Abstract
A large number of genes have been positively selected and recruited to participate in various phases of the inflammatory response triggered by microbial stimuli. Because of the complexity of the response, the many phases in which it is deployed, and the many “flavors” in which it appears (depending on quality and intensity of the stimulus as well as the target organ), very elaborated mechanisms evolved to ensure that the expression of the induced genes is carefully and precisely organized so that each gene is expressed in response to specific stimuli and with kinetics and intensities that suit the peculiar function of its product(s). Data accumulated in recent years have strengthened the concept that chromatin is an essential substrate at which multiple signals are integrated to promote a correctly choreographed expression of the genes involved in inflammatory transcriptional responses. Although the current level of understanding of these mechanisms is far from complete, some concepts and ideas have resisted experimental challenges and now represent accepted paradigms that are the subject of this article.
A gene's chromatin configuration can tailor its response to transcription factors. In inflammatory responses, this ensures that target genes respond appropriately to NF-κB activation.
NF-κB AND THE COMPLEXITY OF INFLAMMATORY TRANSCRIPTIONAL RESPONSES
The ability to efficiently cope with microbial infections is an essential requisite of complex life on earth. Indeed, prototypes of the innate immune system and rudimentary inflammatory responses already appeared in rather primitive multicellular eukaryotes, and extremely more complex and evolved versions of them are now found throughout not only the animal but also the plant kingdom (Janeway and Medzhitov 2002).
The sources of complexity in inflammatory responses, and the ensuing need to efficiently cope with them, include the great diversity of microbial stimuli; the need to adjust the intensity of the response to that of its inducer; the need for innate immune cells to properly interact with and control the activation of the adaptive immune system; the presence of multiple organs with individual properties in terms of exposure to microbial sources, tissue organization, repair capacity, blood, and lymphatic supply; and finally the need to create an efficient interface between inflammatory responses and complex networks of metabolic pathways, so that even massive recruitment of energy sources to fight microbes will not jeopardize metabolic homeostasis. Moreover, several of the molecules and mechanisms invented during evolution to fight microbes were also recruited by nonmicrobial inflammatory responses, such as those triggered by tissue damage. Because of this complexity and of the many contexts in which an inflammatory response may unfold, the products of a huge number of genes have been selected during evolution to participate in one or more phases of inflammation. Although some genes, like the one encoding TNFα, are activated with rather monotonous patterns in almost every type of inflammatory response, several others are induced with complex kinetics and in a highly stimulus-selective and cell-type- or tissue-restricted fashion (Nau et al. 2002). In simple words, even if thousands of genes are regulated at the level of transcription to participate in inflammatory responses, kinetic, spatial, and quantitative parameters of activation are highly gene-specific, as they reflect the specific function of each gene product in the unfolding or control of the response.
The transcription factors of the NF-κB/Rel family directly control the induction of a very large fraction of the inflammatory transcriptome. Moreover, they contribute to other transcriptional programs, like those induced by antigen-receptor triggering in lymphocytes. The almost universal requirement for NF-κB/Rel in the induction of inflammatory genes poses a conundrum: How can we explain such a complexity in transcriptional patterns if only a few master controllers are involved? Data accumulated in recent years showed an essential role of chromatin organization in integrating information from multiple sources to control NF-κB recruitment to the underlying genomic sequences and generate complex and diverse NF-κB-regulated gene expression programs (Natoli et al. 2005).
