Abstract
Mutations in HepA-related protein (HARP) are the only identified causes of Schimke immunoosseous dysplasia (SIOD). HARP has a unique annealing helicase activity in vitro, but the in vivo functional significance remains unknown. Here, we demonstrated that HARP is recruited to stalled replication forks via its direct interaction with Replication protein A (RPA). Cells with HARP depletion displayed increased spontaneous DNA damage and G2/M arrest, suggesting that HARP normally acts to stabilize stalled replication forks. Our data place the annealing helicase activity of HARP at replication forks and propose that SIOD syndrome may be caused by the destabilization of replication forks during cell proliferation.
Keywords: HARP, SMARCAL1, SIOD, replication fork, annealing helicase
HARP (HepA-related protein), also known as SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a-like 1), is a member of the SWI/SNF family of proteins (Eisen et al. 1995; Coleman et al. 2000; Flaus et al. 2006). Mutations in HARP are responsible for an autosomal recessive disorder known as Schimke immunoosseous dysplasia (SIOD), with the diagnostic features of spondyloepiphyseal dysplasia, renal dysfunction, and T-cell immunodeficiency (Schimke et al. 1971; Spranger et al. 1991; Boerkoel et al. 2000). Recent studies suggested that HARP has an unusual biochemical activity as an annealing helicase, which is opposite to helicases that are normally involved in DNA unwinding (Yusufzai and Kadonaga 2008). However, the exact biological function of this HARP annealing helicase activity remains unknown.
In this study, we report that HARP associates with Replication protein A (RPA) in vitro and in vivo. RPA was initially identified as an ssDNA-binding protein that is absolutely required for DNA replication of simian virus 40 (SV40) (for reviews, see Waga and Stillman 1998; Stenlund 2003; Fanning et al. 2006). Human RPA is a stable heterotrimer composed of three subunits—RPA70, RPA32, and RPA14 (also named as RPA1, RPA2, and RPA3)—that are conserved among eukaryotes. RPA is essential for DNA replication, repair, and recombination, and DNA damage signaling pathways in eukaryotic cells (Zou and Elledge 2003; Binz et al. 2004; Stauffer and Chazin 2004b; Fanning et al. 2006). The functions of RPA in these diverse processes depend on its ssDNA-binding activity and its ability to interact with multiple proteins involved in these pathways (for reviews, see Fanning et al. 2006; Zou et al. 2006). Therefore, RPA can be considered as an adaptor protein that facilitates various biochemical reactions that occur at or involve ssDNA. The interaction between HARP and RPA suggest that HARP may function during replication and/or DNA repair. Indeed, our subsequent studies indicate that HARP is involved in the protection of stalled replication forks and thus provide a plausible mechanism for the development of SIOD syndrome.
Results and Discussion
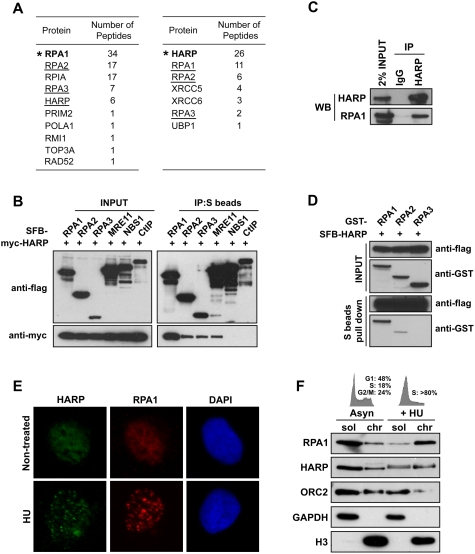
In an effort to identify new RPA-associated proteins, we performed tandem affinity purification (TAP) using soluble or chromatin fraction prepared from 293T cells stably expressing triple-epitope-tagged (S-protein, Flag, and streptavidin-binding peptide) RPA1 (SFB-RPA1). Mass spectrometry analysis revealed several RPA1-associated proteins in addition to RPA2 and RPA3 (Fig. 1A). Several of them are known RPA-binding proteins. These include POLA1/PRIM2 (Dornreiter et al. 1992), TOP3A/RMI1 (Brosh et al. 2000), and RAD52 (Fig. 1A; Sugiyama and Kowalczykowski 2002). Interestingly, we also identified a novel RPA-binding protein as HARP.
