Abstract
In stimulating maturation of colonic carcinoma cells, the short chain fatty acid butyrate, and 1α, 25-dihydroxyvitamin D3, were shown to attenuate transcription of the cyclin D1 gene, giving rise to truncated transcripts of this locus. Moreover, a sequence which is highly conserved in the human, mouse, rat, and dog genome was found in the 4 kb long intron 3 of the human cyclin D1 gene, and is capable of forming a hairpin structure similar to that of microRNA precursors. The expression of this sequence is also decreased by the attenuation. Thus, the transcriptional attenuation at the cyclin D1 locus not only down-regulates the expression of this key gene in mucosal cell maturation and tumorigenesis, but may also abrogate the generation of a molecule that encompasses this conserved sequence in cyclin D1 intron 3.
Induction of cell maturation, especially by physiological agents that are likely to exhibit low toxicity, is an important approach to chemoprevention of cancer. For example, the short chain fatty acid butyrate, derived by fermentation of dietary fiber and a principal energy source for colonic epithelial cells, is a physiological regulator of colonic cell maturation. Upon treatment with butyrate, colonic epithelial cells that have activated the ras signaling cascade are sensitized to respond to butyrate (Klampfer et al., 2004), and undergo cell cycle arrest, differentiation along the absorptive cell lineage with simultaneous repression of secretory cell differentiation, and eventual apoptosis (Hague et al., 1993; Augenlicht et al., 1995; Heerdt et al., 1994; Augenicht et al., 2003). Moreover, at least the apoptotic response appears to be linked to mitochondrial metabolism of butyrate (Augenlicht et al., 1999).
This response of colonic cells to butyrate is mediated by a cascade of changes in gene expression that monotonically increases in complexity as a function of time following treatment (Mariadason et al., 2000). Among these changes is an increase in β-catenin-Tcf complex formation and signaling (Bordonaro et al., 1999). This is counter-intuitive to the affects of butyrate as a chemoprevention agent, since elevated β-catenin-Tcf signaling maintains intestinal cells in a proliferative, progenitor cell phenotype (van de Wetering et al., 2002). For example, an important target positively regulated by β-catenin-Tcf signaling is the c-myc gene (He et al., 1998), a gene whose down-regulation we have identified as tightly linked to controlling the reprogramming of intestinal epithelial cells in vivo during their migration and maturation along the crypt-villus axis (Mariadason et al., 2005). However, we and others have demonstrated that elevated transcriptional initiation of c-myc by butyrate is abrogated by transcriptional attenuation of elongation of the transcript, resulting in decreased steady state levels of c-myc expression, consistent with the cell cycle arrest stimulated by the short-chain fatty acid (Heruth et al., 1993; Keene et al., 1999; Wilson et al., 2002).
Cyclin D1 is another gene that is transcriptionally induced by β-catenin-Tcf signaling (Shtutman et al., 1999; Tetsu and McCormick, 1999). Moreover, cyclin D1 plays a direct role in intestinal tumorigenesis, since its genetic inactivation reduces intestinal tumor formation in mice that inherit a mutation in the Apc gene (Hulit et al., 2004). Here we utilized a unique method of transcription site imaging to demonstrate that butyrate also attenuates transcription of the cyclin D1 gene. Moreover, this attenuation is also seen in response to vitamin D3, another physiological regulator of colonic cell maturation.
Materials and Methods
Cell lines
SW837 (CCL-235) rectal and Dld-1 (CCL-221) colon adenocarcinoma cells (American Type Culture Collection) were grown at 37°C, 5% CO2 in RPMI 1640 medium, supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 units/ml streptomycin, 0.25 μg/ml amphotericin (0.25 μg/ml), 100 μM non-essential amino acids, 1 mM sodium pyruvate, and 10 mM HEPES buffer solution, either in 175 cm2 flasks, or seeded onto glass coverslips (for FISH analysis) coated with 0.5% gelatin in 6-well plates. Stimulation was with 5 × 10−3 M butyrate, or 10−7 M 1 α,25-dihydroxyvitamin D3, at 60% confluency for 0–24 h.
