phlh-2::mCherry::his-11 transgenic animals were crossed with each of the Phlh-x::GFP animals to determine co-expression (indicated by white arrowheads).
(C) Co-expression matrix of HLH-2 and its partners using a controlled vocabulary. Yellow indicates temporal expression; green depicts spatial expression.
Together, our observations identify specificity and promiscuity in the spatiotemporal expression network, both from the bHLH and from the tissue standpoint (visualized in integrated network below).
Co-Expression Analysis of HLH-2 Heterodimers
We used a dual-reporter approach to determine where and when HLH-2 and each of its partners are co-expressed because these involve most of the heterodimers we identified. We created a transgenic C. elegans strain that carries a Phlh-2::mCherry::his-11 construct that drives expression of a red fluorescent protein (mCherry) in the nucleus of cells where Phlh-2 is active. Phlh-2 exhibits broad activity in the embryo and its activity becomes more restricted in larvae and adults, consistent with previous HLH-2 immunofluorescence data (Figure 3B, Supplemental Figure S4 and Supplemental Table S4)(Krause et al., 1997).
We crossed the Phlh-2::mCherry::his-11 transgenic animals with relevant Phlh::GFP lines (corresponding to HLH-2 partners), resulting in double transgenic animals. When the two hlh promoters are active in the same cell, these cells appear with a green cytoplasm and yellow nucleus in a merged fluorescence image (Figures 3B, 3C, Supplemental Figure S4).
HLH-2 and most of its partners are first expressed at the comma stage of embryogenesis (Supplemental Figure S4), which is associated with the onset of cellular differentiation. This is in agreement with observations that orthologs of HLH-2 partners are important regulators of cell lineage commitment and differentiation (Massari and Murre, 2000). However, there is some temporal specificity as some HLH-2 dimers are expressed only during embryogenesis and in the first larval stage (e.g. HLH-2/HLH-3) whereas others are expressed throughout the lifetime of the animal (e.g. HLH-2/HLH-8). As has been observed for other organisms, we found that the HLH-2 partners exhibit a more tissue-restricted expression pattern as compared to HLH-2 (Massari and Murre, 2000)(Supplemental Table S4). Post-hatching, most HLH-2 heterodimers are expressed only in a subset of tissues, including neurons, the vulva, some hypodermal cells and distal tip cells (Figure 3C).
In summary, we observed broader, or “tissue-promiscuous”, activity for several bHLH promoters, including those that correspond to the bHLH proteins that interact with multiple partners, and we observed more restricted, or “tissue-restricted”, activity for others. Conversely, we observed that some tissues express many, whereas others express few, bHLH genes.
DNA Binding Specificity Analysis of Homo- and Heterodimeric TFs
bHLH TFs bind DNA as obligatory homo- or heterodimers and are classically described as recognizing E-box sequences (CANNTG)(Massari and Murre, 2000). Previously, a handful of DNA sequences that can be bound by seven of the known C. elegans bHLH dimers had been identified (Harfe et al., 1998; Krause et al., 1997; Ooe et al., 2007; Portman and Emmons, 2000; Powell-Coffman et al., 1998; Yuan et al., 1998). However, in those studies only one or a few of all possible E-boxes were considered, and no experiments were done to determine the comprehensive DNA binding preferences of all C. elegans bHLH dimers.
We used PBMs assays to comprehensively identify the sequence preferences of the bHLH dimers (Berger et al., 2006, Berger et al., 2008; Zhu et al., 2009). We first tested each available bHLH TF individually in PBM assays, as a GST fusion protein and obtained DNA binding profiles for MXL-3, HLH-1, HLH-11 HLH-25, HLH-26, HLH-27, HLH-29, HLH-30 and REF-1, demonstrating that these proteins can bind DNA without protein partners, presumably as homodimers. Proteins that yielded sequence-specific DNA binding profiles in PBM assays but that were not detected as interacting with any bHLH protein by Y2H assays (e.g. HLH-25) may dimerize in a DNA-dependent manner (Peirano and Wegner, 2000).
Importantly, none of the bHLH proteins that participate in heterodimeric interactions exhibited significant sequence-specific DNA binding on their own (Figure 4A, Supplemental Figures S5 and S6). This presented a convenient strategy to determine the DNA binding profiles of heterodimeric TFs, by incubating the DNA microarrays simultaneously with a GST-fusion bHLH protein that did not bind to DNA on its own, and a FLAG-tagged partner protein with subsequent detection using a fluorophore-conjugated anti-GST antibody. We examined each of the bHLH heterodimers identified by our Y2H screen in this manner.
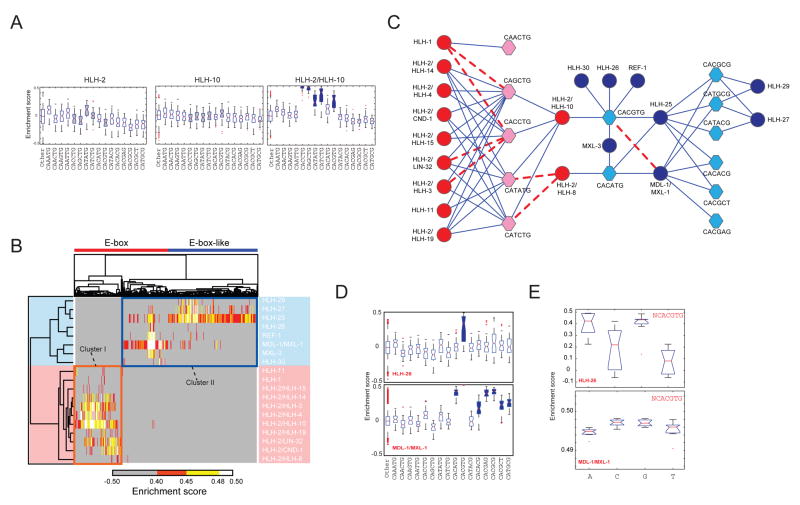
Figure 4. PBM Analysis of C. elegans bHLH Dimers.
