Abstract
Naphthalimides, particularly amonafide and 2-(2-dimethylamino)-6-thia-2-aza-benzo[def]chrysene-1,3-diones (R16), have been identified to possess anticancer activities and to induce G2-M arrest through inhibiting topoisomerase II accompanied by Chk1 degradation. The current study was designed to precisely dissect the signaling pathway(s) responsible for the naphthalimide-induced cell cycle arrest in human colon carcinoma HCT116 cells. Using phosphorylated histone H3 and mitotic protein monoclonal 2 as mitosis markers, we first specified the G2 arrest elicited by the R16 and amonafide. Then, R16 and amonafide were revealed to induce phosphorylation of the DNA damage sensor ataxia telangiectasia-mutated (ATM) responding to DNA double-strand breaks (DSBs). Inhibition of ATM by both the pharmacological inhibitor caffeine and the specific small interference RNA (siRNA) rescued the G2 arrest elicited by R16, indicating its ATM-dependent characteristic. Furthermore, depletion of Chk2, but not Chk1 with their corresponding siRNA, statistically significantly reversed the R16- and amonafide-triggered G2 arrest. Moreover, the naphthalimides phosphorylated Chk2 in an ATM-dependent manner but induced Chk1 degradation. These data indicate that R16 and amonafide preferentially used Chk2 as evidenced by the differential ATM-executed phosphorylation of Chk1 and Chk2. Thus, a clear signaling pathway can be established, in which ATM relays the DNA DSBs signaling triggered by the naphthalimides to the checkpoint kinases, predominantly to Chk2,which finally elicits G2 arrest. The mechanistic elucidation not only favors the development of the naphthalimides as anticancer agents but also provides an alternative strategy of Chk2 inhibition to potentiate the anticancer activities of these agents.
Introduction
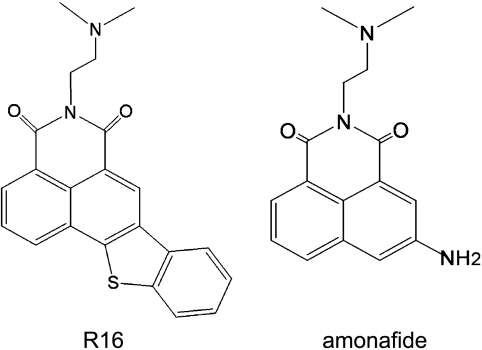
Naphthalimides are important anticancer compounds that bind to DNA by intercalation, showing potent anticancer activity against human cancers preclinically and clinically [1–6]. However, early naphthalimides, for example, amonafide (Figure 1), although effective in its phase 2 clinical trials when administered either alone or in combination, suffered from the dose-limiting bone marrow toxicity caused by the toxic N-acetyl-amonafide generated through N-acetylation by N-acetyltransferase 2 (NAT2). Especially unacceptably, this hematotoxicity was highly variable and unpredictable owing to NAT2, the polymorphic enzyme with differential activity between individuals [1,4–6]. To get rid of such unpredictable thus unacceptable toxicity, persistent efforts have been put into eliminating the site on naphthalimides catalyzed by NAT2, which resulted in the emergence of a new generation of naphthalimide analogs, represented by UNBS5162 [2,3] and 2-(2-dimethylamino)-6-thia-2-aza-benzo[def]chrysene-1,3-diones (R16; Figure 1) [7]. These new-generation naphthalimide analogs not only remove the potential unpredictable toxicity risks of the early ones but also retain their anticancer activity. Moreover, some of them such as UNBS5162 are conferred to distinct mechanisms of action [3]. Currently, UNBS5162 is undergoing phase 1 clinical trials in cancer patients with advanced solid tumors or lymphoma [2,3], which makes it reasonable to expect that this class of compounds enter clinical anticancer use.
Figure 1.
Chemical structures of amonafide and R16.
Although UNBS5162 uniquely antagonizes CXCL chemokine expression [3], the naphthalimides amonafide and R16 inhibit DNA topoisomerase II (Top2) and trap Top2 cleavable complexes [7]. Also, like other Top2 inhibitors, both amonafide and R16 induce DNA double-strand breaks (DSBs) and arrest cells at the G2-M phase [7]; however, unlike etoposide (VP16) and adriamycin (ADR), they exhibit significant anti-multidrug resistance abilities [7]. In addition, we have shown that R16 induces degradation of Chk1 through the ubiquitin-proteasome pathway and impairs the function of this kinase [8]. We are therefore inspired to further dissect molecular signaling pathway(s) that are used in inducing cell cycle arrest, so as to have a clear understanding of naphthalimides as promising anticancer candidates and to find out manipulable strategies to achieve better efficiency in their potential clinical uses.