TWO CLASSES OF NF-κB-REGULATED GENES DEFINED BY DIFFERENTIAL CHROMATIN CONFIGURATIONS AND INDUCTION KINETICS
Genes activated in response to inflammatory cues are a highly heterogeneous set whose products participate in several biological processes occurring sequentially or in parallel during inflammation (Nau et al. 2002), including the following: Chemoattractants (chemokines) promoting the recruitment of inflammatory cells to the peripheral sites of inflammation or recruiting cells of the adaptive immune system for antigen presentation; general inflammatory cytokines (like TNFα and interleukin 1β) that mainly contribute to response amplification and immune cell activation; antiapoptotic and antioxidant proteins that enable cells to survive both to the infection and to toxic products accumulating in the infiltrate as a consequence of cell activation and death (mainly of neutrophils); enzymes producing mediators that control vascular permeability and endothelial growth factors controlling inflammatory neoangiogenesis; proteins or enzymes involved in microbial recognition and killing; and cytokines instructing the differentiation of T lymphocytes (like IL12, which promotes Th1 differentiation of CD4+ T lymphocytes). Although all of these genes are required for different components of the response, they are not activated simultaneously. For instance, in LPS-stimulated human dendritic cells (DCs), expression of chemokines attracting T cells in the T-dependent areas of the lymph node (like ELC/CCL19) is postponed some hours as compared to the induction of a chemokine-like IL8/Cxcl8, which recruits neutrophils to peripheral inflammatory sites (Saccani et al. 2002). Therefore, DCs express IL8 in the periphery shortly after the encounter with the microbe and ELC/Ccl19 only after migration to the lymph node.
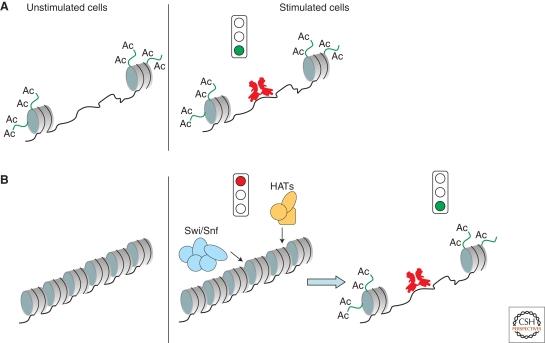
An initial clue on the mechanistic bases of kinetic complexity in NF-κB-dependent gene expression came from the analysis of NF-κB recruitment to target genes using chromatin immunoprecipitation (ChIP) (Saccani et al. 2001). Although entry of NF-κB in the nucleus after a stimulus like TNFα or LPS is massive, fast, and synchronous, kinetics of recruitment to target genes was found to be asynchronous and complex. Although some genes were bound by NF-κB without any detectable delay after its nuclear entry (“fast” genes), others were bound (and activated) with much slower kinetics, in some cases even hours after NF-κB nuclear translocation (“slow” genes). Slow genes include primary genes activated with slow kinetics but not requiring new protein synthesis, and secondary genes (requiring new protein synthesis for activation) (Ramirez-Carrozzi et al. 2006). Delayed recruitment of NF-κB to slow target genes does not depend on an overall lower affinity of their promoters for NF-κB, because both fast and slow genes bear high-affinity DNA-binding sites (Saccani et al. 2001). Conversely, chromatin configuration at the two groups of genes is clearly different. Before stimulation, fast genes display a chromatin landscape typical of genes poised for immediate activation, including high levels of histone H3 and H4 acetylation, and trimethylation of lysine 4 of histone H3 (H3K4me3, a histone modification specifically enriched at active or poised transcription start sites, TSS) (Saccani et al. 2001). Moreover, the promoters of fast NF-κB targets are constitutively accessible to nucleases, thus indicating an overall “open” and accessible organization (Fig. 1) (Ramirez-Carrozzi et al. 2006). Conversely, slow genes in unstimulated cells are associated with hypoacetylated histones and are negative for H3K4me3. In response to activation, both acetylation and H3K4 trimethylation progressively increase, with a kinetics that apparently precedes NF-κB recruitment (Fig. 1) (Saccani et al. 2001; Kayama et al. 2008). In any case, the final levels of acetylation and H3K4me3 usually do not reach those of fast genes. Importantly, changes in histone modifications are paralleled by an increase in accessibility to nucleases, thus suggesting a progressive decompaction of the locus. Decompaction seems to be an overall inefficient process, occurring in only a fraction of the cells, thus often leading to a mosaic expression of the corresponding gene (Weinmann et al. 2001). According to these data, the kinetic behavior of NF-κB target genes reflects the chromatin configuration of the nucleosomes in the vicinity of/overlapping the NF-κB binding sites. On these premises, we have proposed a classification of NF-κB target genes based on chromatin configuration-dependent differential kinetics of NF-κB recruitment. According to this classification, we indicate the “fast” genes as those with constitutive and immediate accessibility (CIA) and the “slow” ones as those with regulated and late accessibility (RLA). The properties of these two groups of genes are summarized in Table 1 (some of them are further discussed later).