(A) Box plots of enrichment score (ES) distribution of HLH-2, HLH-10 and HLH-2/HLH-10 binding to E-boxes and E-box-related sequences. E-boxes bound preferentially (AUC ≥ 0.85, Q < 0.001) by HLH-2/HLH-10 are indicated in blue (right panel). The corresponding E-boxes are colored gray in the single protein box plots for comparison (left and middle panel). In each box plot, the central bar indicates the median, the edges of the box indicate the 25th and 75th percentiles, the whiskers extend to the most extreme data points not considered outliers, and individual points that are plotted correspond to outliers.
(B) Clustergram of all bHLH dimers that yielded DNA binding profiles at a PBM ES ≥ 0.40. Orange box – cluster I; blue box – cluster II.
(C) bHLH DNA binding network. bHLH dimers are indicated in circles, E- and E-box-like sequences are indicated in hexagons. Red – cluster I; blue – cluster II. Blue lines –novel interactions; dashed red lines – previously reported interactions.
(D) Box plots of ES distribution of HLH-26 and MDL-1/MXL-1 binding to E-boxes and E-box-like sequences. Note: the box plot for CACGTG bound by the MDL-1/MXL-1 heterodimer is barely visible because of its narrow range and high ES.
(E) Box plots of ES distribution of nucleotides flanking CACGTG when bound by HLH-26 or MDL-1/MXL-1.
We obtained DNA binding profiles for 9 homodimers and 10 heterodimers, including most heterodimers involving HLH-2, two class IV dimers, and five out of six REF-1 family proteins (Class VI) (Supplemental Figure S7 and S8, see below). We did not detect any sequence-specific DNA binding by the bHLH-PAS class of dimers, even though these readily form heterodimers in the Y2H system. It is possible that sequence-specific DNA binding by members of this class requires ligands or post-translational modifications (Crews, 1998).
Two Clusters of DNA-Binding Specificities in the C. elegans bHLH Family
The PBM-derived 8-mer data span the full affinity range of DNA binding preferences (Berger et al., 2006). We calculated enrichment scores (ESs) from the PBM signal intensities for all possible 8-mers, and for each bHLH dimer that yielded sequence-specific DNA binding, and derived position weight matrices (PWMs) for each dimer (Supplemental Table S5, Supplemental Figure S8). We imposed a conservative threshold (ES ≥ 0.40) to identify significantly bound 8-mers. We then hierarchically clustered both the dimers and the 8-mers and found that the bHLH proteins can be grouped into two clusters corresponding to different bHLH classes: Cluster I contains HLH-2 and its partners, HLH-1 and HLH-11, and cluster II contains class III, IV, and VI bHLH proteins (Figure 4B).
As expected, HLH-2-containing dimers (cluster I) exhibit a strong preference for E-box sequences (CANNTG) (Massari and Murre, 2000). Surprisingly, however, cluster II dimers, in addition to binding a few E-boxes, also bind multiple non-E-box sequences. These resemble E-boxes, but contain a C or A in the fifth position and a G or T in the sixth position of the binding site (CAYRMK). These “E-box-like sequences” include the reported CACGCG binding site of Drosophila Hairy, and N-boxes (CACNAG), which are bound by Drosophila Enhancer of Split (Davis and Turner, 2001).
We determined the statistical significance of the preference of each bHLH dimer for E-box and E-box-like sequences as compared to all other 8-mers (Supplemental Figure S7). As shown in Figure 4A, neither HLH-2 nor HLH-10 alone can bind significantly to any E-box or E-box-like sequence. However, when combined, they can bind five different sequences. Figure 4C shows that the bHLH DNA binding network also displays degrees of specificity and promiscuity. For instance, only HLH-1 homodimers can bind CAA-containing E-boxes (Supplemental Figures S7). Some E-boxes and E-box-like sequences are preferred by relatively few dimers, whereas others are bound by many dimers. For example, CACATG is bound by only four dimers, but CACCTG is bound by ten distinct dimers. Conversely, some bHLH dimers bind few E-boxes or E-box-like sequences whereas others bind many: HLH-30 only binds CACGTG, but HLH-2/HLH-10 binds five different E-boxes (Figure 4C). This demonstrates that there is specificity and promiscuity in the bHLH DNA binding network, both from the view of the proteins and at the level of their DNA binding sequences.
Flanking Nucleotides Contribute to bHLH DNA Binding Specificity
The PBM ES of a particular DNA sequence bound by a dimer is a reflection of relative DNA binding affinities (Berger et al., 2006). We noticed that the ES distribution for 8-mers corresponding to a particular dimer/sequence combination varied greatly. For instance, both HLH-26 and MDL-1/MXL-1 bind CACGTG E-boxes, but HLH-26 does so with a broad ES range and MDL-1/MXL-1 with a very narrow ES range (Figure 4D). This suggests that, in contrast to MDL-1/MXL-1, not all CACGTG E-boxes are bound equally well by HLH-26. We considered the possibility that differences may be due to effects of nucleotides flanking the core CACGTG E-box. Indeed, flanking nucleotides have been reported previously to contribute to bHLH dimer DNA binding (Fisher and Goding, 1992; Walhout et al., 1998). However, the effects of nucleotides flanking the E-box and E-box-like sequences had not been analyzed systematically for most bHLH TFs. Since each bHLH monomer may directly contact the flanking nucleotide immediately 5′ of the E-box (Ellenberger et al., 1994; Fisher and Goding, 1992), we examined the influence of this position on relative DNA binding preferences. We found that for the MDL-1/MXL-1 dimer each of the four possible nucleotides flanking the CACGTG core sequence is recognized approximately equally well; the ES for each relevant 8-mer is between 0.49 and 0.50 (Figure 4E). However, HLH-26 exhibits a strong preference for a 5′ (median 8-mer ES > 0.40), and disfavors a 5′ T (median 8-mer ES < 0.10) and, to a lesser extent, a 5′ C (0 ≤ ES ≤ 0.40)(Figure 4E).