Figure 1.
HARP associates with RPA complex and is recruited to stalled replication forks in response to replication stress. (A) TAP was performed using 293T cells stably expressing tagged RPA1 or HARP. The data from mass spectrometry analysis are shown in the tables. (B) The association of HARP with RPA complex was confirmed by coimmunoprecipitation with overexpressed proteins. 293T cells were transfected with plasmids encoding myc-tagged wild-type HARP together with plasmids encoding SFB-tagged RPA1, RPA2, RPA3, MRE11, NBS1, or CtIP. Cells were lysed 24 h after transfection. Coimmunoprecipitation was carried out using S-protein beads and immunoblotting was performed using antibodies as indicated. (C) Association of endogenous HARP with RPA1 in 293T cells was performed by coimmunoprecipitation using anti-HARP antibody. (D) HARP binds strongly to RPA1. The in vitro binding assay was performed using the baculovirus expression system. Sf9 cells were coinfected with baculoviruses expressing indicated constructs. Pull-down experiments were performed using S-protein beads and immunoblotting was carried out using indicated antibodies. (E) HARP localizes at stalled replication forks in response to replication stress. U2OS cells were mock-treated or treated with 5 mM HU for 6 h. Immunostaining experiments were performed using anti-HARP and anti-RPA1 antibodies. (F) HARP accumulates in chromatin fraction following HU treatment. Asynchronized or HU-treated U2OS cells were subjected to fractionation. Soluble (sol) and chromatin (chr) fractions were separated and immunoblotted with indicated antibodies. Cell cycle patterns are shown on top.
To ensure that HARP indeed associates with RPA, we performed reverse TAP using a cell line stably expressing tagged HARP, and identified RPA1, RPA2, and RPA3 as major HARP-associated proteins (Fig. 1A). These data strongly suggest that HARP is a bone fide RPA-binding protein. We confirmed the in vivo interaction of myc-tagged HARP with SFB-tagged RPA1, RPA2, and RPA3 (Fig. 1B). MRE11, NBS1, and CtIP were included as controls, among which MRE11 has been reported to be associated with RPA complex (Robison et al. 2004; Olson et al. 2007). In addition, we performed a coimmunoprecipitation experiment in 293T cells and verified the in vivo interaction between endogenous HARP and RPA1 (Fig. 1C). To explore whether the formation of the RPA–HARP complex might be regulated following DNA damage, especially in normal cells, we performed coimmunoprecipitation using human mammary epithelial cells (HMEC). As shown in Supplemental Figure S1A, the interaction of HARP with RPA did not change following hydroxyurea (HU) treatment or ionizing radiation (IR), suggesting that this interaction is likely to be constitutive.
Overexpression experiments indicate that HARP binds more strongly to RPA1 than the other two RPA components (Fig. 1B). Using insect Sf9 cells and the baculovirus expression system, we confirmed that HARP indeed interacts tightly with RPA1 (Fig. 1D), but we also detected an interaction between HARP and RPA2, suggesting that HARP may associate with more than one component within the RPA complex.