Figure 1.
Differential chromatin states at NF-κB-dependent genes. In the upper part of the figure, a gene is shown with an open and accessible chromatin structure (A), whereas in the bottom part, a gene is depicted with a closed chromatin configuration requiring Swi/Snf-dependent chromatin remodeling to enhance accessibility to NF-κB and transcriptional activation (B). NF-κB is shown in red.
Table 1.
Properties of NF-κB regulated genes with different chromatin configurations
| Constitutively and immediately accessible genes | Genes with regulated and late accessibility | |
|---|---|---|
| Timing of mRNA induction after stimulus | <30 min | >60 min (to several hours) |
| Kinetics of NF-κB recruitment | Immediate (minutes after nuclear entry) | Delayed (>60 min) |
| Requirement for chromatin remodeling | No | Yes |
| Requirement for new protein synthesis | No | Yes/No depending on the gene |
| Constitative H3/H4 acetylation | Yes | No |
| Constitative H3K4me3 | Yes | No |
An essential and still open question is whether these two different chromatin configurations are written in the underlying DNA sequence or are epigenetically determined.
GENETIC DISSECTION OF CHROMATIN REMODELING EVENTS AT NF-κB-REGULATED GENES
The data described previously established a clear correlation between alternative chromatin configurations and differential kinetics of induction of NF-κB target genes. Subsequent data from Stephen Smale and colleagues provided formal evidence that chromatin remodeling is required for the activation of essentially all RLA genes (Ramirez-Carrozzi et al. 2006). “Chromatin remodeling” generically refers to all of the alterations of histone/DNA contacts performed by multimolecular ATP-dependent machines (Becker and Horz 2002). The Swi/Snf chromatin remodeling complexes contain the essential ATPase subunits Brg1 and Brm, which are largely interchangeable: Simultaneous depletion of both ATPases by RNA interference abrogated the induction of all RLA genes, including both the late primary genes and the secondary genes (Ramirez-Carrozzi et al. 2006). Impaired induction was associated with lack of remodeling, as indicated by the inability of LPS stimulation to increase accessibility of chromatin at their promoters to restriction enzymes.
Interestingly, the basal and repressed state was actively enforced by an opposing chromatin remodeling complex, Mi2/Nurd. Cells depleted of the helicase component of Mi2/Nurd (Mi2β/Chd4) showed normal kinetics of gene activation but greatly enhanced levels of induction, possibly because of unopposed Swi/Snf-mediated remodeling. In the interpretation of these data, it should be considered that apparently remodeling is an incomplete event, often occurring in only a fraction of the stimulated cells: One possibility is that the relative levels or activity of Swi/Snf versus Mi2/Nurd in individual cells tilt the balance towards activation or repression and that in the absence of Mi2/Nurd, the actual fraction of cells expressing RLA genes is increased.
Additional genetic analyses of NF-κB-dependent gene induction in LPS-stimulated cells have uncovered further regulatory layers (Kayama et al. 2008). LPS signals through Toll-like receptor 4 (TLR4) via two main adapters, MyD88 and Trif (Kawai and Akira 2007). In LPS-stimulated Myd88−/− macrophages NF-κB activation is only marginally impaired, because of compensation by TRIF. These minimal alterations of NF-κB activation are surprisingly associated with a profound defect in the expression of most or all secondary genes (Kayama et al. 2008). Defective inducibility is associated with an absolute loss of nucleosome remodeling, absent increase in H3K4 trimethylation or histone acetylation, and finally impaired or absent recruitment of NF-κB. Therefore, NF-κB recruitment to secondary LPS-induced genes is directly controlled by remodeling events triggered by MyD88. Conversely, activation of all fast/CIA genes occurs normally in MyD88-deficient cells.
IkBζ, a distant paralog of IkBα encoded by the Nfkbiz gene, is a transcriptional coregulator whose expression is rapidly induced by NF-κB, thus behaving as an early primary response gene (Haruta et al. 2001).