We found that most bHLH proteins exhibit preferences at the 5′ flanking nucleotide position (Supplemental Figure S9). We found that most dimers disfavor a 5′ T; this observation is similar to what has been reported for the yeast bHLH homodimer Pho4p (Fisher and Goding, 1992). However, there are exceptions: HLH-11 and MDL-1/MXL-1 heterodimer both tolerate a 5′ T, and HLH-30 actually favors a 5′ T (Supplemental Figure S9).
In summary, we identified both prominent and subtle differences in E-box or E-box-like sequence recognition and flanking site preferences between different bHLH dimers, which likely contribute to target site selection and gene regulation in vivo.
Functional Annotation of Putative bHLH Target Genes
We reasoned that we could harness the DNA binding specificity data to identify candidate target genes for each bHLH dimer, and then use these genes to initiate functional annotation of the dimers by searching for over-represented Gene Ontology (GO) categories. To do so, we took full advantage of the PBM data by considering sequences that capture E-box or E-box-like core sequences as well as flanking nucleotide preferences (Figure 5A). The highest level of sequence conservation of gene regulatory regions within related nematode species lies in the 500 bp upstream of transcription initiation sites (Castillo-Davis et al., 2004). Therefore, we searched this genomic region for all predicted C. elegans genes for the different bHLH binding sequences to identify candidate bHLH target genes. We calculated a cumulative ES for each gene, with respect to each of the bHLH dimers, to identify genes with either single “high-affinity” binding sites, or with multiple “lower affinity” binding sites, or a combination of both. We then identified over-represented GO annotation terms associated with these putative target genes, and, hence, with the relevant bHLH dimer (Supplemental Table S6, Supplemental Table S7).
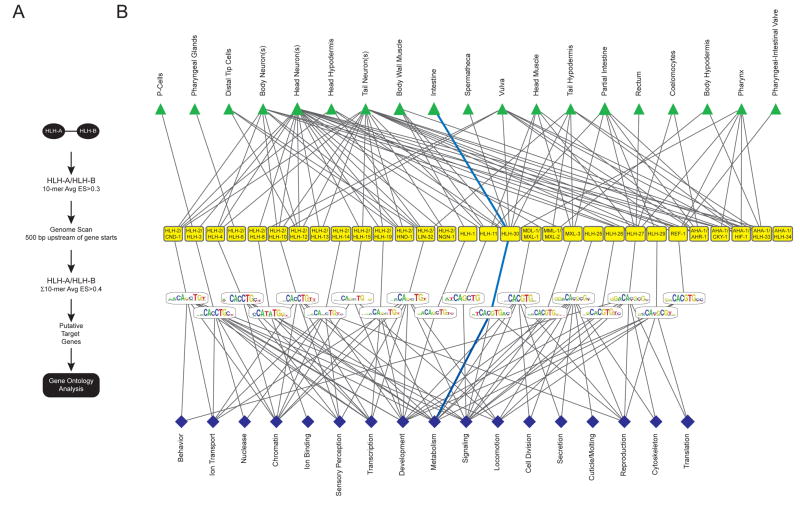
Figure 5. An Integrated bHLH Network.
(A) Flow diagram describing how GO annotations were obtained (see Experimental Procedures for details).
(B) Integrated bHLH network that combines dimerization, spatiotemporal expression, DNA binding specificities and GO categories. The blue lines depict a “network path” connecting the intestine to the “metabolism” GO category through HLH-30.
We identified multiple enriched GO terms, including Molecular Function terms associated with transcription and signaling, and Biological Process terms associated with development and metabolism. Some of the annotations we obtained are in agreement with what was previously known, either in C. elegans or for orthologs in other organisms. For instance, the connection of MDL-1/MXL-1 to “cell division” is evolutionarily conserved with the orthologous human dimer MAD/MAX (Yuan et al., 1998). However, the majority of functional annotations are novel.
An Integrated bHLH Dimerization, DNA binding and Expression Network
We assembled all separately measured functional bHLH parameters into the first integrated network for any TF family, combining dimerization, spatiotemporal expression patterns, DNA binding specificities, and enriched GO annotations of candidate target genes (Figure 5B).
As discussed above, all the nodes, i.e. dimers, tissues and DNA binding sequences, exhibit specificity and promiscuity in this network. In addition, we observed specificity and promiscuity for the different GO categories: some are associated with few bHLH dimers, whereas others are associated with many. For instance, “cell division” is associated only with MDL-1/MXL-1 and HLH-25, whereas “development” is associated with 11 different dimers (Figure 5B). Conversely, some bHLH dimers are associated with few categories, whereas others are associated with many; HLH-1 is connected solely to “development”, but HLH-25 is connected to nine different GO terms. However, it is important to note that “development” can be divided into “embryonic development”, “larval development” and several other terms that exhibit only partial overlap between different bHLH dimers. Similarly, “signaling”, “metabolism” and “reproduction” can be divided into more specific terms that enable the further differentiation between distinct bHLH dimers (Supplemental Figure S10).
Network Validation of HLH-30
To assess the validity of our integrated network, we focused on HLH-30, for which we had a viable deletion mutant available [hlh-30(tm1978)]. HLH-30 is strongly expressed in the intestine and weakly in other tissues (Figure 6A). This enables the identification of downstream target genes by expression profiling in vivo (i.e. this would be more difficult for bHLH TFs that exhibit more restricted expression patterns). RNAi knockdown of hlh-30 leads to a reduced fat phenotype (Ashrafi et al., 2003). Our integrated bHLH network contains a unique path that connects HLH-30 to the intestine, the main organ of fat storage, and to the GO categories “metabolism”, “reproduction” and “signaling” (Figure 5B). HLH-30 specifically binds CACGTG E-boxes (Figure 6B), and favors a flanking 5′ T (Figure 6C). This leads to the prediction that HLH-30 regulates (fat) metabolism in the intestine by binding target genes that contain HLH-30-bound CACGTG E-boxes in their promoter.