Cell cycle checkpoints ensure the orderly and timely progression of some critical events such as DNA replication and chromosome segregation in cells. More importantly, cell cycle arrest may play a critical role in mediating the chemotherapeutic effects of Top2 inhibitors [9]. In response to DNA DSBs, checkpoint signaling pathways are activated to arrest cells at G1/S, S, or G2/M transitions, thus allowing time to repair the damage or, in the case of unrepairable lesions, to execute apoptosis. Ataxia telangiectasia-mutated (ATM) and ATM-Rad3-related (ATR) have been well documented as two apical protein kinases in DNA damage response pathways [10]. Generally, ATM primarily responds to ionizing radiation-induced DNA DSBs, whereas ATR is activated by stalled replication resulting from UV and some genotoxic drugs [11]. Two structurally unrelated but functionally similar protein serine/threonine kinases Chk1 and Chk2 are the substrates of ATM and ATR. Chk2 is phosphorylated by ATM at Thr68 and thus activated [12], whereas Chk1 activation requires phosphorylation at Ser317 and Ser345 catalyzed by ATM and/or ATR [13,14]. The activated checkpoint kinases phosphorylate their substrates such as Cdc25C that regulate the activity of the Cdc2-cylcin B1 complex, a cyclin-dependent kinase complex functioning as an ultimate target of the G2 checkpoint signaling pathway [15].
The present study used amonafide and R16 to define the underlying mechanism by which the naphthalimides induce cell cycle arrest of tumor cells. We first used phosphorylated histone H3 antibody and the mitosis-specific monoclonal antibody mitotic protein monoclonal 2 (MPM-2) as mitosis markers to define that the naphthalimides arrested HCT116 cells at the G2 phase but not at the M phase. Then using pharmacological inhibitors and the small interference RNA (siRNA) technology, we dissected for the first time how amonafide and R16 activated the ATM-Chk2 pathway to arrest cells at the G2 phase. The current findings greatly extend our previous study by elucidating the detailed molecular mechanism of amonafide- and R16-driven cell cycle arrest, which will possibly benefit the clinical trials of the naphthalimides for cancer therapy.
Materials and Methods
Reagents
R16 was synthesized, and its purity was more than 99% [7]. The parental compound amonafide was obtained from the Drug Synthesis and Chemistry Branch, Division of Cancer Treatment, National Cancer Institute (Bethesda, MD). VP16, ADR, camptothecin, hydroxyurea (HU), and caffeine were purchased from Sigma (St Louise, MO). All these compounds except caffeine were dissolved in dimethyl sulfoxide as stock solutions of 10 mM (R16, amonafide, and camptothecin) or 20 mM (VP16 and ADR). Caffeine (80 mM) was dissolved in sterilized water. The stock solutions were kept frozen in aliquot at -20°C and thawed immediately before each experiment.
Cell Culture
Human colorectal carcinoma HCT116 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in McCoy 5A medium (Life Technologies, Grand Island, NE) supplemented with 10% heat-inactivated fetal calf serum (Life Technologies), l-glutamine (2 mM), penicillin (100 IU/ml), streptomycin (100 µg/ml), and HEPES (10 mM, pH 7.4) at 37°C and 5% CO2 in a humid environment.
Flow Cytometry
The distribution of HCT116 cells at different stages in the cell cycle was estimated by flow cytometric DNA analyses. Briefly, 5 x 105 cells were incubated overnight in six-well plates in McCoy 5A medium containing 10% FBS, then treated with or without various concentrations of compounds for indicated times, in untransfected cells or in transfected cells. Cells were harvested and washed with phosphate-buffered saline (PBS; pH 7.4), fixed with 70% ethanol/30% PBS at 4°C. Followed by PBS washing, the pellet was dissolved in RNaseA solution (20 µg/ml) and incubated at 37°C for 15 minutes, stained with propidium iodide (Sigma) for 30 minutes in the dark at room temperature. For each sample at least 1 x 104 cells were analyzed using a FACS-Calibur cytometer (Becton Dickinson, San Jose, CA), and the percentage of cells in each cell cycle phase was calculated using the CELLQUEST and ModFITLT software packages (Becton Dickinson).
Neutral Single-Cell Gel Electrophoresis Assays
DNA DSBs were evaluated using neutral single-cell gel electrophoresis (NSCGE) assays as previously described [16,17]. Briefly, after treatments with the indicated agents, cells were harvested, mixed with low-melting point agarose, layered onto microscope slides precoated with normal-melting point agarose, then solidified, lysed, equilibrated, electrophoresed, and then stained with 4′,6-diamidino-2-phenylindole. Cells were viewed using an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan).
Western Blot Analysis
Standard Western blot analyses were done by using the following appropriate primary antibodies to phospho-Chk1 (Ser345), γ-H2AX, phosphorylated histone H3, and histone H3 (Cell Signaling Technology, Beverly, MA); Chk1 and cyclin B1 (Santa Cruz Biotech, Santa Cruz, CA); phospho-Chk2 (Thr68; R&D systems, Inc, Minneapolis, MN); phospho-ATM (Ser1981) and ATM (Rockland Immunochemicals, Gilbertsville, PA); ATR (Calbiochem, La Jolla, CA); and MPM-2 (Upstate, Lake Placid, NY). Proteins were visualized with peroxidase-coupled secondary antibodies (Calbiochem; diluted 1:2000), using ECL-plus kit from Amersham Biosciences (Buckinghamshire, UK) for detection.
