Abstract
We examined the emergence of CXCR4 (i.e., X4) tropism in 67 male human immunodeficiency virus type 1 (HIV-1) seroconverters from the Multicenter AIDS Cohort Study (MACS) who were selected to reflect the full spectrum of rates of HIV-1 disease progression. A mean of 10 serial samples per donor were evaluated by a laboratory-validated, commercially available assay to determine phenotypic coreceptor use. A total of 52% of men had dual- or mixed-tropic HIV-1 detected at 1 or more of the time points tested. Use of X4 by HIV-1 was detected more frequently among men who developed AIDS (defined as a CD4+ T cell count of < 200 cells/μL and/or an AIDS-defining illness)≤11 years after seroconversion than among those who did not (P = .005), as well as among men who exhibited a total T cell count decline (i.e., a CD3+ inflection point), compared with those who did not (P = .03). For men in whom both X4 virus and an inflection point were detected, emergence of X4 virus preceded the inflection point by a median of 0.83 years. The median CD4+ T cell count at first detection of X4 viruses before the onset of AIDS was 475 cells/μL. We conclude that HIV-1 variants that used X4 frequently emerged at high CD4+ T cell counts and may contribute to the decrease in T cell numbers during late HIV-1 infection.
To infect cells, HIV-1 must bind to both the CD4 protein and another transmembrane “coreceptor.” Early in the course of disease, HIV-1 variants usually bind to the chemokine coreceptor CCR5; such variants are termed “R5 viruses.” Viruses using another chemokine coreceptor, CXCR4, termed “X4 viruses,” emerge later in disease progression in ~50% of individuals infected with clade B virus [1], although a higher percentage has been suggested on the basis of studies that used sensitive detection techniques [2]. The presence of X4 viruses has been associated with accelerated disease progression and a decrease in CD4+ T cells [3, 4], but despite descriptions of in vitro cytopathicity of X4 viruses [5-7], no causal in vivo relationship or mechanism for this association has been proven [8, 9]. In 3 studies of coreceptor evolution during untreated HIV-1 infection [3, 4, 10], X4 viruses tended to emerge just before the beginning of the decrease in the total (CD3+) T cell count (i.e., the CD3+ inflection point) that presages the onset of an AIDS-defining illness [11]. The ability of HIV-1 to bind to CXCR4 may facilitate destruction of naive T cells, which preferentially express CXCR4 as opposed to CCR5 [8], and thus may contribute to more rapid depletion of T cells.
The uncertainty surrounding mechanisms of selection and evolution of X4-using HIV-1 and the inability to predict the emergence of these strains have taken on greater importance since the development of CCR5 antagonists as inhibitors of HIV-1 replication [12,13]. For example, in a recent study of monotherapy with a CCR5 antagonist, X4 viruses present in low amounts replaced the R5 population shortly after commencement of therapy [14]. This has led to concern that use of CCR5 antagonists may predispose to selection of CXCR4-tropic viruses and, thus, to faster progression of HIV-1 disease than would occur without blockade of CCR5.
To further characterize the emergence of X4 strains during HIV-1 infection, we undertook a systematic analysis of HIV-1 coreceptor tropism in men from the Multicenter AIDS Cohort Study (MACS; study Web site available at: http://www.statepi.jhsph.edu/macs/macs.html) who were selected for this study in order to reflect the full spectrum of rates of HIV-1 disease progression. Our specific goals were to evaluate (1) the timing of X4 virus emergence with respect to CD3+ T cell decline and development of an AIDS-defining event, (2) the degree to which emergence of X4 viruses correlates with an increased risk of rapid disease progression, and (3) whether determination of the CD4+ T cell count can be an accurate screen for detectable levels of CXCR4-tropic viruses.
SUBJECTS, MATERIALS, AND METHODS
Study population
The MACS is a longitudinal study of the natural history of HIV-1 infection in men who have sex with men that has enrolled 6972 men in Baltimore, Pittsburgh, Chicago, and Los Angeles. A total of 4953 men were enrolled during 1984–1985 [15], 668 during 1987–1991, and 1351 during 2001–2003. Men have been followed up at 6-month intervals with administration of questionnaires, physical examinations, and collection of blood specimens. Some specimens undergo immediate laboratory analysis to determine characteristics such as plasma viral load and T cell subsets, whereas others, including plasma, serum, and cryopreserved peripheral blood mononu-clear cells, are stored for future use.