In the absence of IkBζ, the process leading to the activation of some secondary genes (like IL12b and IL6) is aborted at a specific stage after chromatin remodeling but before recruitment of NF-κB and loading of the preinitiation complex (Yamamoto et al. 2004; Kayama et al. 2008). In IkBζ-deficient cells, accessibility of the DNA associated with promoter nucleosomes is normally increased after stimulation, but NF-κB and Pol II are not loaded. The increase in H3K4me3 that accompanies the induction of these genes is also completely blocked in the absence of IkBζ, consistent with the idea that H3K4 methyltransferases are recruited to TSSs by association with initiating or early elongating forms of RNA Pol II (Kayama et al. 2008). Therefore, chromatin remodeling and downstream events (including NF-κB recruitment and loading of the preinitiation complex) can be genetically dissected. The simplest and most intuitive model that can be proposed on the basis of the data described previously is that remodeling exposes NF-κB binding sites and makes them available for binding. However, the fact that remodeling is not followed by NF-κB recruitment in the absence of IkBζ suggests the existence of additional intermediate events, whose molecular nature is unknown, occurring between remodeling and factor recruitment.
Overall, the following steps can be identified as part of the process leading to the activation of late primary and secondary inflammatory NF-κB-dependent genes. The first step is the induction or activation of factors that promote chromatin remodeling. The identity of such factors has remained elusive until now: A likely possibility is that they are transcription factors rapidly activated by stimulation and acting mainly or only to prepare the target DNA for NF-κB landing. Remodeling itself is operated mainly or exclusively by Swi/Snf complexes. However, although obviously required, remodeling is not sufficient to precipitate downstream events, as it can be dissociated from NF-κB recruitment and gene activation in cells lacking IkBζ. Remodeling is then followed by NF-κB recruitment, Pol II loading, and H3K4 trimethylation of a few nucleosomes surrounding the TSS. It should be considered that some transcription factors have a marked preference for H3K4me3-positive genomic regions (Guccione et al. 2006); therefore, it is possible that initial Pol II recruitment and H3K4 trimethylation trigger a feed-forward mechanism promoting further NF-κB recruitment and maximal gene induction. In any case, NF-κB recruitment to hundreds of inflammatory genes is the last step of a complex multistep program whose final aim is to relieve NF-κB binding sites from the negative control exerted by chromatin. Coordinated control of chromatin remodeling and NF-κB persistence in the nucleus in response to stimulation may also underlie selectivity in gene induction in response to alternative stimuli or to stimuli of different duration (Hoffmann et al. 2002). For instance, a short-lasting NF-κB activation is by itself incompatible with the induction of RLA genes, as NF-κB will be extruded from the nucleus before remodeling is completed and kB sites made accessible. Moreover, stimulus specificity in gene activation may arise as a consequence of the differential ability of alternative stimuli with similar NF-κB activation potential to trigger the signaling pathways leading to chromatin remodeling.
THE MOLECULAR BASES OF DIFFERENTIAL CHROMATIN ACCESSIBILITY TO NF-κB PROTEINS
The data discussed in the previous sections indicate that chromatin remodeling precedes, and is required for, NF-κB recruitment to a large fraction of NF-κB-dependent genes, which basically include all target genes except the early primary ones. However, although in vivo biochemical data demonstrate that chromatin accessibility to nucleases at regions surrounding NF-κB binding sites increases in response to stimulation and before NF-κB binding, they do not provide any clue about the nature of the chromatin configurations that are nonpermissive for NF-κB binding. In this section, I discuss how the fundamental unit of chromatin organization, the nucleosome, impacts on NF-κB interaction with cognate binding sites.
The general paradigm is that the packaging of DNA with core histones to generate nucleosomes, and the further folding of the nucleosomal chain into higher order fibers with various degrees of compaction, imposes a steric hindrance to transcription factor binding (Kornberg and Lorch 1999). However, transcription factors bind DNA using a large variety of structural motifs (Garvie and Wolberger 2001): Therefore, their ability to bind nucleosomal sites shows a continuous distribution, with some factors completely unable to contact a site contained in a nucleosome, and some others able to bind nucleosomal and naked sites with a comparable affinity. The TATA-box binding protein (TBP) lies at one end of this distribution, as it has no measurable affinity towards nucleosomal sites (Imbalzano et al. 1994). The heat shock factor (HSF1) can bind nucleosomal elements but the affinity is 100-fold lower than that for naked sites (Taylor et al. 1991). Finally, the glucocorticoid receptor is representative of those factors able to bind with similar affinity naked and nucleosomal DNA (Li and Wrange 1995).