RPA is the major ssDNA-binding protein in eukaryotic cells that accumulates along stretches of ssDNA generated at or near stalled replication forks. A physical interaction between HARP and RPA as demonstrated above raises the possibility that HARP may colocalize with RPA at ssDNA regions in the cell. In fact, discrete foci of Flag-tagged HARP were readily detected in cells following HU treatment (data not shown). In order to verify that the endogenous protein can also localize to focus structure upon replication stress, we generated a polyclonal anti-HARP antibody. As shown in Figure 1E, HARP could form distinct nuclear foci following HU treatment, suggesting that HARP localizes to stalled replication forks. In addition, just like RPA1, HARP also accumulated in chromatin fractions in HU-treated cells (Fig. 1F). However, the chromatin loading of HARP protein following IR was not observed (Supplemental Fig. S2), indicating that HARP mainly functions in response to replication stress. We also observed a mobility shift of HARP that resulted from HARP phosphorylation following HU treatment (Supplemental Fig. S3A). HARP phosphorylation could be inhibited by preincubation of cells with caffeine, an inhibitor of ATM and ATR kinases. This inhibition was not observed in cells pretreated with DNA-PK inhibtor NU7441 or ATM inhibitor KU55933 (Supplemental Fig. S3B), suggesting that HU-induced HARP phosphorylation depends on ATR or a combination of ATR and other PIKKs.
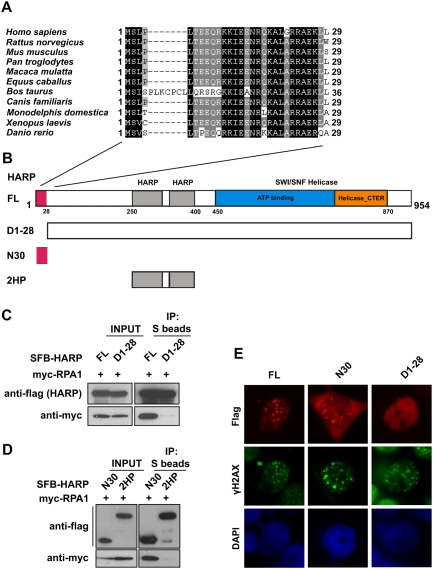
When aligning the human HARP protein sequence with its orthologs from other species, we found that HARP has three distinct conserved regions (Fig. 2A,B; Supplemental Fig. S4): (1) a region of 28 amino acids at the very N terminus (N28); (2) two tandem HARP domains (2HP), each of which contains ∼60 residues (residues 239–307 and residues 331–400); and (3) a SWI/SNF helicase domain (420–890) that is defined by a region of ∼400–500 amino acids that contains seven motifs (I, Ia, II, III, IV, V, and VI) that are highly conserved among DNA and RNA helicases (Koonin 1993; Hall and Matson 1999). We first generated a series of truncation and internal deletion mutants of HARP and established that ∼250 residues at the very N terminus of HARP are required for RPA binding and its HU-induced focus formation (data not shown). Next, we further defined this region to the N-terminal 28 amino acids, which show significant sequence conservation among species (Fig. 2A). While the full-length HARP bound strongly to RPA, the mutant lacking the N-terminal 28 amino acids (D1-28) failed to do so (Fig. 2C). Moreover, a fragment of HARP that contains the N-terminal 30 amino acids (N30) is sufficient for RPA-binding, whereas the tandem HARP domain (2HP) did not show any affinity for RPA (Fig. 2B,D). Consistently, while the D1-28 mutant of HARP lost its focus-forming ability, a fragment of HARP that contains only the N-terminal 30 residues could form nuclear foci following HU treatment (Fig. 2E). Therefore, we conclude that HARP is likely to be recruited to stalled replication forks via an interaction between its conserved N-terminal 28 amino acids and RPA. Interestingly, the RPA-binding region of HARP described here is very similar to the RPA2-interacting motif found in TIPIN and XPA (Unsal-Kacmaz et al. 2007). However, our data indicate that this region of HARP binds stronger to RPA1 (Fig. 1B,D). To further study the interaction between HARP and RPA, we used the GST-tagged RPA interaction region in HARP (N1-30, GST-HPN30) and a similar region in XPA (N1-45, GST-XPAN45). Both of these recombinant motifs could pull down myc-tagged RPA1 as well as RPA2, although the interaction between XPA-N45 and RPA2 is much stronger than that between the N terminus of HARP and RPA2 (Supplemental Fig. S1B). These data suggest that proteins like HARP and XPA may bind to more than one component in the RPA complex.
Figure 2.