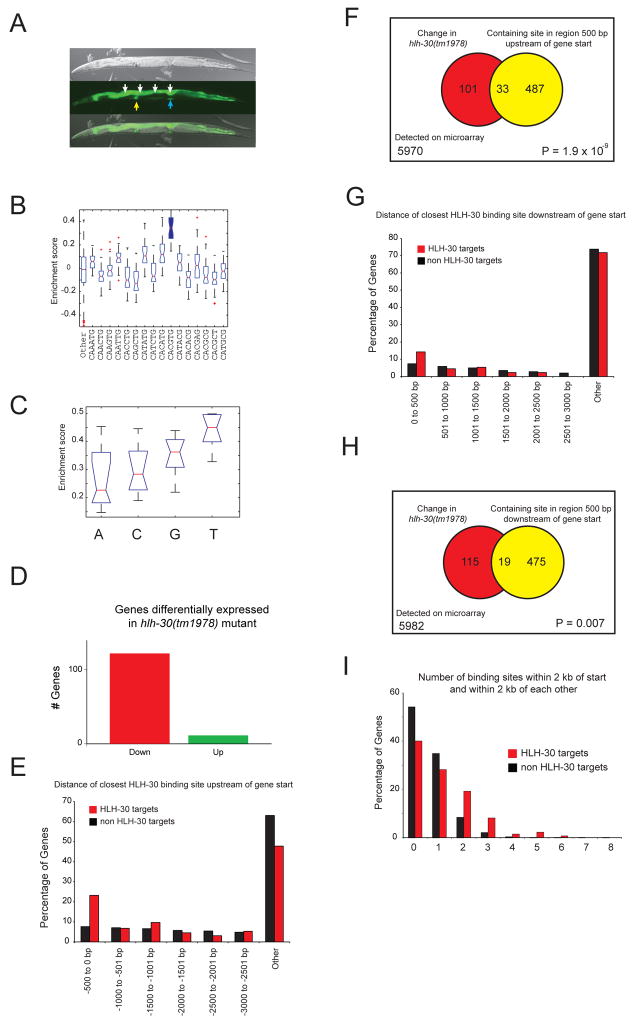
Figure 6. Network validation reveals conserved molecular and biological function of HLH-30.
(A) Phlh-30 drives GFP expression in different tissues, including the intestine (white arrows), spermatheca (yellow arrow) and vulva (blue arrow). Top – DIC image; middle –GFP image; bottom – merged images.
(B) HLH-30 strongly prefers the CACGTG E-box.
(C) HLH-30 strongly favors a 5′king the CACGTG E-box.
(D) HLH-30 activates gene expression. The majority of genes that change significantly in hlh-30(tm1978) mutant animals exhibit reduced expression (red), while the expression of a minority is increased (green).
(E) Distribution of genes for which the location of the closest HLH-30 binding site upstream of the transcriptional start is in the indicated window of distance (in increments of 500 bp).
(F) Venn diagram demonstrating association of gene expression change in hlh-30(tm1978) mutant animals with the region 500 bp upstream of the gene start harboring an HLH-30 binding site.
(G) Distribution of genes for which the location of the closest HLH-30 binding site downstream of the gene start is in the indicated genomic regions (in increments of 500 bp).
(H) Venn diagram demonstrating association of gene expression change in hlh-30(tm1978) mutant animals with the region 500 bp downstream of the gene start harboring an HLH-30 binding site.
(I) HLH-30 targets have two or more HLH-30 binding sites within 2 kb of each other in the region up or downstream of the gene start more often than do non-HLH-30 targets.
To test this prediction, we performed gene expression profiling of wild type and hlh-30(tm1978) mutant animals and compared the resulting expression data. We identified 134 genes that were significantly differentially expressed: 122 exhibited decreased, and 12 exhibited increased expression in the mutant (Figure 6D, Supplemental Table S8). This suggests that HLH-30 is primarily a transcriptional activator, which is in agreement with our observation that it is a strong auto-activator in Y2H assays (Figure 2A). We refer to all genes that change in expression in the hlh-30(tm1978) mutant as “HLH-30 target genes”, although some may change in expression due to indirect effects rather than direct regulation by HLH-30.
HLH-30 target genes more frequently possess an HLH-30 binding site in 500 bp promoter sequences than non-target genes (Figures 6E, F; Fisher’s exact test p = 1.9 × 10−9). The consistency between the PBM-derived and experimentally identified HLH-30 target genes supports our overall approach for identifying candidate bHLH target genes using PBM data. When we searched genomic sequences downstream of the transcriptional start, we also observed an increase in HLH-30 binding sites in targets versus non-targets, albeit less significantly (Figures 6G, H; Fisher’s exact test p = 0.007). Finally, we found that HLH-30 targets significantly more frequently possess multiple HLH-30 binding sites than non-targets (Figure 6I, chi-square test p = 2.2 × 10−16).
Next, we examined the experimentally determined HLH-30 target genes for over-represented GO terms, and found enrichment for various metabolic, as well as aging terms (Supplemental Table S9). Interestingly, the human ortholog of HLH-30, TFE3, has been reported to activate metabolic genes through E-boxes as well (Nakagawa et al., 2006). This suggests that both the molecular and biological functions of HLH-30 are evolutionarily conserved.
We have likely underestimated the number of in vivo HLH-30 target genes because only changes in genes that are broadly or highly expressed can be detected in whole animal gene expression analysis. Thus, it is more difficult to evaluate the association of HLH-30 with the GO term “reproduction”; even though Phlh-30 drives expression in the spermatheca and the vulva (Figure 6A). Nevertheless, the whole animal gene expression analysis does provide support for our overall method and approach.