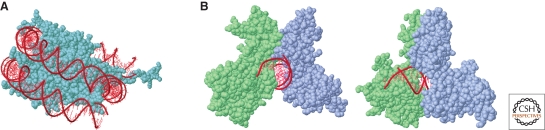
Available structural data are substantially incompatible with the formation of a complex between any NF-κB dimer and high affinity binding sites incorporated in a nucleosome. The Rel homology domain (RHD) of different NF-κB dimers crystallized onto a kB site has a butterfly-like shape, with the wings connected to a cylindrical body of DNA (Chen et al. 1998; Huxford et al. 1999). The amino-terminal domains of the two monomers make base-specific contacts, whereas the carboxy-terminal domains interact with each other and generate the interface between the two subunits. Moreover, they both make nonspecific contacts with the sugar-phosphate backbone of the DNA. The NF-κB dimer does not completely encircle the DNA; however, the DNA-binding cleft delimited by the two amino-terminal domains is clearly too narrow to accommodate the surface of a nucleosome (Fig. 2) (Natoli et al. 2005). Therefore, the ensuing prediction is that nucleosomal sites cannot be bound by NF-κB and that the incorporation of a kB site in a nucleosome is completely sufficient to prevent binding. Surprisingly, experimental data contradict this prediction. Incorporation of kB sites in a nucleosome causes no detectable reduction in the affinity for NF-κB, suggesting that nucleosomes by themselves are transparent to NF-κB (Angelov et al. 2004). Importantly, high affinity binding to nucleosomal sites is independent of the position of the NF-κB binding site relative to the dyad axis of the nucleosome (Angelov et al. 2004). This observation is very relevant in terms of understanding the mechanisms of NF-κB binding to nucleosomes. In fact, when a transcription factor binding site is placed close to the dyad axis of the nucleosome, inhibition of binding is stronger than when the same site is placed in a peripheral location. This difference is because of the spontaneous and transient (on a millisecond time scale) uncoiling of DNA from the periphery of the nucleosome, followed by its quick reassociation with histones (Widom 1998). These “breathing motions” of mono-nucleosomal DNA occur almost exclusively at the peripheral edge of the nucleosome and are strongly reduced or completely absent close to the dyad axis. Although a transient “site exposure” may occur at peripherally located sites and explain their accessibility even to transcription factors encircling DNA, this is not the case for internally located sites. Moreover, it is unclear if and how these breathing motions occur in vivo in the content of nucleosomal chains. In any case, it is safe to conclude that NF-κB binding to nucleosomal sites does not occur as a consequence of the spontaneous uncoiling of DNA from the surface of the nucleosome, as in this case a marked reduction or complete loss of affinity for internally located sites and a marginal or no reduction for peripheral sites would be observed.
Figure 2.
Structural views of the nucleosome and NF-κB. (A) Nucleosome structure showing the DNA surrounding the histone octamer. (B) The structure of an NF-κB dimer (p65/p50) crystallized onto an NF-κB binding site with front (left) and side (right) views. p50 is shown in green, p65 in gray, and DNA in red. (From Natoli et al. Nature Immunol. 6, 439, 2005; Nature Publishing Group.)