The conserved N terminus of HARP is required for RPA1 binding and its foci formation following replication stress. (A) Alignment of N-terminal sequences of HARP from different species. (B) Schematic representation of wild-type HARP and the mutants used in the following study. (C) The 28 amino acids at the N terminus of HARP are required for RPA1 binding. 293T cells were transfected with plasmids encoding myc-tagged RPA1 together with plasmids encoding SFB-tagged full-length HARP (FL) or the D1-28 mutant. Coimmunoprecipitation was carried out using S-protein beads and immunoblotting was performed using antibodies as indicated. (D) The N-terminal fragment (N30) of HARP is sufficient for RPA1 binding. Coimmunoprecipitation was performed similar to that described in C. The tandem HARP domain (2HP) was used as a control. (E) The conserved N terminus of HARP is responsible and sufficient for HARP foci formation following replication stress. 293T cells were transfected with plasmids encoding SFB-tagged full-length HARP (FL), the N30 fragment, or the D1-28 mutant. Immunostaining experiments were performed 6 h after HU treatment using indicated antibodies.
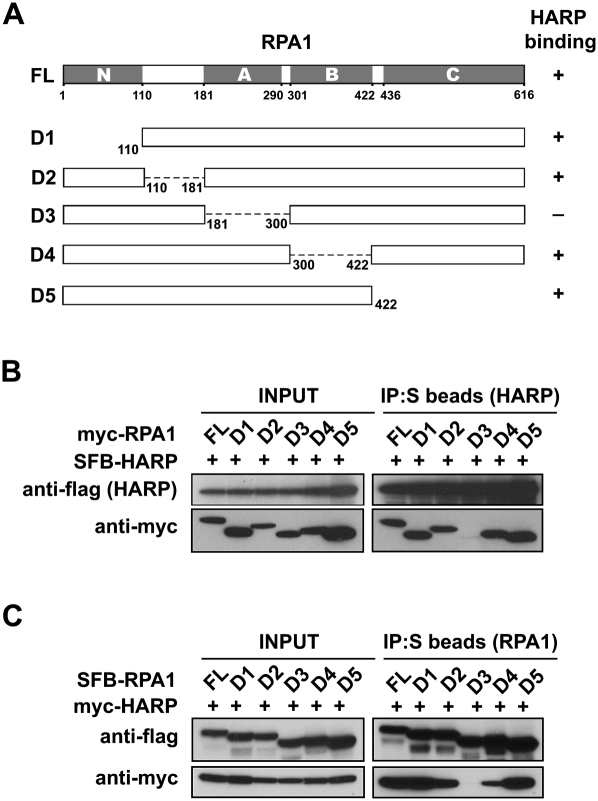
Since HARP binds strongly to RPA1, we generated a series of truncation or internal deletion mutants of RPA1 (Fig. 3A) to further characterize the HARP/RPA1 interaction. RPA1 has four oligonucleotide-binding (OB) folds: N, A, B, and C domains, with A, B, and C domains that have known ssDNA-binding activities. The N and A domains of RPA1 are also involved in protein–protein interactions. The N domain assumes an OB fold, but it lacks the conserved aromatic residues that can confer high-affinity ssDNA-binding activity, and therefore it binds to ssDNA very weakly (Daughdrill et al. 2001; Bochkareva et al. 2005). The A domain of RPA1 binds to ssDNA with an affinity that is higher than the other ssDNA-binding domains (B or C). In addition, the A domain of RPA1 also serves as the major protein–protein interaction domain, which has been reported to interact with many viral or cellular proteins including papillomavirus E1 helicase (Loo and Melendy 2004), XPA (Daughdrill et al. 2003), human Rad51 recombinase (Stauffer and Chazin 2004a), and the RecQ family Werner and Bloom Syndrome helicases (Doherty et al. 2005). As shown in Figure 3B, while wild-type and other mutants of RPA1 could be coimmunoprecipitated with HARP, the D3 mutant, which is deleted of A domain, failed to bind to HARP. The reverse coimmunoprecipitation experiments also confirmed this result (Fig. 3C). Together, these data suggest that RPA1 binds to HARP via its A domain, which is versatile for ssDNA binding as well as protein–protein interaction. To determine whether there is any competition between HARP and ssDNA for RPA1 binding, we purified RPA complex and HARP. In vitro experiments showed that HARP did not affect the binding of RPA to ssDNA (Supplemental Fig. S5).