Participants for the present study were selected from 425 MACS participants who seroconverted before 1995, whose sero-conversion dates were known with an accuracy of ±5 months [16], and who had frequent study visits and unthawed samples available from most visits. Progression of HIV-1 disease was defined in terms of the interval between seroconversion and onset of an AIDS-defining event, defined as development of a CD4+ T cell count of <200 cells/μL on 2 consecutive study visits and/or an AIDS-defining illness. Progression was considered rapid if the interval was ≤5 years, moderate if it was >5 to 9 years, slow if it was >9 to 11 years, and very slow if it was >11 years or if no event was recorded during >11 years of follow-up. The follow-up times for individuals who started highly active anti-retroviral therapy (HAART) before development of AIDS were censored at the date of HAART initiation. If available, plasma specimens obtained at the first postseroconversion time point and every 12 months thereafter were tested. Because of previous studies that linked the emergence of X4 viruses to the onset of CD3+ T cell decline [2, 10], samples from men with an inflection point as defined elsewhere [17] were studied at 6-month intervals for 2 years before and after the inflection point.
HIV-1 phenotypic analysis
Serum that had never been thawed was aliquotted, and 0.5–1 mL was shipped on dry ice to Monogram Biosciences (formerly Virologic; South San Francisco, CA) for rethawing and subsequent analysis of phenotypic coreceptor use by means of the PhenoSense Entry assay (currently marketed as the Trofile assay). This is a laboratory-validated, commercially available assay [18]. Briefly, the plasma or serum specimen is thawed, the viral RNA extracted, and the full-length env genes amplified by PCR. The env genes are cloned into a vector and cotransfected with a luciferase reporter pseudovirus lacking the env gene. The pseudovirions produced are used to infect U87 human cell lines expressing CD4 and either CCR5 or CXCR4. The ability of the pseudovirions to enter the cells is detected by chemiluminescence and expressed as relative light units (RLU), which reflect the distribution of tropisms in the donor's circulating swarm of HIV. The assay does not distinguish between truly dual-tropic viruses and mixed populations of viruses tropic for either CCR5 or CXCR4. It can reliably detect a minority population that consists of as little as 5%–10% of the total viral population [19].
Statistical analysis
Men in whom X4 virus was detected at any time point were compared with men with persisting R5 virus, according to age at seroconversion, race, MACS study site, year of seroconversion, CCR5Δ32 heterozygosity, and early CD4+ T cell counts and log10 plasma HIV RNA loads. Early CD4+ T cell count and viral load were defined as the average of all measurements made during the first 2.5 years following sero-conversion. The statistical significance of differences within the study group and those between the study group and the overall group of MACS seroconverters was determined using χ2 tests for categorical variables and t tests with unequal variances for continuous variables. The overall group was defined as all men who seroconverted before 1995 and whose date of seroconversion was known with an accuracy of ±5 months. Logistic regression models in which the 4 progression groups were incorporated as independent variables were used to examine whether X4 emergence before onset of AIDS (treated as the dependent variable) was related to the rate of progression to AIDS. The rate of progression to AIDS was evaluated according to the following 3 outcomes: the time to achievement of a confirmed CD4+ T cell count of <200 cells/μL, the time to onset of an AIDS-defining illness, and the time to the earlier of these events. Repeated measurement logistic regression was used to examine whether a change in a biomarker (i.e., CD4+ T cell count or plasma HIV-1 RNA concentration) from one visit to the next influenced the emergence of X4 virus at the later visit. Product-limit nonpara-metric survival analyses were used to describe the distribution of time from emergence of X4 virus to development of a CD4+ T cell count of >200 cells/μL and/or an AIDS-defining illness or to censoring at initiation of HAART. We considered the α for statistical significance to be .05.
RESULTS
Characteristics of the study population
The population selected for study consisted of 67 men, who contributed 678 samples for testing. Tropism was determined in 630 samples (93%) and could not be determined for 48 specimens (7%), mainly because of low viral loads (i.e., >1000 copies/mL). On average, samples from 10 time points were tested per patient; more samples (mean, 11) were tested for men with slow disease progression (hereafter, “slow progressors”), and fewer (mean, 7) were tested for those with rapid disease progression (hereafter, “rapid progressors”). Of the 67 men enrolled, 35 (52%) exhibited an X4 virus at any of the time points tested. These men did not differ from the 32 men who had persistent CCR5 tropism with respect to age, seroconversion date, race, or study center (table 1). However, they did have higher early HIV loads, a difference that was of borderline statistical significance (P = .08), and were more likely to be CCR5Δ32 heterozygotes, a difference that was not significant (P = .11).