RNA was isolated from cultures and assayed for steady state level of cyclin D1 RNA transcripts by qRT-PCR using SYBR Green (Applied Biosystems), prior DNase I treatment using the ImProm-II Reverse Transcription System (Promega). cDNA was amplified using the Fast Start High Fidelity PCR-System (Roche), with the following primers, designed by Primer Express Software (ABI): CCND1 exon 1: forward primer: 5′-CCCTCGGTGTCCTACTTCAA-3′; reverse primer: 5′-GGGGATGGTCTCCTTCATCT-3′, exon 1 spanning exon 2: forward primer: 5′-CCCTCGGTGTCCTACTTCAA-3′; reverse primer: 5′-GGGGATGGTCTCCTTCATCT-3′, exon 2: forward primer: 5′-AACAGAAGTGCGAGGAGGAG-3′; reverse primer: 5′-GGATGGAGTTGTCGGTGTAG-3′, exon5: forward primer: 5′-TGCCAGGAGCAGATCGAAG-3′; reverse primer: 5′-TCCTCCTCCTCCTCTTCCTC-3′. Primers specific for human GAPDH were: forward: 5′-TCGGAGTCAACGGATTTGG-3′; reverse: 5′-CAACAATCCACTTTACCAGAGTTAAAA-3′. Transcript expression by qRT-PCR was assayed in duplicate and standardized by GAPDH mRNA expression. Relative levels were quantified by calculating 2−ΔΔCT, where ΔΔCT is the difference in CT (cycle number at which the amount of amplified target reaches a fixed threshold) between target and reference.
Analysis of transcription sites
Transcription sites in interphase nuclei were detected using a method of fluorescent in situ hybridization, as previously described (Femino et al., 1998; Levsky et al., 2002; Wilson et al., 2002). Probes, 50 nucleotides in length complementary to regions of the cyclin D1 gene, were synthesized with deoxythymidine (amino-allyl dT) at defined positions in the sequence, purified by reverse chromatography, and the activated fluorophores Cy3, Cy3.5, and Cy5 (Amersham) were chemically coupled to these primary amines. Each probe was labeled with a total of five identical fluorophore molecules (detailed methods at http://protocols.singerlab.org/). The probe set in Figure 2 was: for the 5′ end of the cyclin D1 gene (exons 1 and 2), hCy D1-1: 5′-CTACGCGCGGATGGTTTCCACTTCGCAGCACAGGAGCTGGTGTTCCATA, hCyD1-2: 5′-TTGACAGGAAGCGGTCCAGGTAGTTCATGGCCAGCGGGAAGACCTCCTCTTCGC ACTA, hCyD1-3:5′-CTTCTTAGAGGCCACGAACATGCAAGTGGCCCCCAGCAGCTGC AGGCGGCTCTA and hCyD1-4: 5′-ATCTTGAGCTTGTTCACCAGGAGCAGCTCCATT TGCAGCAGCTCCTCGGGCCGTC; for the 3′ end of the cyclin D1 gene (exon 5): hCy D1-5:5′-CTGGGTCCACCATGGCTAAGTGAAGCAT-GAGGTATTGTGAAACAGCAACC, hCyD1-6: 5′-GCTGTAAAAACTACTATGATGCTACGCCCCCGATCAGATGAAGTGCCC AG and hCyD1-7: 5′-GAAGCTATTCCAATCATCCCGAATGAGAGTCCTACAGGTACA ACGCCGTG. For β-actin the following probes were used: hBA-1: 5′-GTGAACTTTGG GGGATGCTCGCTCCAAC-CGACTGCTGTCACCTTCACCGT, hBA-2: 5′-TCCTTAGAG AGAAGTGGGGTGGCTTTTAGGATGGCAAGGGACTTCCTGTA and hBA-3: 5′-CTTTT ATTCAACTGGTCTCAAGTCAGTGTACAGGTAAGCCCTGGCTGCCT. Hybridization of these probes, in different combinations to untreated and butyrate treated SW 837 cells, was as described (Wilson et al., 2002). For scoring of transcription sites, images were captured with a Photometrics Cool SNAP HQ digital CCD camera (Roper Scientific, Tucson, AZ) mounted on an AX-70 Provis microscope using a PlanApo 60 ×, 1.4 NA objective and 100 W mercury lamp for epi-illumination (Olympus, Melville, NY). A Uniblitz model VS35S2ZM0 shutter with model VMM-D1 electronic controller (Vincent Associates, Rochester, NY) was