siRNA Transfection
All the siRNA sequences were duplexes of 21-mer RNA with a 2-mer 3′ overhang produced by Genepharma, Co (Shanghai, China). The ATM targeting sequences are 5′-AACATACTACTCAAAGACATT-3′ [18] for ATM siRNA1 and 5′-AAGCAC CAGUCCAGUAUUGGC-3′ [19] for ATM siRNA2, the ATR targeting sequence is 5′-AACCTCCGTGATGTTGCTTGA-3′ [20], the Chk1 targeting sequence is 5′-AAGTTCAACTTGCTGTGAATA-3′ [21], and the Chk2 targeting sequence is 5′-AAGAACCUGAGGACCAAGAAC-3′ [22]. Cells (2 x 105) were incubated overnight in six-well dishes in McCoy 5A medium containing 10% FBS. The medium was replaced with fresh Opti-MEM (Life Technologies) containing siRNA and Oligofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations. After 4 hours, Opti-MEM was changed to McCoy 5A medium containing 10% bovine serum, and incubation was continued for another 24 hours. After exposure to different compounds for indicated times, treated or untreated cells were collected for Western blot analyses for protein expression and for flow cytometry.
Immunofluorescence Analyses
For immunofluorescence analyses, treated or untreated cells growing on coverslips were rinsed with PBS, fixed with 4% paraformaldehyde for 15 minutes and permeabilized with 0.1% Triton X-100 in PBS for 10 minutes. The samples were blocked with TBS/3% BSA for 30 minutes, incubated with the p-ATM (Ser1981) antibody (Rockland Immunochemicals; diluted 1:100) for 60 minutes at room temperature. After three washes with TBS, samples were incubated with fluorescent secondary Alexa Fluor 488 anti-rabbit serum immunoglobulin G (diluted 1:200; Molecular Probes, Eugene, OR) 60 minutes, then washed with TBS and incubated with 4′,6-diamidino-2-phenylindole for 5 minutes. Images were photographed using a Leica TCSSP2 confocal microscope (Leica, Mannheim, German) or an Olympus BX51 fluorescence microscope (Olympus). Quantitation was performed by analyzing at least 100 randomly selected cells per slide.
Results
Naphthalimides Arrest Cell Cycle at the G2 Phase
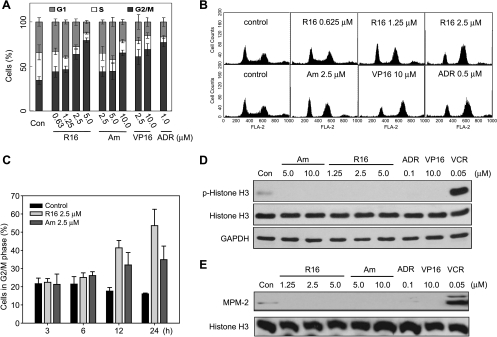
We have demonstrated that both amonafide and R16 trigger significant G2-M arrest in human promyelocytic leukemia HL-60 cells [7]. We have also found that R16 induces the degradation of Chk1 protein in various solid tumor cells including human colon cancer HCT116, rhabdomyosarcoma Rh30, lung cancer A549, and cervical cancer HeLa cells and revealed the involvement of the ubiquitin-proteasome pathway in this action of R16 in HCT116 cells [8]. We thus ask how and why these naphthalimides impact on the cell cycle progression in solid tumor cells. To answer the questions, we used HCT116 cells because the cells have been successfully used to investigate the effect of R16 on Chk1 protein, one of the most important cell cycle checkpoint kinases. Treatments with amonafide or R16 led to prominent G2-M arrest in concentration- and time-dependent manners in HCT116 cells (Figure 2, A–C). To precisely define whether the G2-M arrest is G2 or M phase arrest, we used two well-characterized mitosis markers phosphorylated histone H3 [23] and MPM-2 (a monoclonal antibody interacting with mitosis-specific phosphorylated proteins [24]). As expected, the mitosis inhibitor vincristine dramatically upregulated phosphorylated histone H3 and MPM-2, both of which, however, were undetectable in the cells exposed to all the Top2 inhibitors R16, amonafide, VP16, and ADR at the conditions of effectively arresting cell cycle progression (Figure 2, D and E). The data indicate that the naphthalimides R16 and amonafide arrest cell cycle at the G2 phase not at the M phase in HCT116 cells. Moreover, this result was further confirmed by using human colon cancer HT29 and cervical cancer HeLa cells.
Figure 2.
R16 arrests HCT116 cells at G2 phase. (A and B) R16 and amonafide increased the G2/M population of HCT116 cells in a concentration-dependent manner. HCT116 cells were treated with different compounds at indicated concentrations for 24 hours and then subjected to flow cytometry. The data were expressed as mean ± SD from three independent experiments (A) and the typical histograms were shown (B). (C) R16 and amonafide increased the G2/M population in a time-dependent fashion. HCT116 cells were treated with R16 (2.5 µM) or amonafide (2.5 µM) for the different periods and were then subjected to flow cytometry. The data were expressed as mean ± SD from three independent experiments. (D and E) Treatments with R16 or amonafide did not increase the molecular markers of mitosis arrest phosphorylated histone H3 (p-Histone H3) and MPM-2. HCT116 cells were treated with indicated concentrations of the tested compounds for 24 hours and were then subjected to Western blot analysis. All the Top2 inhibitors (R16, amonafide, VP16, and ADR) did not increase p-Histone H3 (D) and MPM-2 (E) as the mitosis inhibitor vincristine did (D and E). The images were representative of three separate experiments.