Table 1.
Baseline characteristics of the selected study population and of all seroconverters in the Multicenter AIDS Cohort Study (MACS).
| Study population |
|||||
|---|---|---|---|---|---|
| Characteristic | Overal (n = 67) |
Men with R5 at all time points (n = 32) |
Men with X4 at ≥1 time point (n = 35) |
Pa | All MACS seroconverters (n = 425) |
| Age at seroconversion, mean ± SD, years | 34.1 ± 7.2 | 35.1 ± 7.2 | 33.6 ± 7.2 | .32 | 34.6 ± 7.9 |
| Seroconversion date, median (IQR) | 1986.1 (1985.0–1989.1) | 1986.3 (1985.2–1990.0) | 1985.6 (1984.9–1988.9) | .16 | 1986.4 (1985.2–1989.8) |
| Race | .43 | ||||
| White | 58 (86.6) | 26 (81.2) | 32 (91.4) | 364 (85.7) | |
| Black | 5 (7.4) | 3 (9.4) | 2 (5.7) | 23 (5.4) | |
| Other | 4 (6.0) | 3 (9.4) | 1 (2.9) | 38 (8.9) | |
| Study center | .41 | ||||
| Baltimore | 29 (43.3) | 11 (34.4) | 18 (51.4) | 106 (24.9)b | |
| Chicago | 11 (16.4) | 5 (15.6) | 6 (17.1) | 97 (22.8) | |
| Pittsburgh | 6 (9.0) | 3 (9.4) | 3 (8.6) | 111 (26.1) | |
| Los Angeles | 21 (31.3) | 13 (40.6) | 8 (22.9) | 111 (26.1) | |
| CCR5Δ32 heterozygosity | 12 (17.9) | 3 (9.4) | 9 (25.7) | .11 | 73 (17.2) |
| Set point viral load, mean ± SD, log10 copies/mLc | 4.3 ± 0.6 | 4.1 ± 0.7 | 4.4 ± 0.4 | .08 | 4.3 ± 0.7 |
| Early CD4+ T cell count, mean ± SD, cells//μLc | 737 ± 259 | 742 ± 286 | 732 ± 237 | .88 | 665 ± 262d |
NOTE. Data are no. (%) of subjects, unless otherwise indicated. IQR, interquartile range; SD, standard deviation.
Values pertain to differences between the R5 group and the X4 group and were determined by the t test and Fisher exact test for continuous and categorical variables, respectively.
P = .0012 for the distribution by study center for the study population, compared with that for all MACS seroconverters.
Values were calculated on the basis of all measurements performed 0.5–2.5 years after seroconversion.
P < .04 for the CD4+ T cell count for the study population, compared with that for all MACS seroconverters.
Table 1 also compares the study population with the group of all seroconverters from the MACS. The study slightly over-sampled seroconverters enrolled in Baltimore and Los Angeles. The average CD4+ T cell counts during the first 2.5 years after seroconversion were slightly higher in the study sample than in the total seroconverting population (P = .08), but this difference was insignificant after adjustment for higher CD4+ T cell counts in general among men in Baltimore relative to those among men at the other study sites. The other characteristics examined were similar in the study group and all seroconverters in the MACS, as were proportions of rapid progressors (29% vs. 31%, respectively) and men with very slow progression of disease (hereafter, “very slow progressors”; 24% vs. 26%, respectively).
Emergence and detection of X4 virus
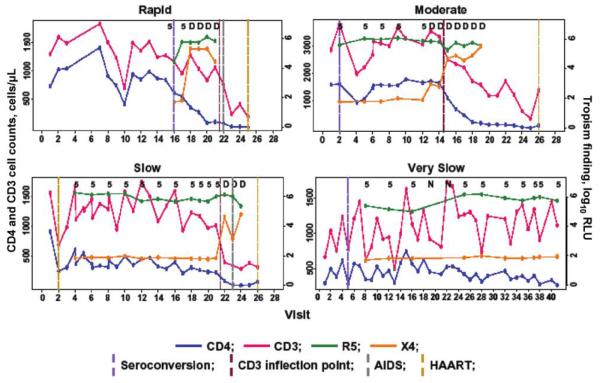
Figure 1 shows representative natural histories of disease progression in subjects from each progression category, in terms of CD4+ T cell counts, CD3+ T cell counts, and luminescence for CCR5- and CXCR4-tropic viruses. Most determinations were clearly positive for one or both types of virus, with few low—and, therefore, potentially equivocal—values. In addition, there was usually a clear increase in the luminescence associated with emergence of X4 virus over several time points. As in previous studies, total (i.e., CD3+) T cell levels fluctuated but were generally lower after the T cell inflection point than at any point before this time.