used for capturing of images in three dimensions. These images were acquired using #31000 (DAPI), #SP-102v1 (Cy3), #SP-103v1 (Cy3.5) and #41008 (Cy5) fluorescence filter sets (ChromaTechnology, Brattleboro, VT). Acquisition and data analysis were performed using IPLab Windows version 3.6 software from Scanalytics (Fairfax, VA). Images acquired using the Cy3, Cy3.5, and Cy5 filters were assigned green, red, and blue colors for visualization. To be scored as a true transcription site, the simultaneous detection of at least two colors was required at a site, independent hybridization events highly unlikely to be attributable to chance. Only sites in non-overlapping nuclei were counted. The scoring was performed with the observer blinded to the identity of the slide; at least 600–1,500 cells were counted per slide in order to maximize reproducibility of results. For acquisition of the data, multiple fields on every slide were photographed, chosen because of their appearance with DAPI staining, as having multiple, but non-overlapping cells, and intact nuclei; their appearance in regards to Cy3, Cy3.5, and Cy5 was not considered in this selection. Different dye combinations were used among experiments to minimize potential error due to differential affinity among the dyes. Transcription sites were scored as three-color (e.g., red/green/blue) or two-color (e.g., red/blue) based on the intensity of signal in the third channel above background. Sites in which intensity of signal of the third color was ambiguous (<5% of nuclei) were not included in the analysis. The number of counted nuclei (DAPI image) was also scored, to determine the relative frequency of occurrence of the sites.
Fig. 2.
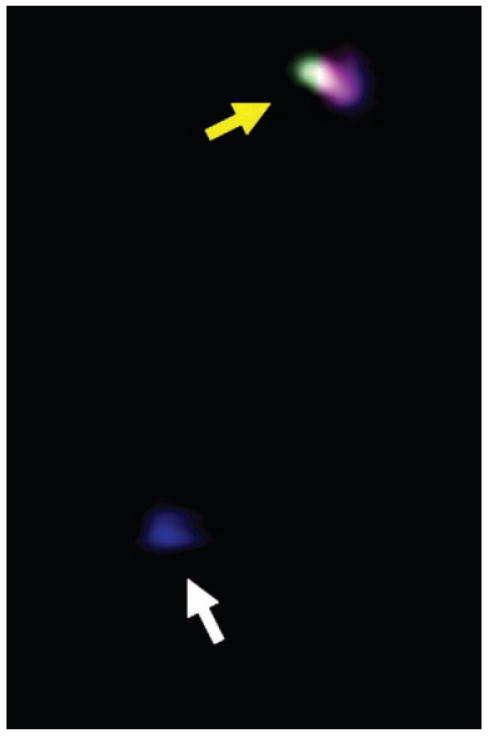
Example of a cyclin D1 transcription site that is labeled with three fluorophores (yellow arrow) or a single fluorophore (white arrow).
Results and Discussion
Consistent with numerous reports, 6 h following treatment of SW837 with 5 mM of the short chain fatty acid butyrate, cells in the G1 phase of the cell cycle increased by 40% (data not shown). Using sets of primers that assay exon 1, exon 1 to 2, exon 2, and exon 5 of the human cyclin D1 gene by qRT-PCR, steady state levels of expression of cyclin D1 mRNA were found to decrease progressively beginning at 2 h post-treatment, reaching approximately 20% of control value by 12 h (Fig. 1A), leading to decreased cyclin D1 protein levels (Fig. 1B). Similar results were seen for DLD1 cells, for which the increase of cells in G1 was 137% following butyrate treatment, again paralleled by a decrease in cyclin D1 steady state levels (not shown). Thus, these cell lines undergo standard cell cycle arrest and coincident decrease in cyclin D1 expression in response to butyrate. We have previously reported that steady state levels of c-myc mRNA and protein, also encoded by a Wnt target gene, also decreased in SW837 cells in response to butyrate. However, interrogation of the minor c-myc RNA fraction localized at the site of transcription in the interphase nucleus revealed that transcriptional attenuation was a mechanism that contributed substantially to this decrease in c-myc steady state levels (Wilson et al., 2002).
Fig. 1.