Figure 3.
RPA1 associates with HARP through its ssDNA-binding domain A. (A) Schematic representation of full-length RPA1 and the mutants used in this study. Their ability to bind to HARP is indicated. (B,C) RPA1 with domain A deletion could not bind to HARP. 293T cells were transfected with plasmids encoding myc-tagged wild-type or mutant RPA1 together with plasmids encoding SFB-tagged HARP. Alternatively, 293T cells were transfected with plasmids encoding SFB-tagged wild-type or mutant RPA1 together with plasmids encoding myc-tagged HARP. Coimmunoprecipitation was carried out using S-protein beads and immunoblotting was performed using antibodies as indicated.
As discussed above, RPA complex is essential for DNA replication, recombination, and repair in eukaryotes. HARP has been shown recently to possess annealing helicase activity. Mutations in the helicase domain identified in SIOD patients abolish or diminish this annealing helicase activity, suggesting that this atypical biochemical activity of HARP is important for its function in vivo, although where and when this annealing helicase activity is used in the cell remains unknown. The observations that HARP can be recruited to stalled replication forks via its interaction with RPA1 led us to propose that the annealing helicase activity of HARP may normally be involved in the stabilization of stalled or stressed replication forks during DNA replication.
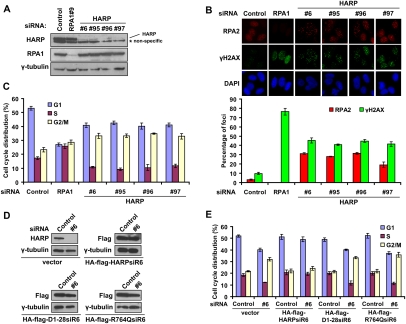
To test this hypothesis, a set of siRNAs specifically targeting HARP were synthesized and introduced into U2OS cells. All four HARP-specific siRNAs and the RPA1 siRNA worked well, since they led to the specific down-regulation of their target genes (Fig. 4A). Moreover, the protein level of HARP remained the same in cells with RPA1 depletion and vice versa, suggesting that although HARP and RPA1 interact, they do not influence each other's stability in cells. If the annealing helicase activity of HARP is normally involved in the stabilization of replication forks, we reasoned that HARP-depleted cells might display longer stretches of ssDNA during DNA replication, which could result in collapsed replication forks and DNA double-stranded breaks. As shown previously (Zou and Elledge 2003; Jazayeri et al. 2006), the appearance of RPA foci could be used as a marker for the emerging ssDNA regions in the cell. Indeed, RPA foci were detected in ∼30% of the cells with HARP knockdown, which is an ∼10-fold increase over that observed in cells transfected with control siRNA (Fig. 4B). The frequency of dsDNA breaks was also higher in cells depleted with HARP, as indicated by the formation of phosphorylated H2AX (γH2AX) foci, which largely overlap with RPA foci (Fig. 4B), and the increase of ATM and CHK2 phosphorylation in HARP-depleted cells (Supplemental Fig. S6).
Figure 4.