DNA DSBs Contribute to G2 Arrest Caused by R16
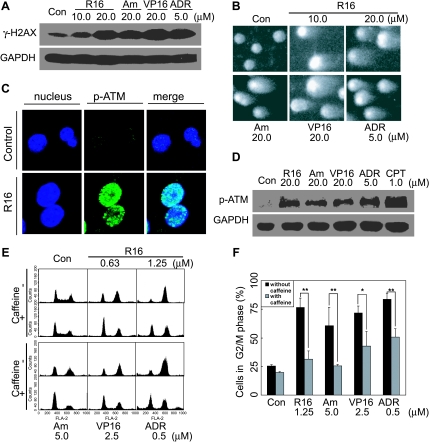
The ability of amonafide and R16 to induce DNA DSBs by inhibiting Top2 has been shown in HL-60 cells [7]. To investigate the mechanism of G2 arrest elicited by naphthalimides, we first validated this ability of amonafide and R16 by detecting the levels of the phosphorylated histone γ-H2AX. HCT116 cells treated with 20 µM R16 or 20 µM amonafide for 2 hours exhibited comparable increased phosphorylation levels of γ-H2AX with those of the cells exposed to the references VP16 or ADR (Figure 3A). This result was further directly confirmed by using the NSCGE (comet assays), a widely used method for measuring cellular DNA DSBs. The exposure to R16 or amonafide for 2 hours produced typical comet tails in HCT116 cells, an evident indicator of DNA DSBs (Figure 3B). Moreover, both amonafide and R16 induced the formation of p-ATM (Ser1981) foci in the treated cells (Figure 3C) and the enhanced levels of phosphorylated ATM (Ser1981; Figure 3D), indicating that the DNA DSBs activated the ATM signaling pathway. More importantly, caffeine, a well-known ATM/ATR inhibitor, effectively prevented the G2 arrest induced by amonafide or R16 (Figure 3, E and F). The data collectively indicate that amonafide and R16 trigger DNA DSBs that contribute to the G2 arrest in HCT116 cells.
Figure 3.
DNA DSBs contribute to R16-mediated G2 arrest. (A and B) R16 and amonafide induced DNA DSBs. HCT116 cells were treated with different compounds at indicated concentrations for 2 hours and were then subjected to Western blot analysis to detect the level of γ-H2Ax (A) or to NSCGE assays to detect the broken DNA (comet tails; B). (C) R16 (10 µM for 2 hours) induced the formation of p-ATM foci in HCT116 cells detected by immunofluorescence assays. (D) R16 and amonafide (at indicated concentrations for 2 hours) increased the phosphorylation of ATM detected by Western blot analysis. (E and F) The ATM/ATR inhibitor caffeine (2 mM, 30 minutes) abrogated R16- and amonafide-induced (24 hours) G2 arrest in HCT116 cells detected by flow cytometry. The data were expressed as the typical histograms (E) or as mean ± SD from three independent experiments (F), *P < .05; **P < .01.
ATM Is Indispensable for R16-Driven G2 Arrest
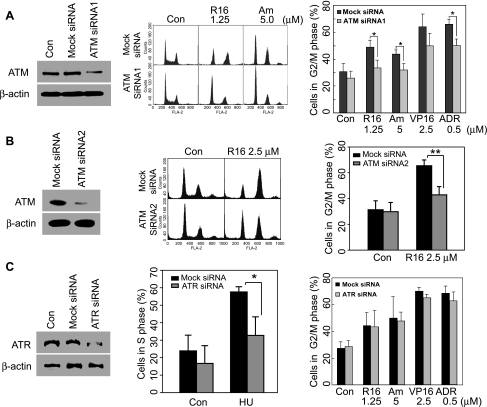
Both ATM and ATR have been reported to activate cell cycle checkpoints and relay signals to the downstream kinases including Chk1 and Chk2. To examine whether the G2 arrest induced by naphthalimides is dependent of ATM and ATR, we knocked down ATM and ATR with their respective specific siRNA. ATM-targeting siRNA1 downregulated the protein expression of ATM in transfected HCT116 cells, and at the same time, apparently abated R16-induced G2 arrest, as indicated by the fact that ATM siRNA1 decreased G2 population from 49.02% to 33.56% in response to the treatment with R16 (Figure 4A). In amonafide-treated cells, similar abrogation was observed (Figure 4A). At the same time, neither the level of ATM nor the cell cycle distribution was affected in mock siRNA-transfected cells. In addition, another ATM siRNA, ATM siRNA2, was used to further confirm the indispensability of ATM in R16-induced G2 arrest. Silencing ATM with ATM siRNA2 (100 nM) effectively attenuated the G2 population in R16 (2.5 µM, 24 hours)-treated cells, from 65.44% to 42.87% (Figure 4B).
Figure 4.