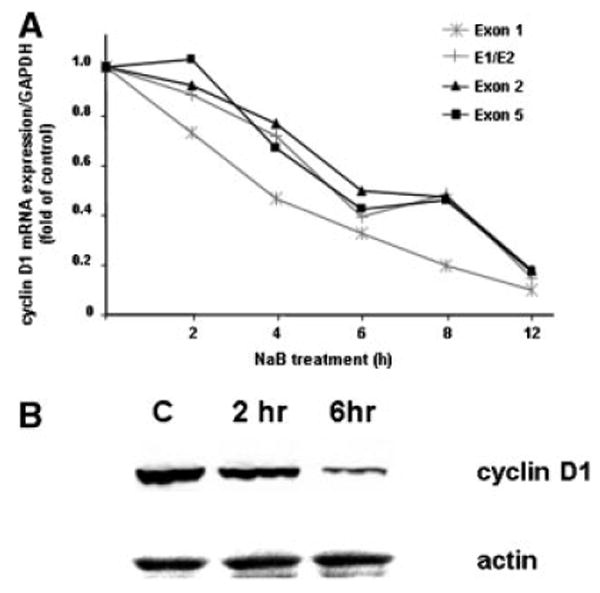
Decreased expression of cyclin D1 in SW837 cells treated with 5 mM butyrate. A: RNA was isolated from cultures and assayed by qRT-PCR for steady state of transcripts of the cyclin D1 gene using primers that amplified exon 1, exon 1/2, exon 2 and exon 5; (B) Western blot analysis of protein.
To investigate whether transcriptional attenuation was also triggered at the cyclin D1 locus by butyrate, we utilized the same fluorescence based method we previously reported to image nuclear transcription sites of the cyclin D1 gene within individual cells (Femino et al., 1998; Levsky et al., 2002; Wilson et al., 2002). This method takes advantage of the fact that the highest concentration of any specific mRNA sequence is at the site of synthesis, and thus fluorescent probes hybridizing to these molecules emit a bright flash of light localized to the cognate gene in the nucleus. Oligonucleotide probes, which recognized exon 1 or exon 2 of the growing cyclin D1 RNA transcripts were labeled with two spectrally distinct fluorophores; these interrogated initiation of cyclin D1 transcription. The 3′ end of the RNA encoded by exon 5 was evaluated using probes labeled with a third distinct fluorophore. These exon 5 probes could only hybridize when full length, or almost completed transcripts, were present at the site of transcription. Therefore the detection of three colors at the nuclear site of transcription indicated the presence of full-length transcripts at the cyclin D1 locus, and the detection of a two-colored site indicated initiation, but attenuation, of cyclin D1 transcription. We have published images of such differentially labeled transcription sites (Wilson et al., 2002), and an example of a single color and three color cyclin D1 transcription site is shown in Figure 2. As described in Methods, the site showing only a single color would not have been scored as a true transcription site. Figure 3A shows the distribution of transcription sites detected in control, untreated SW837 or DLD-1 cells. For each cell line, we detected no active cyclin D1 transcription sites in approximately 70% of the nuclei examined, consistent with the fact that even expressed loci cycle between active transcription and no transcription, especially in an unsynchronized cell population. For SW837 cells, those active sites generating only initiated (i.e., two color) transcripts were approximately equivalent to those that generated full-length (three color) transcripts (Fig. 3A). For DLD-1 cells, there was a bias towards a larger fraction of active transcription sites generating full-length transcripts (Fig. 3A).
Fig. 3.