HARP depletion results in spontaneous DNA damage and G2/M cell cycle arrest. (A) Knockdown efficiency in U2OS cells transfected with indicated RPA1 and HARP siRNAs was confirmed by immunoblotting. (B) RPA and γH2AX foci are greatly increased in HARP-depleted cells. U2OS cells were transfected with indicated siRNAs. Seventy-two hours later, cells were subjected to immunostaining using indicated antibodies. The quantification of foci-positive cells was performed by counting a total of 200 cells per sample. (C) HARP depletion induces G2/M accumulation. Seventy-two hours after siRNAs transfection, U2OS cells were fixed and subjected to cell cycle analysis. The percentage of cells in different cell cycle phases was determined. Data were presented as mean ± SD from three different experiments. (D) U2OS derivative cell lines stably expressing siRNA-resistant wild-type HARP (HA-Flag-HARPsiR6), D1-28 mutants (HA-Flag-D1-28siR6), and R764Q mutants (HA-Flag-R764QsiR6) were generated. The endogenous and exogenous HARP expression was confirmed by immunoblotting using indicated antibodies and extracts were prepared from cells transfected with indicated siRNAs. (E) The D1-28 and R764Q mutants could not restore HARP function in vivo. Seventy-two hours after siRNA treatment, the indicated stable cells were fixed and subjected to cell cycle analysis. The percentage of cells in different cell cycle phases was determined. Data are presented as mean ± SD from three different experiments.
These results suggest that when the annealing helicase HARP is removed from the cell, prolonged ssDNA coated with RPA will be generated that would be cleaved by nucleases and thus would result in the collapse of replication forks and spontaneous DNA breaks. In this sense, HARP depletion should interfere with normal cell cycle progress. Inconsistent with our hypothesis, cells with HARP depletion started to accumulate in G2/M, probably in response to DNA breaks generated during DNA replication (Fig. 4C; see also Supplemental Fig. S7A for FACS profiles). At the same time, we also observed a slight decrease in S phase, suggesting that fewer cells with HARP depletion would enter and continue DNA replication (Fig. 4C).
As we showed above, RPA is involved in the recruitment of HARP to stressed or stalled replication forks, and HARP is required for the maintenance of the stability of replication forks. Therefore, it would be interesting to further test whether the RPA-binding ability and the annealing helicase activity of HARP are both required for its function in vivo. We generated siRNA-resistant HA-Flag-tagged wild-type, D1-28 mutant, and R764Q mutant HARP constructs so that we were able to express exogenous HARP when the endogenous HARP was depleted by siRNA. The D1-28 mutant is deficient in RPA1 binding and also loses its ability to form nuclear foci upon replication stress (Fig. 2C,E). The R764Q mutation is located in the conserved ATPase region of HARP and is derived from patients with a severe form of SIOD (Boerkoel et al. 2002). The R764Q mutant has no detectable annealing helicase activity (Yusufzai and Kadonaga 2008). U2OS derivative cell lines stably expressing siRNA-resistant wild-type HARP, D1-28 mutants, and R764Q mutants were generated. The expression of endogenous and exogenous HARP or HARP mutants were confirmed by immunoblotting in these cells transfected with control or HARP siRNAs (Fig. 4D). Following the depletion of endogenous HARP, we observed a G2/M accumulation that was rescued by the expression of siRNA-resistant wild-type HARP. Interestingly, neither the D1-28 mutant nor the R764Q mutant could restore the normal cell cycle progression (Fig. 4E; see also Supplemental Fig. S7B for FACS profiles). Similarly, we also used RPA and γH2AX foci formation as a readout of HARP function. As shown in Supplemental Figure S8, following the depletion of endogenous HARP, RPA and γH2AX foci formation were greatly enhanced. This increase was suppressed by the expression of siRNA-resistant wild-type HARP but not the D1-28 and R764Q mutants. These data suggested that both RPA1 binding and the annealing helicase activity of HARP are required for its function during DNA replication.