Since in vitro mono-nucleosomal kB sites are as accessible to NF-κB proteins as the naked ones, the requirement for chromatin remodeling for NF-κB binding and activation of all RLA genes in vivo cannot be accounted for by the simple wrapping of kB sites around histone octamers. Two possibilities can be envisioned: First, although nucleosomes by themselves are transparent to NF-κB, compacted nucleosomal chains may create an efficient barrier to binding. In this case, remodeling would not result in the simple movement of one or a few nucleosomes or in the release of DNA from the nucleosomal surface, but it would consist in a more extensive reconfiguration of chromatin at the promoters and TSSs of RLA genes. The second possibility is that the nucleosomes controlling NF-κB recruitment have special properties that make them able to efficiently block the interaction of NF-κB with cognate sites. Nucleosomes in the vicinity of TSSs are extensively modified by a plethora of post-translational modifications (Kouzarides 2007). Some of them alter the intrinsic ability of nucleosomes to make internucleosomal interactions and generate compact fibers. For instance, acetylation of the amino-terminal histone tails neutralizes their positive charge and reduces the attractive interactions with the negatively charged DNA (both on the same and on adjacent nucleosomes) (Kornberg and Lorch 1999). Severely hypoacetylated nucleosomes bearing tails folded back on the nucleosomal DNA may be efficient at occluding kB sites, and nucleosome remodeling may act to release DNA from the nucleosomal surface. An alternative possibility is that the incorporation of histone variants in place of canonical core histones may affect the accessibility of NF-κB binding sites. When incorporated in nucleosomes, the macroH2A variant (which contains an extended carboxy-terminal macro domain), can completely block access to centrally located but not peripheral kB sites in vitro (Angelov et al. 2003). This effect depends on the presence of the macro domain, which is also required for inhibition of chromatin remodeling and transcriptional activation by macroH2A. In a specific case, this macroH2A-dependent mechanism of kB site occlusion has been shown to occur in vivo (Agelopoulos and Thanos 2006). However, the genomic distribution of macroH2A-containing nucleosomes is still debated, and it is unknown whether macroH2A is commonly incorporated in nucleosomes in the vicinity of TSSs and used for dynamic regulatory purposes.
A piece of information that is still unavailable and is likely necessary to address several of the issues discussed previously, is a high-resolution map of nucleosome distribution (and modifications associated with individual nucleosomes) at the regulatory regions of NF-κB-dependent genes, including promoters, TSS, and distant enhancers. The current understanding of the rules dictating nucleosome positions is rather primitive and therefore algorithms with a strong predictive ability are not available yet (Segal et al. 2006). However, it is easy to predict that the exponential progress in this area, mainly fueled by high-throughput multiparallel next-generation sequencing technologies, will lead to the solution of this problem in a relatively short time, in spite of the complexity and high content in repeats of mammalian genomes. The main issue is to understand to what extent the underlying genomic sequence controls the position of individual nucleosomes and how well-positioned nucleosomes impact on the accessibility of DNA binding sites. Definitive maps at nucleotide resolution have just been reported in yeast (Mavrich et al. 2008a) and Drosophila (Mavrich et al. 2008b), and in both cases they are consistent with the idea that only nucleosomes in the vicinity of/surrounding TSSs are well-positioned, in a manner possibly dictated by the DNA sequence. Positioned nucleosomes at these locations then act as physical barriers imposing the position to adjacent nucleosomes (Mavrich et al. 2008a). Such barrier effect is progressively lost, moving away from the positioned nucleosomes. In the specific case of the transcriptional responses controlled by NF-κB, only anecdotic reports are available to date, and in some cases they are compatible with the idea that nucleosomes may physically occlude NF-κB binding sites. For instance, in macrophages, a positioned nucleosome encompassing the cis-regulatory elements required for the induction of Il12b gene transcription is quickly remodeled in response to activation (Weinmann et al. 1999). Although Il12b induction requires cRel, remodeling is cRel independent (Weinmann et al. 2001). Therefore, it is possible to envision a nucleosome-mediated sequential mechanism of cooperation between early acting transcription factors and NF-κB: Factors binding to repressive chromatin in the promoter (or at a distant regulatory site in an accessible conformation) drive chromatin decompaction and/or nucleosomal changes, eventually making the promoter accessible to NF-κB.
RESTRICTING THE SEARCH SPACE FOR NF-κB: ANOTHER ROLE FOR NUCLEOSOMAL INCORPORATION OF NF-κB SITES?