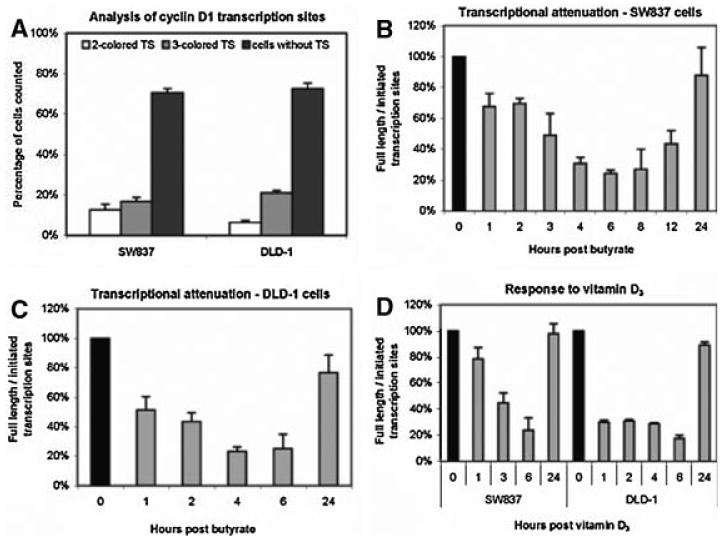
FISH analysis for detection of active cyclin D1 transcription sites (A) FISH analysis of cyclin D1 transcription sites in untreated, growing SW837 cells. Sites were detected in nuclei using two differentially labeled probes complementary to the 5′ end of the cyclin D1 mRNA, and a third probe complementary to the 3′ end. Data were calculated as percent of nuclei containing transcript that were two colored (initiated) or three colored (completed) or free of detectable, active transcript sites, mean ± standard error; (B) The ratio of nuclei that contained three color (full length) to two color (initiated) cyclin D1 transcription sites as a function of time following treatment of SW837 cells with 5 mM butyrate; (C) similar to 2B, but ratios of nuclei containing full length to initiated transcripts as a function of time following treatment of DLD-1 cells with 5 mM butyrate; (D) similar to 2B, but ratios of nuclei containing full length to initiated transcripts as a function of time following treatment of either SW837 (left) or DLD-1 (right) cells with 10−7 M vitamin D3.
The frequency of cell nuclei containing active cyclin D1 transcription sites (either 2- or 3-colored) remained constant following butyrate treatment of SW837 cells (28 ± 3%—not shown). However, the ratio between cells with cyclin D1 loci generating full length transcripts in SW837 cells to those in which transcription was initiated, but not completed, decreased as a function of time. The ratio of full-length to initiated transcription sites decreased by 30% relative to untreated cells within 1 h of butyrate stimulation (Fig. 3B). This decrease reached 50% relative to control by 3 h (P < 0.05), with a maximal decrease of 78% at 6 h, and a recovery thereafter (Fig. 3B). Similar results were seen for DLD-1 cells, although the decrease appeared to be more rapid (∼50% decrease by 1 h), reaching a 75% decrease in the ratio of full length to initiated sites at 6 h post-butyrate treatment, relative to control cells (Fig. 3C).
Treatment of colon carcinoma cells with 10−7 M 1α, 25-dihydroxyvitamin D3 also induces a cell cycle arrest (Diaz et al., 2000). Figure 3D illustrates that vitamin D3 also decreased the ratio of full length to initiated transcription sites within an hour in both SW837 and DLD-1 cells, respectively, with a maximum decrease again at 6 h, although again, the decrease appeared to be more rapid in DLD-1 cells than in SW837.
Transcriptional attenuation of the cyclin D1 gene therefore makes a substantial contribution to the reduced steady state levels of cyclin D1 that accompany the cell cycle arrest of colon carcinoma cells induced by butyrate or 1α, 25-dihydroxyvitamin D3.
We also investigated sequence elements of the primary transcript of the cyclin D1 gene in relation to the transcriptional attenuation. Using the pre-computed multiple species alignments provided by the UCSC Genome Browser (Karolchik et al., 2003), we found two regions of pronounced sequence conservation between the human and mouse genome in the 4 kb long intron 3 of the mouse cyclin D1 gene. We then used the Zuker's Mfold program to predict the most favorable secondary structure (Zuker, 2003). The first sequence (encircled area, Fig. 4A) forms a longer hairpin structure that is highly conserved between human, mouse, rat, and dog cyclin D1 loci, although for the dog, the sequence is present in intron 2 rather than intron 3, which is the longest intron in the dog, as is intron 3 in the other species (Fig. 4B) (Altschul et al., 1997). qRT-PCR analysis shows that this sequence is transcribed, and, like exons 1–5, is decreased in expression by butyrate (not shown). However, although it has a secondary structure similar to known microRNA precursors (Bartel, 2004), and its high conservation across species suggests functional significance, we have not found evidence that it exists as an independent non-coding small RNA molecule.
Fig. 4.