Multi-Parameter Analysis of bHLH TFs
To examine the overall extent to which bHLH TFs differ from each other we compared all possible 861 bHLH-bHLH pairs. We derived a Similarity Score (SS) for each pair and for each parameter (Figure 7A), clustered the bHLH TFs and dimers according to these scores, and visualized these as heat maps, resulting in one heat map per parameter (Supplemental Figure S11). Figure 7B shows a summary of the entire parameter analysis. We observed that for each parameter the majority of the pairs have a low SS. For instance, more than 80% of the bHLH-bHLH pairs share fewer than 25% of their target genes (Figure 7B). We observed the lowest degree of divergence in spatial expression; however, this is likely because not all expression could be resolved to the level of individual cells (see below).
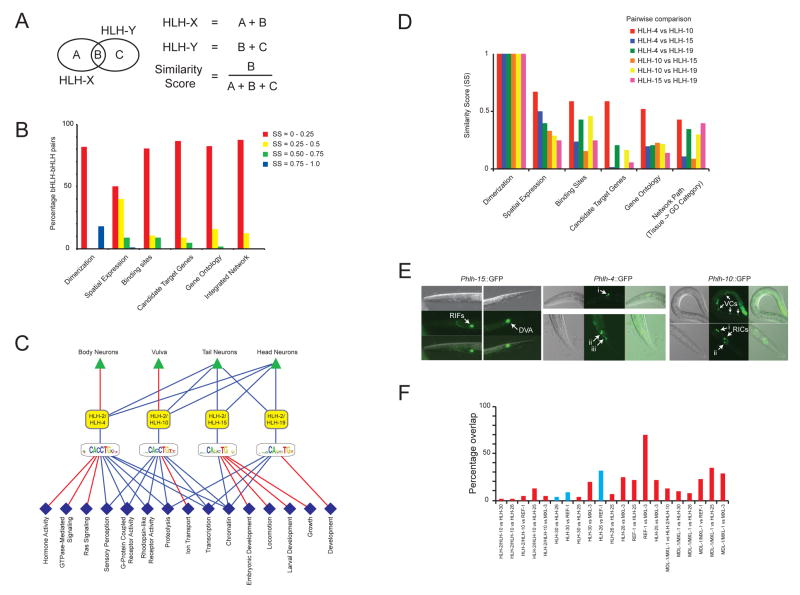
Figure 7. Most bHLH proteins differ from each other in multiple functional parameters.
(A) For each bHLH-bHLH pair we calculated a Similarity Score (SS) for each functional TF parameter as indicated.
(B) Integrated parameter overlap analysis of all bHLH-bHLH pairs and dimer pairs (see Supplemental Figure S12 for individual parameter analysis). SSs were binned into four groups as indicated.
(C) Sub-networks of bHLH proteins with the highest degree of similarity. Red lines –unique functional parameters; blue lines – shared functional parameters. Dark blue diamonds – Molecular Function; light blue diamonds - Biological Process.
(D) Individual similarity scores for all bHLH-bHLH pairs shown in (C).
(E) Detailed analysis of neuronal expression conferred by Phlh-15, Phlh-4 and Phlh-10. Phlh-4::GFP: i) two sensory head neurons (one bilaterally symmetric pair) of the lateral ganglion, likely AWA or AWB; ii) three pairs of tail neurons of the lumbar ganglion, likely PVQ, PVC, PVW, and/or LUA; iii) two tail neurons (likely a bilaterally symmetric pair) of the lumbar ganglion with processes to the tail. Phlh-10::GFP: i) two interneurons (one bilaterally symmetric pair) of the retrovesicular ganglion, likely RIF or RIG; ii) two sensory head neurons (one bilaterally symmetric pair) of the lateral ganglion, likely AWA or AWB.
(F) Percentage overlap of candidate target genes comparing bHLH dimers that can bind CACGTG E-boxes. Blue bars indicate comparisons in which both dimers exclusively bind CACGTG, red indicates comparisons in which one or both dimers can also bind other E-boxes or E-box-like sequences.
Several bHLH-bHLH pairs are more similar in one or more parameter than most other pairs. A sub-network of the most similar bHLH TFs is shown in Figure 7C. These all share HLH-2 as their dimerization partner and, for clarity, heterodimers are depicted as single nodes. The parameter comparisons among these dimers are provided in Figure 7D. Several observations can be made from this analysis. First, several tissues and GO categories can be connected by paths that go through these different dimers. For instance, head neurons can be connected to sensory perception via both HLH-2/HLH-4 and HLH-2/HLH10. We refer to such similar connections as “network paths”. In fact, we found that HLH-4 and HLH-10 share ~40% of their network paths in the integrated network (SS = 0.43, Figure 5B). This suggests that they may be highly similar in various TF parameters. Indeed, they share more than 50% of each of the parameters measured (SS = 0.52 – 0.67, Figure 7D). HLH-15 and HLH-19 also share ~40% of their network paths in the integrated network (SS = 0.4, Figure 5B). These two dimers connect head and tail neurons to chromatin. Surprisingly, in this case they are quite divergent in each of the individual parameters. In fact, they share fewer than 10% of their predicted target genes (SS = 0.06, Figure 7C). This means that HLH-4 and HLH-10 may regulate an overlapping set of target genes in head neurons to control sensory perception, whereas HLH-15 and HLH-19 may regulate different sets of chromatin genes in (developing) head neurons. The annotation “head neurons” is very broad as there are ~200 different neurons comprising this category. Therefore, we further refined the expression annotations of HLH-4, HLH-10 and HLH-15 (the expression of HLH-19 diminishes after the animals hatch and could not be annotated in more detail). We found that HLH-4 and HLH-10 may be expressed in a similar set of neurons, whereas the expression of HLH-15 is clearly distinct (Figure 7D). This supports the hypothesis that HLH-4 and HLH-10 may share target genes in the same cell(s).