Depletion of ATM but not ATR abates the G2 arrest elicited by R16 and amonafide. (A and B) ATM knockdown rescued HCT116 cells from the G2 arrest triggered by R16 and amonafide. Cells were transfected with 100 nM ATM siRNA1 (A) or 100 nM ATM siRNA2 (B) for 24 hours before being exposed to R16 or amonafide for the following 24 hours. Then the cell cycle distribution was analyzed by flow cytometry. Left panel indicates the efficiency of ATM depletion; middle panel, typical histograms; right panel, mean ± SD from three separate experiments. *P < .05; **P < .01. (C) ATR knockdown with ATR siRNA (100 nM) did not prevent the G2 arrest induced by R16 and amonafide although it effectively abated the S arrest elicited by HU. HCT116 cells were transfected with 100 nM ATR siRNA for 24 hours before the treatment with different compounds at the indicated concentrations for the following 24 hours. Left panel indicates the efficiency of ATR depletion; middle panel, S arrest and HU; right panel, G2/M arrest and R16, amonafide, VP16, and ADR. The data were expressed as mean ± SD from three independent experiments; *P < .05.
Furthermore, the role of ATR, a kinase related to ATM, in R16- or amonafide-caused G2 arrest was investigated. Silencing ATR with ATR-specific siRNA decreased the level of ATR protein in HCT116 cells but imposed minimal effects on the cell cycle distribution in R16- and amonafide-treated cells (Figure 4C). In contrast, transfection with the same ATR siRNA attenuated the S arrest induced by HU (0.5 mM, 24 hours) in HCT116 cells (Figure 4C), revealing the sufficient reduction of ATR function.
Collectively, these data demonstrate that G2 arrest driven by R16 and amonafide is ATM-dependent in HCT116 cells.
R16-Induced G2 Arrest Depends on Chk2
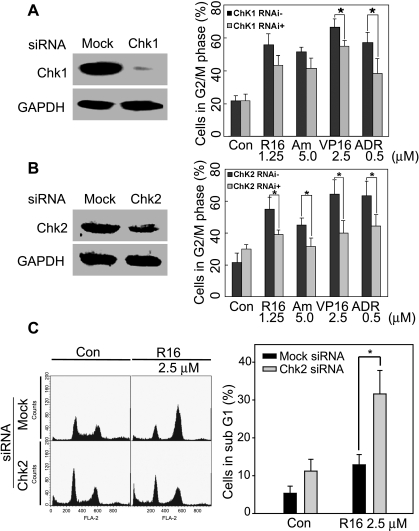
As immediate substrates of ATM, the cell cycle checkpoint kinases Chk1 and Chk2 are responsible for relaying the cell cycle effects of ATM [12]. To investigate the contribution of Chk1 and Chk2 to R16- and amonafide-triggered G2 arrest, we depleted Chk1 and Chk2 with their corresponding specific siRNA, respectively (Figure 5, A and B), and then examined the cell cycle progression in HCT116 cells. Silencing Chk1 slightly eased the G2 arrest elicited by R16 and amonafide (P > .05; Figure 5A). In contrast, depletion of Chk2 reversed R16- and amonafide-induced G2 arrest with significant statistical difference (P < .05; Figure 5B). However, knockdown of either Chk1 or Chk2 statistically significantly diminished the increment of G2-M population induced by the other two classic Top2 inhibitors VP16 and ADR (Figure 5, A and B). Meanwhile, the cells bearing downregulated Chk2 were more susceptible to the treatment with R16. As demonstrated in Figure 5C, at the concentration of 2.5 µM, R16 caused more sub-G1 population (31.60%) in Chk2-depleted cells than in the mock siRNA-transfected cells (12.88%). These data suggest a predominant role of Chk2 over Chk1 in the process of amonafide- and R16-elicited G2 arrest.
Figure 5.
Depletion of Chk2 but not Chk1 prevents the G2 arrest induced by R16 and amonafide. (A) Chk1 knockdown with its siRNA (100 nM) did not decrease the G2 arrest triggered by R16 and amonafide statistically significantly although reducing the G2 arrest triggered by the other two Top2 inhibitors VP16 and ADR. Left panel indicates the efficiency of Chk1 depletion; right panel, mean ± SD from three separate experiments. *P < .05. (B and C) Silencing of Chk2 diminished G2 arrest but potentiated apoptosis induction by R16 significantly. HCT116 ells were treated with the compounds at the indicated concentrations for 24 hours after Chk2 silencing with its siRNA (100 nM, 24 hours). Then the cells were subjected to flow cytometry. The percentage of G2 population (B) or apoptotic cells (C) were expressed as mean ± SD from three separate experiments. *P < .05. B: Left panel indicates the efficiency of Chk2 depletion; right panel, mean ± SD of G2 population (%). *P < .05. C: Left panel indicates typical histograms to show the sub-G1 population (apoptotic cells); right panel, mean ± SD of apoptotic cells (%). *P < .05.
Chk2 and Chk1 Are Differentially Phosphorylated by ATM in Response to the Naphthalimides
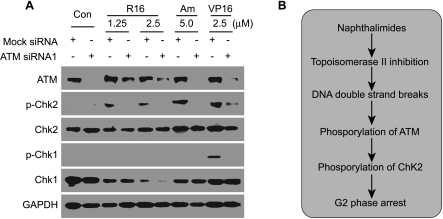
To further characterize the differential contribution of Chk1 and Chk2 to the naphthalimide-elicited G2 arrest, we compared the phosphorylation of Chk1 and Chk2. In all the groups treated with different Top2 inhibitors (R16, amonafide, or VP16) for 24 hours, Chk2 protein was phosphorylated as indicated by the elevation of p-Chk2 levels, which was antagonized effectively by the pretreatment with ATM siRNA1 (Figure 6A). Under the same experimental treatment, however, phosphorylated Chk1 was undetectable in response to both R16 and amonafide, although Chk1 was normally phosphorylated by the treatment with VP16, which was also prevented by the pretreatment with ATM siRNA1 (Figure 6A). In addition, we further confirmed that the naphthalimides induced the degradation of Chk1 that has been reported in our previous article [8]. Our data indicate that Chk2 and Chk1 are differentially phosphorylated by ATM in response to the naphthalimides.