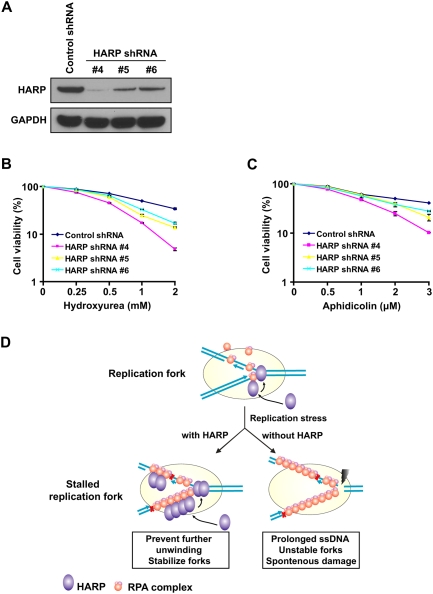
Together, we propose that, in response to spontaneous or replication stress, replication forks would be stalled and allow the formation of ssDNA regions that are coated with RPA. Through its interaction with RPA, HARP is recruited to these stalled replication forks and exerts its annealing helicase activity to prevent further separation of dsDNA and thus stabilize stalled replication forks. However, in the absence of HARP and its annealing activity, extensive ssDNA regions would be generated at stalled replication forks, which could be easily attacked by nucleases that lead to fork collapse and the emergence of DNA breaks (Fig. 5D). Actually, by using HeLa cells stably expressing HARP shRNA, we showed that HARP insufficiency causes hypersensitivity to replication stress, such as HU and aphidicolin (Fig. 5A–C), Similar results were obtained using U2OS cells (Supplemental Fig. S9A,B). We did not detect any obvious hypersensitivity to IR in HeLa or U2OS cells after HARP depletion (Supplemental Fig. S9C), suggesting that HARP may not play a major role in response to DNA double-strand breaks, which is consistent with the result that HARP did not show any significant relocalization to chromatin fractions following IR treatment (Supplemental Fig. S2).
Figure 5.
HARP insufficiency causes hypersensitivity to replication stress. (A) Knockdown efficiency of HARP in HeLa cells stably expressing the indicated shRNA was confirmed by immunoblotting. (B) Survival curves in response to increasing doses of HU for indicated cell lines. (C) Survival curves in response to increasing doses of aphidicolin for indicated cell lines. Cell survival assays were performed according to the Materials and Methods. Data are presented as mean ± SD from three different experiments. (D) A proposed model of HARP function at replication forks. See the text for details.
Materials and methods
See the Supplemental Material for additional materials and methods.
Antibodies
Anti-HARP antibodies were raised by immunizing rabbits with MBP-HARP fusion proteins containing residues 1–300 and residues 654–954 of HARP. Antisera were affinity-purified using the AminoLink plus Immobilization and purification kit (Pierce). Another anti-HARP antibody was obtained from Bethyl Laboratories. Anti-γ-H2AX was described previously (Huen et al. 2007). The anti-myc and anti-GST antibodies were obtained from Santa Cruz Biotechnologies. Anti-γ-tubulin and anti-Flag (M2) were obtained from Sigma. Anti-RPA1 antibody was purchased from EMD Chemicals. Anti-RPA2 was from Abcam. Anti-H3 was obtained from Upstate Biotechnologies Cell Signaling. Anti-ORC2 and anti-GAPDH antibodies were obtained from Millipore.
Cell culture, transfection, siRNAs, and shRNAs
HeLa, 293T, and U2OS cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. Plasmid transfection was performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. All siRNA duplexes were purchased from Dharmacon Research. The sequence of RPA1 siRNA #9 was CCCUAGAACUGGUUGACGAUU, and the sequences of HARP siRNAs were #6, GCUUUGACCUUCUUAGCAAUU, #95, CUGAUUCAAGAGAAGAUUAdTdT, #96, GCUUUGACCUUCUUAGCAAdTdT, and #97, GGCUCUCACUGGAAUCUCUdTdT. The siRNA-resistant wild-type and mutant HARP constructs were generated by changing 8 nucleotides in the HARP siRNA #6 targeting region (C1584T, T1587C, C1590T, C1591T, T1593G, C1594T, T1596G, and C1599T substitutions). The siRNA transfection was performed using Oligofectamine (Invitrogen) following the manufacturer's instruction. Transfection was repeated twice with an interval of 24 h to achieve maximal RNAi effect. Control shRNA and HARP shRNA target sets were purchased from Open Biosystems, and stable clones of HeLa cells were made by retroviral infection followed by puromycin selection for 2 wk.