HLH-4 and HLH-15 confer different loss-of-function phenotypes: RNAi of hlh-15 results in high fat content (Ashrafi et al., 2003), but no other detectable phenotype, and RNAi of hlh-4 results in slow growth and protruding vulva (Simmer et al., 2003). These two TFs share almost 25% of their DNA binding sites (SS = 0.24) but less than 5% of their candidate target genes (SS = 0.01), most likely because HLH-2/HLH-4 has a broader DNA binding specificity than HLH-2/HLH15. In addition, HLH-4 and HLH-15 are expressed in distinct neurons (Figure 7E). This indicates that the functional divergence of these two bHLH TFs is likely accomplished by relatively small changes in spatiotemporal expression and DNA binding specificities.
Even though the bHLH TFs shown in Figure 7C exhibit a relatively high degree of similarity, there are also important differences that can contribute to TF divergence. For instance, of the four bHLH dimers shown, only one is expressed in the vulva (HLH-2/HLH-10). Similarly, only two of the dimers are expressed in later stages of development (HLH-2/HLH-4 and HLH-2/HLH-10), whereas the other two are exclusively expressed during embryogenesis and in the first larval stage (Figure 3B, Supplemental Figure S4).
Finally, we analyzed molecular and functional divergence among a set of bHLH dimers that can all bind the CACGTG E-box. Three of these dimers exclusively bind this E-box (HLH-30, HLH-26 and REF-1) whereas the others (HLH-2/HLH-10, MXL-3, HLH-25 and MDL-1/MXL-1) also bind other E-box and/or E-box-like sequences (Figure 4C). Interestingly, we find little overlap between these different dimers in their candidate target genes (Figure 7F). This indicates that several of these dimers may utilize multiple different E-box and E-box-like sequences in their target genes and that target genes may discriminate bHLH dimers by harboring different combinations of E-box and E-box-like sequences. Even for dimers that exclusively bind the CACGTG E-box, we find little overlap in their candidate target genes. Indeed, HLH-30 favors a flanking T, HLH-26 favors an A or G and REF-1 disfavors a T, indicating that flanking nucleotides may play an important role in functional TF divergence. Finally, the pair that shares the largest proportion of predicted target genes, REF-1 and MXL-3, exhibits non-overlapping spatiotemporal expression patterns, which likely contributes to their functional divergence (Supplemental Table S4).
DISCUSSION
We present the first integrated network for any TF family that provides connections between proteins, the tissues in which they are expressed, the DNA sequences they preferentially bind, their candidate target genes and enriched GO categories associated with these target genes.
Several observations indicate that our individual TF parameter datasets are of high quality, and most importantly, each of the different datasets validate each other. For instance, PBM assays with five combinations of bHLH proteins that did not heterodimerize in Y2H assays did not yield any specific DNA binding motifs (Supplemental Figure S6). This indicates that PBM validates Y2H, and vice versa. Similarly, the observation that bHLH proteins that dimerize are more likely co-expressed than those that do not dimerize validates the Y2H data. See Supplemental Materials for further discussion of the quality of the individual data types.
The integrated bHLH network is likely not yet complete. For instance, we used only bHLH promoter activity, and did not include other potential regulatory sequences. In addition, we did not annotate bHLH expression in males or dauers, or under different conditions. Finally, for future models of gene regulation it will be important to incorporate expression levels of different bHLHs in different cell types, because protein levels will determine the binding to high or low affinity binding sites, and, hence the selection of tissue-specific target genes.
Previously, two other integrated networks were reported for C. elegans genes. The first connects genes involved in early embryogenesis by protein-protein interactions, phenotypes and expression profiles (Gunsalus et al., 2005). The second is a probabilistic network that used various data types and that can be used to predict genetic interactions (Lee et al., 2007). Although powerful, neither network focused on TFs or provided interactions between proteins, DNA sequences, and tissues or cell types, and therefore could not address the question of divergence in paralogous TF families.
A priori, we reasoned that paralogous TFs could attain functional specificity by individualizing a single molecular parameter. However, we found a spectrum of differences among the TFs in all parameters; some bHLH TFs are relatively similar in one or more parameters, whereas others are highly divergent. This is reflected by the observation of both specificity and promiscuity in the integrated network; some nodes (e.g. DNA sequences, tissues) are highly connected to many bHLH TFs, and others are not. Considering all the parameters measured, most bHLH TFs differ substantially from each other. There are several relatively similar bHLH TFs that exhibit only limited divergence in one or more TF parameters. However, we found that a minor difference in DNA binding specificity, either in the core E-box or E-box-like sequence, or in the flanking nucleotides, can result in little overlap in candidate target genes.
Even though many paralogous TFs have distinct biological functions, there are also examples of redundant TF paralogs. For example, members of mammalian ETS family of TFs can function partially redundantly by binding to overlapping sets of target genes (Hollenhorst et al., 2007). Similarly, FLH TFs in C. elegans can redundantly regulate microRNA expression (Ow et al., 2008). Finally, in C. elegans, paralogous TFs such as paired homeodomains can function in modules in the context of neuronal regulatory networks (Vermeirssen et al., 2007). Future systematic studies of genetic interactions will reveal the extent of genetic redundancy within TF families.
In addition to enabling studies of TF divergence, this integrated network is also useful for generating specific hypotheses, as demonstrated by our gene expression profiling analysis of hlh-30 mutant animals. Moreover, each of the individual data types provides a first comprehensive catalog of dimers, expression patterns and binding sites for a metazoan TF family. These data will be useful for gaining insight into the molecular determinants of the interactions in which the various bHLH proteins participate.
The integrated bHLH network confirms previously reported features for the bHLH family, including a promiscuous role in dimerization, DNA binding specificity and expression for the E/Daughterless homolog, HLH-2, and more specific roles for its dimerization partners (Massari and Murre, 2000). AHA-1 and HLH-2, both of which dimerize with multiple bHLH proteins, are auto-activators in Y2H assays whereas most of their dimerization partners are not. Based on these observations, we propose that the bHLH dimerization hubs may confer the transcriptional activation activity to the different dimers, whereas their dimerization partners may contribute specificity in DNA binding.