Figure 6.
R16 induces differential phosphorylation of Chk2 and Chk1. (A) Chk2 phosphorylation triggered by R16 was blocked by ATM siRNA1 (100 nM). HCT116 cells were treated with R16, amonafide, or VP16 at the indicated concentrations for 24 hours after being transfected with 100 nM ATM siRNA1 for 24 hours. Then the cells were subjected to Western blot analysis for Chk1, Chk2, p-Chk1, and p-Chk2. The images were representative of three independent experiments. (B) Schematic presentation of the possible mechanistic link between the inhibition of Top2 and G2 arrest elicited by the naphthalimides.
Discussion
Both amonafide and R16 are distinguished from other naphthalimide analogs for their anticancer activity [4,5,7]. However, the molecular mechanisms remained to be fully elucidated. We have revealed in detail that both R16 and amonafide target human Top2, thus generating DNA DSBs in HL-60 cells and leading to G2-M arrest [7]. We have also demonstrated that R16 induces degradation of Chk1 through the ubiquitin-proteasome pathway, which contributes to its anticancer activity [8]. In the present study, we further specified their cell cycle arrest precisely at the G2 phase and, more importantly, established the possible molecular link between DNA DSBs and G2 arrest elicited by R16 and amonafide.
Correctly defining cell cycle arrest by anticancer drugs is important because it helps clarify the mechanisms of their anticancer activities. Moreover, specificity of anticancer drugs in cell cycle arrest has been taken as a key basis for their combination therapy [25]. Using the mitosis-specific marker phosphorylated histone H3 and MPM-2 [24], we demonstrated that few cells were at the M phase in R16- and amonafide-treated groups HCT116 cells, which corroborated the G2 arrest elicited by these two compounds.
The G2 checkpoint is often activated by DNA DSBs lesions. So we further confirmed that R16 and amonafide induced DNA DSBs in HCT116 cells by revealing that both of the compounds enhanced cellular levels of the phosphorylated histone γ-H2AX and caused the generation of “comet tails” [26], as they did in HL-60 [7]. After the generation of DNA DSBs, ATM, the main sensor of DNA DSBs, was activated by phosphorylation at its serine 1981 in R16- or amonafide-treated cells. The pan phosphoinositide 3-kinase inhibitor caffeine and its specific ATM siRNA (including ATM siRNA1 and ATM siRNA2), but not ATR siRNA, rescued the G2 arrest triggered by R16 and amonafide, showing a causal link between ATM activation and G2 arrest in the treated cancer cells. Although both the checkpoint kinases Chk1 and Chk2 are the immediate substrates of ATM [12,14], our data show that ATM differentially activates them by phosphorylation and that these two kinases also differentially contribute to the G2 arrest induced by R16 and amonafide in HCT116 cells. Thus, our data reveal the mechanism whereby R16 and amonafide induce G2 arrest and the existence of a mechanistic link between the naphthalimide-caused DNA DSBs signals and the activation of the ATM-Chk2 pathway leading to the G2 arrest (Figure 6B).
This signaling pathway to G2 arrest used by R16 as well as amonafide is noticeably different from that used by other classic Top2 inhibitors such as VP16 and ADR. Both VP16 and ADR activate Chk1 and Chk2 similarly by phosphorylation and subsequently lead to G2 arrest [27,28]. In contrast, R16 as well as amonafide differentially phosphorylates/activates Chk1 and Chk2, consequently, resulting in G2 arrest in a manner predominantly dependent on Chk2 than on Chk1. Such differences seem to derive primarily from differential degradation of Chk1 protein: the naphthalimides induce degradation of Chk1 through the ubiquitin-proteasome pathway (Figure 6A) [8], whereas the classic Top2 inhibitors such as VP16 do not (Figure 6A). Noticeably, these differences are of potential clinical importance. Inhibitors of Chk1 and Chk2 have been intensively investigated to be used to potentiate anticancer efficacy of DNA-damaging agents including Top2 inhibitors or to circumvent drug resistance to these agents [29–31]. Our data strongly suggest that both Chk1 and Chk2 inhibitors could be used to sensitize tumor cells to the classic Top2 inhibitors as reported [31–33]; however, only Chk2 inhibitors could be suitable for the combination with the naphthalimides owing to Chk1 degradation resulting from treatments with R16 and amonafide and to additional adverse effects possibly deriving from the administration of Chk1 inhibitors.