Cell cycle analysis
HeLa or U2OS cells were harvested, washed with PBS, and fixed with ice-cold 70% ethanol overnight. Cells were washed in PBS and treated for 30 min at 37°C with RNase A (200 μg/mL) and followed with propidium iodide (25 μg/mL), and were analyzed on a FACScan flow cytometer (Becton Dickinson). The percentage of cells in different cell cycle phases was calculated using WinMDI analysis software.
Acknowledgments
We thank all members of the Chen laboratory, especially Dr. Michael S.Y. Huen, for their advice and technical assistance. This work was supported by grants from the National Institutes of Health (to J.C.). J.C is a recipient of an Era of Hope Scholar award from the Department of Defense and is a member of the Mayo Clinic Breast SPORE program.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1836409.
Supplemental material is available at http://www.genesdev.org.
References
- Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, O'Neill S, Andre JL, Benke PJ, Bogdanovic R, Bulla M, Burguet A, Cockfield S, Cordeiro I, Ehrich JH, et al. Manifestations and treatment of Schimke immuno-osseous dysplasia: 14 new cases and a review of the literature. Eur J Pediatr. 2000;159:1–7. doi: 10.1007/s004310050001. [DOI] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- Coleman MA, Eisen JA, Mohrenweiser HW. Cloning and characterization of HARP/SMARCAL1: A prokaryotic HepA-related SNF2 helicase protein from human and mouse. Genomics. 2000;65:274–282. doi: 10.1006/geno.2000.6174. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Ackerman J, Isern NG, Botuyan MV, Arrowsmith C, Wold MS, Lowry DF. The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res. 2001;29:3270–3276. doi: 10.1093/nar/29.15.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughdrill GW, Buchko GW, Botuyan MV, Arrowsmith C, Wold MS, Kennedy MA, Lowry DF. Chemical shift changes provide evidence for overlapping single-stranded DNA- and XPA-binding sites on the 70 kDa subunit of human replication protein A. Nucleic Acids Res. 2003;31:4176–4183. doi: 10.1093/nar/gkg451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty KM, Sommers JA, Gray MD, Lee JW, von Kobbe C, Thoma NH, Kureekattil RP, Kenny MK, Brosh RM., Jr Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J Biol Chem. 2005;280:29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- Dornreiter I, Erdile LF, Gilbert IU, von Winkler D, Kelly TJ, Fanning E. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Matson SW. Helicase motifs: The engine that powers DNA unwinding. Mol Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Melendy T. Recruitment of replication protein A by the papillomavirus E1 protein and modulation by single-stranded DNA. J Virol. 2004;78:1605–1615. doi: 10.1128/JVI.78.4.1605-1615.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E, Nievera CJ, Liu E, Lee AY, Chen L, Wu X. The Mre11 complex mediates the S-phase checkpoint through an interaction with replication protein A. Mol Cell Biol. 2007;27:6053–6067. doi: 10.1128/MCB.00532-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- Schimke RN, Horton WA, King CR. Chondroitin-6-sulphaturia, defective cellular immunity, and nephrotic syndrome. Lancet. 1971;2:1088–1089. doi: 10.1016/s0140-6736(71)90400-4. [DOI] [PubMed] [Google Scholar]
- Spranger J, Hinkel GK, Stoss H, Thoenes W, Wargowski D, Zepp F. Schimke immuno-osseous dysplasia: A newly recognized multisystem disease. J Pediatr. 1991;119:64–72. doi: 10.1016/s0022-3476(05)81040-6. [DOI] [PubMed] [Google Scholar]
- Stauffer ME, Chazin WJ. Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J Biol Chem. 2004a;279:25638–25645. doi: 10.1074/jbc.M400029200. [DOI] [PubMed] [Google Scholar]
- Stauffer ME, Chazin WJ. Structural mechanisms of DNA replication, repair, and recombination. J Biol Chem. 2004b;279:30915–30918. doi: 10.1074/jbc.R400015200. [DOI] [PubMed] [Google Scholar]
- Stenlund A. Initiation of DNA replication: Lessons from viral initiator proteins. Nat Rev Mol Cell Biol. 2003;4:777–785. doi: 10.1038/nrm1226. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]