Our data and methods provide a framework for similar studies of other C. elegans TF families and of TF families in other organisms, including humans. Similar studies will likely be useful for other protein families, such as kinases, in the context of other types of regulatory networks. Such studies of paralogous genes, including comparisons of integrated networks across species, may provide further insights into the molecular features underlying the evolution of gene families.
EXPERIMENTAL PROCEDURES
Y2H Assays
Y2H assays were performed as described (Walhout and Vidal, 2001) using Gateway-compatible bHLH clones (Supplemental Materials).
Generation of pDEST-mCherry::his-11
The mCherry ORF was PCR-amplified from pAA64 plasmid DNA (generously provided by A. Audhya, Oegema Lab, University of California, San Diego). The resulting amplicon was Gateway cloned into pDONR-221 to generate mCherry-Entry. A PCR fusion strategy created an mCherry::his-11 fusion ORF. The his-11 ORF was amplified from pJH4.52 (generously provided by K. Hagstrom, University of Massachusetts Medical School, Worcester) using a his11-specific forward primer and an att-B2 Gateway-tailed reverse primer. PCR amplification was carried out for 15 cycles to minimize the introduction of mutations. A similar PCR reaction was used to amplify the mCherry ORF using the same att-B1 tailed forward primer and an mCherry-specific reverse primer carrying a his-11-specific tail at the 5′ end of the primer. Both PCR products were simultaneously Gateway cloned into pDONR221. The resulting plasmid contained the mCherry ORF fused in frame to the his-11 ORF. This fragment was then cloned by a Multisite Gateway LR reaction into pDEST-DD03 (Dupuy et al., 2004) along with Phlh-2. The resulting Phlh-2::mCherry::his-11 Destination clone was used directly in microparticle bombardment to create transgenic C. elegans. Primer sequences used are provided in the Supplemental Materials.
C. elegans Transgenesis
Transgenic C. elegans were generated as described (Reece-Hoyes et al., 2007). Double transgenic animals were generated by crossing males that carry Phlh-2::mCherry::his-11 constructs into Phlh::GFP carrying hermaphrodites. Each transgenic line carrying a Phlh::GFP fusion was independently verified by PCR using promoter-specific primers (primer sequences are available upon request).
Protein Binding Microarray Experiments
Microarray design, preparation and PBM experiments were performed and analyzed as described (Berger et al., 2006, Supplemental Materials).
Binding Site Annotation, Mapping and Prediction of bHLH Target Genes
Target genes were predicted by initially calculating for each dimer the average 8-mer enrichment score (AvgES) within all 10-mers that contained an E-box (NN-E-box, N-E-box-N, E-box-NN)(similar for E-box-like sequence). For each bHLH dimer, genomic sequences 500 bp upstream of each WBGene (referred to as transcriptional start) were scanned with the corresponding set of 10-mers with AvgES ≥ 0.3. Each gene was scored by summing the AvgES of all 10-mers found in the 500 bp upstream sequence. All genes having a Sum of AvgESs ≥ 0.4 were considered for analysis of functional category enrichment using the GoMiner algorithm (http://discover.nci.nih.gov/gominer/). For the HLH-30 target gene analysis we mapped the genomic coordinates of all HLH-30 10-mers with an AvgES ≥ 0.3. We uploaded this information as GFF files into our Bio::DB::GFF Database (Stein et al., 2002), and queried this database to calculate relative distances between binding sites and the beginning of a gene.
Parameter Overlap Analysis
For each pair-wise bHLH-bHLH parameter comparison Similarity Scores (SS) were calculated as follows:
For instance, when bHLH-X binds 10 target genes and bHLH-Y binds 20 target genes, and they have 5 target genes in common, the SS would be 5/25 = 0.2. Heat maps were created by clustering the HLHs based on their SSs. The clustering heat maps depicting parameter comparisons were performed using MultiExperiment Viewer version 4.0 (Saeed et al., 2003).
Supplementary Material
Acknowledgments
We thank M. Berger for technical assistance and advice with PBMs and data analysis, S. Jaeger and R.P. McCord for statistical advice, assistance and help with Matlab, T. Siggers for genome scanning assistance, and members of the Walhout and Bulyk laboratories and J. Dekker for advice and critical reading of the manuscript. Special thanks to Shohei Mitani for the hlh-30(tm1978) mutant C. elegans strain and Maria Ow for outcrossing this strain. This work was supported by NIH grants DK068429 to A.J.M.W., GM084491 to M.A., and R01 HG002966 and R01 HG003985 to M.L.B.
Footnotes
Additional information on Experimental Procedures is available in the Supplemental Materials. PBM data are available at http://thebrain.bwh.harvard.edu/pbms/webworksW/ and in the UniPROBE database (Newburger and Bulyk., 2009).