A major and long-standing issue in transcriptional regulation is to understand how transcription factors locate relevant binding sites in the genome (von Hippel and Berg 1989; Halford and Marko 2004). This issue is particularly relevant for inducible factors like NF-κB, which must be able to contact binding sites in a very short time after nuclear entry: Induction of some NF-κB target genes occurs almost instantly after NF-κB nuclear translocation, even if the genomic search space is immense. The kB site is conventionally represented as G−5G−4G−3R−2N−1N0Y+1Y+2 C+3C+4 (R = purine, N = any nucleotide, Y = pyrimidine) or by position weight matrixes showing the probability that each position in the site is occupied by a given nucleotide. A definitive estimate of how frequently kB sites appear in mammalian genomes is unavailable and it is expected that a fraction of them will be species-specific. The 256 variants of the most restrictive kB site consensus sequence in which the first (G−5G−4G−3) and the last (C+3C+4) nucleotides are fixed, are found at over 11,000 locations on the smallest human chromosome (chr22), accounting for about 1/67th of the human genome (Udalova et al. 2002). This leads to an estimate of a minimal number of human genomic kB sites in the order of magnitude of almost 106. The requirement for nucleosome remodeling machines for NF-κB binding and activation of some NF-κB target genes suggests, as described above, that incorporation in chromatin may efficiently prevent NF-κB binding. It can be inferred that a simple way to reduce the number of kB sites competing for available nuclear NF-κB molecules is to wrap a fraction of them (including nonfunctional sites and sites belonging to NF-κB target genes with restricted/tissue-specific expression) in nonaccessible chromatin contexts.
Whether this is really the case is not yet known. However, the advent of ChIP sequencing should allow this point to be directly addressed, and specifically, it should allow the fraction of identifiable genomic sites that are in fact contacted by NF-κB in a given cell type and/or experimental condition to be detected. Initial data obtained with high throughput approaches (Schreiber et al. 2006; Lim et al. 2007) have not specifically addressed this point and therefore no definitive conclusion can be safely drawn at this stage.
It should also be considered that the average residence time of NF-κB on high affinity binding sites in vivo is in the time scale of seconds (Bosisio et al. 2006): Therefore, binding to nonfunctional genomic sites will not lead to the sequestration of NF-κB molecules, and exploiting the insulating properties of chromatin to reduce the representation of binding-competent kB sites may in fact not be necessary.
FUTURE PERSPECTIVES
Our current understanding of the interplay between NF-κB, chromatin organization of the genomic information, and transcriptional output allows the definition of some features of the NF-κB-dependent transcriptional responses that are in part (or largely) chromatin-regulated:
– the kinetic complexity of induction of NF-κB target genes;
– the stimulus-specific induction of target genes;
– the cooperative effects (mediated by nucleosomes) between NF-κB and partner transcription factors.
The definitive understanding of the mechanistic bases of the interaction between chromatin and NF-κB will require:
– the generation of comprehensive nucleosome occupancy maps at NF-κB target genes and the definition of the underlying sequence determinants;
– the identification of the transcription factor synergizing with NF-κB at removing the chromatin-dependent hindrance to gene activation;
– the identification of the nuclear enzymatic activities that are required to overcome the block imposed by chromatin to the activation of NF-κB target genes. The possibility that some of these enzymes may represent drug targets is rather obvious and a scenario in which inhibition of these enzymes may lead to therapeutic down-regulation of selected NF-κB target genes is less futuristic than it may seem.