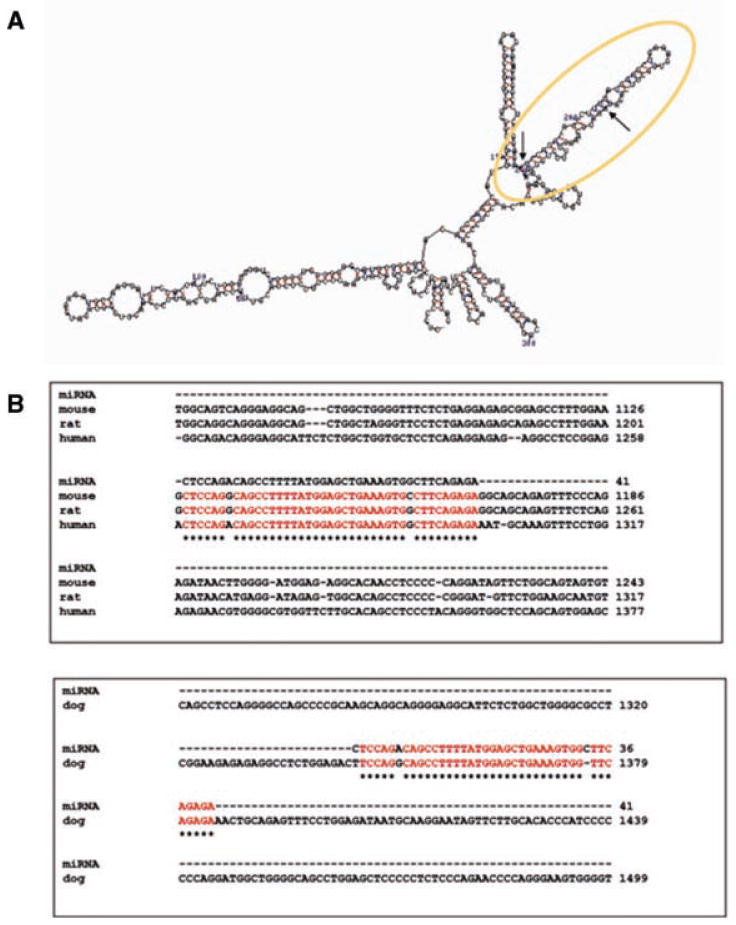
A: Predicted secondary structure of a sequence in intron 3 (1,260–1,300 bp) of the human cyclin D1 gene, a portion of which is highly conserved between the human and mouse genome; (B) sequence homologies within intron 3 (human, mouse, rat) and intron 2 (dog) of the cyclin D1 gene.
In conclusion, we have now demonstrated that transcriptional attenuation triggered by butyrate or vitamin D3 plays an important role in the down regulation of two key genes that regulate colonic cell maturation and transformation—c-myc and cyclin D1 (Wilson et al., 2002), and data above—therefore eliminating the increased steady state levels of c-myc and cyclin D1 which might be expected from the elevation in Wnt signaling. Recent data (Daroqui and Augenlicht, unpublished work) demonstrate that this mechanism of transcriptional attenuation appears to be involved in regulating the expression of a significant percent of genes for which expression is altered in the extensive reprogramming initiated by butyrate (Mariadason et al., 2000). In this regard, it has been reported recently that a high percentage of developmentally regulated sequences in embryonic stem cells are initiated in transcription but fail to produce full length transcripts by a mechanism associated with a unique pattern of histone acetylation (Guenther et al., 2007), which likely influences chromatin structure and interaction with other transcription factors. It is important to note that butyrate, an HDAC inhibitor, is present at very high concentrations in the lumen of the colon, where SCFAs are generated by fermentation of fiber and serve as the principal energy source for colonic epithelial cells. Therefore, butyrate, like vitamin D3, is a physiological agent that provides an extrinsic signal important in establishing and maintaining colonic mucosal homeostasis. Thus, transcriptional attenuation may be a common mechanism involved in normal, well integrated, finely balanced programs of cell maturation in both embryogenesis and in response to physiological inducers of cell maturation in the adult, such as butyrate and vitamin D3 in the intestine.
Acknowledgments
Contract grant sponsor: National Cancer Institute; Contract grant numbers: U54 CA100926, R21 CA123473, R33 CA83208, PO13330.