Y2H data and expression patterns are available in EDGEdb: http://edgedb.umassmed.edu (Barrasa et al., 2007). Supplemental figures and tables are provided in a separate PDF file.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Barrasa MI, Vaglio P, Cavasino F, Jacotot L, Walhout AJM. EDGEdb: a transcription factor-DNA interaction database for the analysis of C. elegans differential gene expression. BMC Genomics. 2007;8:21. doi: 10.1186/1471-2164-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL, Achaz G. Cis-regulatory and protein evolution in orthologous and duplicate genes. Genome Res. 2004;14:1530–1536. doi: 10.1101/gr.2662504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Krause M, Sepanski M, Fire A. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development. 1994;120:1631–1641. doi: 10.1242/dev.120.6.1631. [DOI] [PubMed] [Google Scholar]
- Crews ST. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- Deplancke B, Mukhopadhyay A, Ao W, Elewa AM, Grove CA, Martinez NJ, Sequerra R, Doucette-Stam L, Reece-Hoyes JS, Hope IA, et al. A gene-centered C. elegans protein-DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Dupuy D, Li Q, Deplancke B, Boxem M, Hao T, Lamesch P, Sequerra R, Bosak S, Doucette-Stam L, Hope IA, et al. A first version of the Caenorhabditis elegans promoterome. Genome Res. 2004;14:2169–2175. doi: 10.1101/gr.2497604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher F, Goding CR. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. Embo J. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove CA, Walhout AJ. Transcription factor functionality and transcription regulatory networks. Molecular BioSystems. 2008;4:309–314. doi: 10.1039/b715909a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus KC, Ge H, Schetter AJ, Goldberg DS, Han JD, Hao T, Berriz GF, Bertin N, Huang J, Chuang LS, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- Hallam S, Singer E, Waring D, Jin Y. The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development. 2000;127:4239–4252. doi: 10.1242/dev.127.19.4239. [DOI] [PubMed] [Google Scholar]
- Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev. 1998;12:2623–2635. doi: 10.1101/gad.12.16.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI, Garcia Delgado C, Gonzalez-Ramos M, Kline AD, Jabs EW. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- Jiang H, Guo R, Powell-Coffman JA. The Caenorhabditis elegans hif-1 gene encodes a bHLH-PAS protein that is required for adaptation to hypoxia. Proc Natl Acad Sci U S A . 2001;98:7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Park M, Zhang JM, Yuan J, Harfe BD, Xu SQ, Greenwald I, Cole MD, Paterson BM, Fire A. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. 1997;124:2179–2189. doi: 10.1242/dev.124.11.2179. [DOI] [PubMed] [Google Scholar]
- Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2007;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Reece-Hoyes J, Ambros V, Walhout AJ. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–2015. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa Y, Shimano H, Yoshikawa T, Ide T, Tamura M, Furusawa M, Yamamoto T, Inoue N, Matsuzaka T, Takahashi A, et al. TFE3 transcriptionally activates hepatic IRS-2, participates in insulin signaling and ameliorates diabetes. Nat Med. 2006;12:107–113. doi: 10.1038/nm1334. [DOI] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37(Database issue):D77–82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooe N, Saito K, Oeda K, Nakatuka I, Kaneko H. Characterization of Drosophila and Caenorhabditis elegans NXF-like factors, putative homologs of mammalian NXF. Gene. 2007;400:122–130. doi: 10.1016/j.gene.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Ow MC, Martinez NJ, Olsen P, Silverman S, Barrasa MI, Conradt B, Walhout AJM, Ambros VR. The FLYWCH transcription factors FLH-1, FLH-2 and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev. 2008;22:2520–2534. doi: 10.1101/gad.1678808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirano RI, Wegner M. The glial transcription factor Sox10 binds to DNA both as a monomer and dimer with different functional consequences. Nucleic Acids Res. 2000;28:3047–3055. doi: 10.1093/nar/28.16.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett CL, Breen KT, Ayer DE. A C. elegans Myc-like network cooperates with semaphorin and Wnt signaling pathways to control cell migration. Dev Biol. 2007;310:226–239. doi: 10.1016/j.ydbio.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman DS, Emmons SW. The basic helix-loop-helix transcription factors LIN-32 and HLH-2 function together in multiple steps of a C. elegans neuronal sublineage. Development. 2000;127:5415–5426. doi: 10.1242/dev.127.24.5415. [DOI] [PubMed] [Google Scholar]
- Powell-Coffman JA, Bradfield CA, Wood WB. Caenorhabditis elegans orthologs of the aryl hydrocarbon receptor and its heterodimerization partner the aryl hydrocarbon receptor nuclear translocator. Proc Natl Acad Sci U S A. 1998;95:2844–2849. doi: 10.1073/pnas.95.6.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner SM, Ciribilli Y, Inga A, Borlak J. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Human Molecular Genetics. 2008;17:1397–405. doi: 10.1093/hmg/ddn027. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, Walhout AJM. A compendium of C. elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJ, Vidal M, Hope IA. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evolutionary Biology. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PVE, Kamath FS, Fraser AG, Ahringer J, Plasterk RHA. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biology. 2003;1:77–84. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai KK, Nishiwaki K. bHLH transcription factors regulate organ morphogenesis via activation of an ADAMTS protease in C. elegans. Dev Biol. 2007;308:562–571. doi: 10.1016/j.ydbio.2007.05.024. [DOI] [PubMed] [Google Scholar]
- Tan K, Feizi H, Luo C, Fan SH, Ravasi T, Ideker TG. A systems approach to delineate functions of paralogous transcription factors: role of the Yap family in the DNA damage response. Proc Natl Acad Sci U S A. 2008;105:2934–2939. doi: 10.1073/pnas.0708670105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V, Barrasa MI, Hidalgo C, Babon JAB, Sequerra R, Doucette-Stam L, Barabasi AL, Walhout AJM. Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res. 2007;17:1061–1071. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout AJM. Unraveling Transcription Regulatory Networks by Protein-DNA and Protein-Protein Interaction Mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- Walhout AJM, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- Walhout AJM, van der Vliet PC, Timmers HTM. Sequences flanking the E-box contribute to cooperative binding by c-Myc/Max heterodimers to adjacent binding sites. Biochim Biophys Acta. 1998;1397:189–201. doi: 10.1016/s0167-4781(97)00227-3. [DOI] [PubMed] [Google Scholar]
- Walhout AJM, Vidal M. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods. 2001;24:297–306. doi: 10.1006/meth.2001.1190. [DOI] [PubMed] [Google Scholar]
- Yuan J, Tirabassi RS, Bush AB, Cole MD. The C. elegans MDL-1 and MXL-1 proteins can functionally substitute for vertebrate MAD and MAX. Oncogene. 1998;17:1109–1118. doi: 10.1038/sj.onc.1202036. [DOI] [PubMed] [Google Scholar]
- Zhu C, Byers K, McCord R, Shi Z, Berger M, Newburger D, Saulrieta K, Smith Z, Shah M, Radhakrishnan M, et al. High-resolution DNA binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.