Of note, a recently reported naphthalimide analog UNBS5162 has been shown to be a pan-antagonist of CXCL chemokine expression, to interfere in vivo with amino acid metabolism, and to trigger proautophagic and senescence-like effects [2,3], which are significantly different from the mechanisms of action of amonafide and R16. This is interesting because most of the CXCL chemokines can promote angiogenesis, and thus, it is understandable that UNBS5162 displays antiangiogenic properties in vivo in hormone-refractory prostate cancer models [2,3]. Because UNBS5162 and R16 fall into the same class in their chemical structures, to further examine whether R16 affects the CXCL chemokines and whether UNBS5162 impacts the Top2-DNA-cell cycle axis may also be beneficial to fully understanding their modes of action.
In summary, our present study demonstrates that the naphthalimides R16 and amonafide induce DNA DSBs, then trigger the ATM-activated Chk2-executed pathway and finally lead to G2 phase arrest in HCT116 cells while resulting in Chk1 degradation. These features are different from the classic Top2 inhibitors such as VP16. Apparently, such differences give new insights into the mechanisms of the cell cycle arrest triggered by Top2 inhibitors on one hand, and sufficiently understanding these mechanisms forms the crucial basis for the safe, effective combination of inhibitors of Chk1 or Chk2 with those different Top2 inhibitors in potential clinical settings on the other.
Abbreviations
- R16
2-(2-dimethylamino)-6-thia-2-aza-benzo[def]chrysene-1,3-diones
- Top2
topoisomerase II
- DNA DSBs
DNA double-strand breaks
- VP16
etoposide
- ADR
adriamycin
- PBS
phosphate-buffered saline
- Chk
checkpoint kinase
- ATM
ataxia telangiectasia-mutated
- ATR
ATM and Rad3-related
Footnotes
This work was supported by grants from the National Natural Science Foundation of China (nos. 30772588 and 30721005) and the Science and Technology Commission of Shanghai Municipality (no. 08DZ1980200).
References
- 1.Ingrassia L, Lefranc F, Kiss R, Mijatovic T. Naphthalimides and azonafides as promising anti-cancer agents. Curr Med Chem. 2009;16:1192–1213. doi: 10.2174/092986709787846659. [DOI] [PubMed] [Google Scholar]
- 2.Quaquebeke EV, Mahieu T, Dumont P, Dewelle J, Ribaucour F, Simon G, Sauvage S, Gaussin JF, Tuti J, Yazidi ME, et al. 2,2,2-Trichloro-N-({2-[2-(dimethylamino)ethyl]-1,3-dioxo-2,3-dihydro-1H-benzo[de]isoquinolin-5-yl}carbamoyl)acetamide (UNBS3157), a novel nonhematotoxic naphthalimide derivative with potent antitumor activity. J Med Chem. 2007;50:4122–4134. doi: 10.1021/jm070315q. [DOI] [PubMed] [Google Scholar]
- 3.Mijatovic T, Mahieu T, Bruyère C, Nève ND, Dewelle J, Simon G, Dehoux MJM, Aar E, Haibe-Kains B, Bontempi G, et al. UNBS5162, a novel naphthalimide that decreases CXCL chemokine expression in experimental prostate cancers. Neoplasia. 2008;10:573–586. doi: 10.1593/neo.08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erba HP, Rizzieri DA, O'Donnell MR, Lundberg AS, Ajami AM, Rampersad AD, Capizzi RL. Amonafide and ara-C treatment for secondary acute myeloid leukemia (sAML) J Clin Oncol. 2007;25:7065. [Google Scholar]
- 5.Kuhn JG, Burris HA, Jones SF, Hein DW, Willcutt NT, Greco FA, Thompson DS, Meluch AA, Schwartz RS, Brown DM. Phase I/II dose-escalation trial of amonafide for treatment of advanced solid tumors: genotyping to optimize dose based on polymorphic metabolism. J Clin Oncol. 2007;25:2503. [Google Scholar]
- 6.Ratain MJ, Mick R, Berezin F, Janisch L, Schilsky RL, Williams SF, Smiddy J. Paradoxical relationship between acetylator phenotype and amonafide toxicity. Clin Pharmacol Ther. 1991;50:573–579. doi: 10.1038/clpt.1991.183. [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Huang M, Yang F, Chen Y, Miao ZH, Qian XH, Xu YF, Qin YX, Luo HB, Shen X, et al. R16, a novel amonafide analogue, induces apoptosis and G2-M arrest via poisoning topoisomerase II. Mol Cancer Ther. 2007;6:484–495. doi: 10.1158/1535-7163.MCT-06-0584. [DOI] [PubMed] [Google Scholar]
- 8.Feng JM, Zhu H, Zhang XW, Ding J, Miao ZH. Proteasome-dependent degradation of Chk1 kinase induced by the topoisomerase II inhibitor R16 contributes to its anticancer activity. Cancer Biol Ther. 2008;7:1726–1731. doi: 10.4161/cbt.7.11.6728. [DOI] [PubMed] [Google Scholar]
- 9.Bhonde MR, Hanski ML, Notter M, Gillissen BF, Daniel PT, Zeitz M, Hanski C. Equivalent effect of DNA damage-induced apoptotic cell death or long-term cell cycle arrest on colon carcinoma cell proliferation and tumour growth. Oncogene. 2006;25:165–175. doi: 10.1038/sj.onc.1209017. [DOI] [PubMed] [Google Scholar]