Literature Cited
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augenicht L, Shi L, Mariadason J, Laboisse C, Velcich A. Repression of MUC2 gene expression by butyrate, a physiological regulator of intestinal cell maturation. Oncogene. 2003;22:4983–4992. doi: 10.1038/sj.onc.1206521. [DOI] [PubMed] [Google Scholar]
- Augenlicht LH, Velcich A, Heerdt BG. Short-chain fatty acids and molecular and cellular mechanisms of colonic cell differentiation and transformation. Adv Exp Med Biol. 1995;375:137–148. doi: 10.1007/978-1-4899-0949-7_12. [DOI] [PubMed] [Google Scholar]
- Augenlicht LH, Anthony GM, Chruch TL, Edelmann W, Kucherlapati R, Yang KY, Lipkin M, Heerdt BG. Short chain fatty acid metabolism, apoptosis and Apc initiated tumorigenesis in the mouse gastrointestinal mucosa. Cancer Res. 1999;59:6005–6009. [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bordonaro M, Mariadason JM, Aslam F, Heerdt BG, Augenlicht LH. Butyrate induced cell cycle arrest and apoptotic cascade in colonic carcinoma cells: Modulation of the B-catenin-Tcf pathway, and concordance with effects of sulindac and trichostatin, but not curcumin. Cell Growth Differ. 1999;10:713–720. [PubMed] [Google Scholar]
- Diaz G, Paraskeva C, Thomas M, Binderup L, Hague A. Apoptosis is induced by the active metabolite of vitamin D3 and its analogue EB1089 in colorectal adenoma and carcinoma cells: Possible implication for prevention therapy. Cancer Res. 2000;60:2304–2312. [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hague A, Manning AM, Hanlon KA, Huschtscha LI, Hart D, Paraskeva C. Sodium butyrate induces apoptosis in human colonic tumor cell lines in a p53-independent pathway: Implications for the possible role of dietary fibre in the prevention of large-bowel cancer. Int J Cancer. 1993;55:498–505. doi: 10.1002/ijc.2910550329. [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Heerdt BG, Houston MA, Augenlicht LH. Potentiation by specific short-chain fatty acids of differentiation and apoptosis in human colonic carcinoma cell lines. Cancer Res. 1994;54:3288–3294. [PubMed] [Google Scholar]
- Heruth DP, Zirnstein GW, Bradley JF, Rothberg PG. Sodium butyrate causes an increase in the block to transcriptional elongation in the c-myc gene in SW837 rectal carcinoma cells. J Biol Chem. 1993;268:20466–20472. [PubMed] [Google Scholar]
- Hulit J, Wang C, Li Z, Albanese C, Rao M, Di Vizio D, Shah S, Byers SW, Mahmood R, Augenlicht LH, Russell R, Pestell RG. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol Cell Biol. 2004;24:7598–7611. doi: 10.1128/MCB.24.17.7598-7611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene RG, Mueller A, Landick R, London L. Transcriptional pause, arrest and termination sites for RNA polymerase II in mammalian N- and c-myc genes. Nucleic Acids Res. 1999;27:3173–3182. doi: 10.1093/nar/27.15.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klampfer L, Huang J, Sasazuki T, Shirasawa S, Augenlicht L. Oncogenic Ras promotes butyrate-induced apoptosis through inhibition of gelsolin expression. J Biol Chem. 2004;279:36680–36688. doi: 10.1074/jbc.M405197200. [DOI] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- Mariadason JM, Corner GA, Augenlicht LH. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: Comparison with trichostatin A, sulidac, and curcumin and implications for chemoprevention of colon cancer. Cancer Res. 2000;60:4561–4572. [PubMed] [Google Scholar]
- Mariadason J, Nicholas C, L'Italien K, Zhuang M, Smartt H, Heerdt B, Yang W, Corner G, Wilson A, Klampfer L, Arango D, Augenlicht L. Gene expression profiling of intestinal cell epithelial cell maturation along he crypt-villus axis. Gastroenterology. 2005;128:1081–1088. doi: 10.1053/j.gastro.2005.01.054. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the B-catenin/LEF-1 pathway. Proc Nat Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. B-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batle E, Coudreuse D, Haramis AP, Tjon-Pon-Fong M, Moerer P, van den Born M, Soete G, Pals S, Eilers M, Medema R, Clevers H. The b-catenin/TCF-4 complex imposes a crypt progenitor phenotype on coloretal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Wilson AJ, Velcich A, Arango D, Kurland AR, Shenoy SM, Pezo RC, Levsky JM, Singer RH, Augenlicht LH. Novel detection and differential utilization of a c-myc transcriptional block in colon cancer chemoprevention. Cancer Res. 2002;62:6006–6010. [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]