Footnotes
Editors: Louis M. Staudt and Michael Karin
Additional Perspectives on NF-κB available at www.cshperspectives.org
REFERENCES
- Agelopoulos M, Thanos D 2006. Epigenetic determination of a cell-specific gene expression program by ATF-2 and the histone variant macroH2A. EMBO J 25:4843–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D, Lenouvel F, Hans F, Muller CW, Bouvet P, Bednar J, Moudrianakis EN, Cadet J, Dimitrov S 2004. The histone octamer is invisible when NF-κB binds to the nucleosome. J Biol Chem 279:42374–42382 [DOI] [PubMed] [Google Scholar]
- Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S 2003. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 11:1033–1041 [DOI] [PubMed] [Google Scholar]
- Becker PB, Horz W 2002. ATP-dependent nucleosome remodeling. Annu Rev Biochem 71:247–273 [DOI] [PubMed] [Google Scholar]
- Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G 2006. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-κB-dependent gene activity. Embo J 25:798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G 1998. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature 391:410–413 [DOI] [PubMed] [Google Scholar]
- Garvie CW, Wolberger C 2001. Recognition of specific DNA sequences. Mol Cell 8:937–946 [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B 2006. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol 8:764–770 [DOI] [PubMed] [Google Scholar]
- Halford SE, Marko JF 2004. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res 32:3040–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta H, Kato A, Todokoro K 2001. Isolation of a novel interleukin-1-inducible nuclear protein bearing ankyrin-repeat motifs. J Biol Chem 276:12485–12488 [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D 2002. The IκB-NF-κB signaling module: Temporal control and selective gene activation. Science 298:1241–1245 [DOI] [PubMed] [Google Scholar]
- Huxford T, Malek S, Ghosh G 1999. Structure and mechanism in NF-κ B/I κ B signaling. Cold Spring Harb Symp Quant Biol 64:533–540 [DOI] [PubMed] [Google Scholar]
- Imbalzano AN, Kwon H, Green MR, Kingston RE 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481–485 [DOI] [PubMed] [Google Scholar]
- Janeway CA Jr, Medzhitov R 2002. Innate immune recognition. Annu Rev Immunol 20:197–216 [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S 2007. TLR signaling. Semin Immunol 19:24–32 [DOI] [PubMed] [Google Scholar]
- Kayama H, Ramirez-Carrozzi VR, Yamamoto M, Mizutani T, Kuwata H, Iba H, Matsumoto M, Honda K, Smale ST, Takeda K 2008. Class-specific regulation of pro-inflammatory genes by MyD88 pathways and IκBζ. J Biol Chem 283:12468–12477 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285–294 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- Li Q, Wrange O 1995. Accessibility of a glucocorticoid response element in a nucleosome depends on its rotational positioning. Mol Cell Biol 15:4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CA, Yao F, Wong JJ, George J, Xu H, Chiu KP, Sung WK, Lipovich L, Vega VB, Chen J, et al. 2007. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Mol Cell 27:622–635 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF 2008a. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. 2008b. Nucleosome organization in the Drosophila genome. Nature 453:358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Saccani S, Bosisio D, Marazzi I 2005. Interactions of NF-κB with chromatin: The art of being at the right place at the right time. Nat Immunol 6:439–445 [DOI] [PubMed] [Google Scholar]
- Nau GJ, Richmond JF, Schlesinger A, Jennings EG, Lander ES, Young RA 2002. Human macrophage activation programs induced by bacterial pathogens. Proc Natl Acad Sci 99:1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev 20:282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G 2001. Two waves of nuclear factor κB recruitment to target promoters. J Exp Med 193:1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G 2002. p38-Dependent marking of inflammatory genes for increased NF-κB recruitment. Nat Immunol 3:69–75 [DOI] [PubMed] [Google Scholar]
- Schreiber J, Jenner RG, Murray HL, Gerber GK, Gifford DK, Young RA 2006. Coordinated binding of NF-κB family members in the response of human cells to lipopolysaccharide. Proc Natl Acad Sci 103:5899–5904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442:772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor IC, Workman JL, Schuetz TJ, Kingston RE 1991. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: Differential function of DNA-binding domains. Genes Dev 5:1285–1298 [DOI] [PubMed] [Google Scholar]
- Udalova IA, Mott R, Field D, Kwiatkowski D 2002. Quantitative prediction of NF-κB DNA-protein interactions. Proc Natl Acad Sci 99:8167–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel PH, Berg OG 1989. Facilitated target location in biological systems. J Biol Chem 264:675–678 [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665–675 [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Mitchell DM, Sanjabi S, Bradley MN, Hoffmann A, Liou HC, Smale ST 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat Immunol 2:51–57 [DOI] [PubMed] [Google Scholar]
- Widom J 1998. Structure, dynamics, and function of chromatin in vitro. Annu Rev Biophys Biomol Struct 27:285–327 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, Kaisho T, Kuwata H, Takeuchi O, Takeshige K, et al. 2004. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430:626–630 [DOI] [PubMed] [Google Scholar]