- 10.Yang J, Yu Y, Hamrick HE, Duerksen-Hughes PJ. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogenesis. 2003;24:1571–1580. doi: 10.1093/carcin/bgg137. [DOI] [PubMed] [Google Scholar]
- 11.McGowan CH, Russell P. The DNA damage response: sensing and signaling. Curr Opin Cell Biol. 2004;16:629–633. doi: 10.1016/j.ceb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gatei M, Sloper K, Sorensen C, Syljuäsen R, Falck J, Hobson K, Savage K, Lukas J, Zhou BB, Bartek J, et al. Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J Biol Chem. 2003;278:14806–14811. doi: 10.1074/jbc.M210862200. [DOI] [PubMed] [Google Scholar]
- 15.Poon RY, Chau MS, Yamashita K, Hunter T. The role of Cdc2 feedback loop control in the DNA damage checkpoint in mammalian cells. Cancer Res. 1997;57:5168–5178. [PubMed] [Google Scholar]
- 16.Olive PL, Wlodek D, Banath JP. DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 1991;51:4671–4676. [PubMed] [Google Scholar]
- 17.Lu HR, Zhu H, Huang M, Chen Y, Cai YJ, Miao ZH, Zhang JS, Ding J. Reactive oxygen species elicit apoptosis by concurrently disrupting topoisomerase II and DNA-dependent protein kinase. Mol Pharmacol. 2005;68:983–994. doi: 10.1124/mol.105.011544. [DOI] [PubMed] [Google Scholar]
- 18.Zhou N, Xiao H, Li TK, Nur-E-Kamal A, Liu LF. DNA damage-mediated apoptosis induced by selenium compounds. J Biol Chem. 2003;278:29532–29537. doi: 10.1074/jbc.M301877200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang YW, Otterness DM, Chiang GG, Xie WL, Liu YC, Mercurio F, Abraham RT. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19:607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR regulates fragile site stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 21.Hu B, Han SY, Wang X, Ottey M, Potoczek MB, Dicker A, Huebner K, Wang Y. Involvement of the Fhit gene in the ionizing radiation-activated ATR/CHK1 pathway. J Cell Physiol. 2005;202:518–523. doi: 10.1002/jcp.20139. [DOI] [PubMed] [Google Scholar]
- 22.Kramer A, Mailand N, Lukas C, Syljuasen RG, Wilkinson CJ, Nigg EA, Bartek J, Lukas J. Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 23.Gurley LR, D'Anna JA, Barham SS, Deaven LL, Tobey RA. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- 24.Davis FM, Tsao TY, Fowler SK, Rao PN. Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA. 1983;80:2926–2930. doi: 10.1073/pnas.80.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gandara D, Narayan S, Lara PN, Jr, Goldberg Z, Davies A, Lau DH, Mack P, Gumerlock P, Vijayakumar S. Integration of novel therapeutics into combined modality therapy of locally advanced non-small cell lung cancer. Clin Cancer Res. 2005;11:5057–5062. doi: 10.1158/1078-0432.CCR-05-9012. [DOI] [PubMed] [Google Scholar]
- 26.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 27.Deming PB, Flores KG, Downes CS, Paules RS, Kaufmann WK. ATR enforces the topoisomerase II-dependent G2 checkpoint through inhibition of Plk1 kinase. J Biol Chem. 2002;277:36832–36838. doi: 10.1074/jbc.M206109200. [DOI] [PubMed] [Google Scholar]
- 28.Siu WY, Lau A, Arooz T, Chow JPH, Ho HTB, Poon RYC. Topoisomerase inhibitors differentially activate DNA damage checkpoints through ataxia-telangiectasia mutated-dependent and -independent mechanisms. Mol Cancer Ther. 2004;3:621–632. [PubMed] [Google Scholar]
- 29.O'Connor MJ, Martin NM, Smith GC. Targeted cancer therapies based on the inhibition of DNA strand break repair. Oncogene. 2007;26:7816–7824. doi: 10.1038/sj.onc.1210879. [DOI] [PubMed] [Google Scholar]
- 30.Hotte SJ, Oza A, Winquist EW, Moore M, Chen EX, Brown S, Pond GR, Dancey JE, Hirte HW. Phase I trial of UCN-01 in combination with topotecan in patients with advanced solid cancers: a Princess Margaret Hospital Phase II Consortium study. Ann Oncol. 2006;17:334–340. doi: 10.1093/annonc/mdj076. [DOI] [PubMed] [Google Scholar]
- 31.Monks A, Harris ED, Vaigro-Wolff A, Hose CD, Connelly JW, Sausville EA. UCN-01 enhances the in vitro toxicity of clinical agents in human tumor cell lines. Invest New Drugs. 2000;18:95–107. doi: 10.1023/a:1006313611677. [DOI] [PubMed] [Google Scholar]
- 32.Castedo M, Perfettini JL, Roumier T, Yakushijin K, Horne D, Medema R, Kroemer G. The cell cycle checkpoint kinase Chk2 is a negative regulator of mitotic catastrophe. Oncogene. 2004;23:4353–4361. doi: 10.1038/sj.onc.1207573. [DOI] [PubMed] [Google Scholar]
- 33.Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy—two sides of the same coin? Nat Rev Cancer. 2007;7:925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]