Figure 1.
Representative longitudinal CD4+ and CD3+ T cell counts and HIV tropism patterns, by rate of progression to AIDS, in a population of men from the Multicenter AIDS Cohort Study (MACS). Progression to AIDS was defined as a CD4+ cell count of >200 cells/μL on 2 consecutive study visits and/or onset of an AIDS-defining illness and was considered rapid if the time since seroconversion was ≤5 years, moderate if the time was >5 to 9 years, slow if the time was >9 to 11 years, and very slow if the time was >11 years or if no event was recorded during >11 years of follow-up. Viral tropism was determined by means of the Trofile assay (Monogram Biosciences). A value of <102.1 relative light units (RLU) is considered negative; fluctuations below this value do not change the interpretation of the results. D, mixed CCR5-CXCR4 tropism; N, no result; vertical lines, events relative to semiannual MACS study visits; 5, CCR5 tropism only.
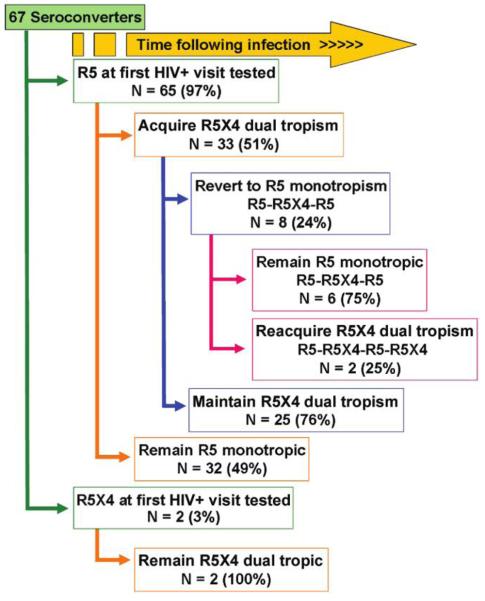
Figure 2 shows the individual patterns of viral coreceptor use over time among the 67 men studied. For all 35 men in whom X4 virus was detected, use of CCR5 coreceptors by HIV-1 was concomitantly detected in the X4 virus–positive specimens. (As mentioned, the assay used does not distinguish between viruses that can bind both coreceptors and mixtures of monotropic viruses.) Of these men, 27 (77%) continued to exhibit X4 viruses, whereas 8 (23%) had at least 1 reversion to “pure” R5 viral populations (i.e., populations in which X4 viruses made up less than 5%–10% of the total viral population).
Figure 2.
Individual patterns of X4 virus emergence among 67 men from the Multicenter AIDS Cohort Study.
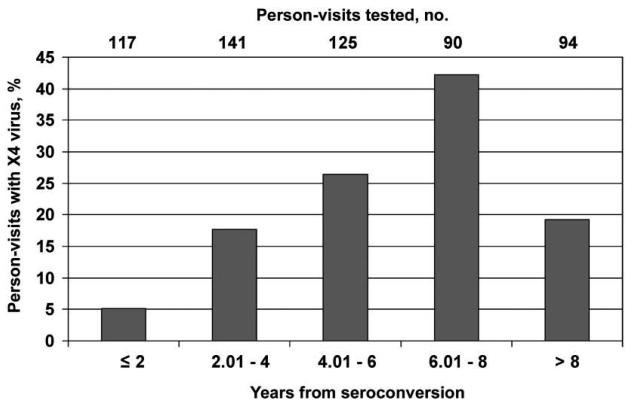
The likelihood of X4 virus emergence increased with the time since seroconversion for up to 8 years (figure 3). The earliest emergence of X4 viruses was ~15 months after seroconversion, observed in 2 men; for one of them, the sample yielding this finding was the earliest sample for which tropism could be determined. Four men (6.0%) developed a detectable level of X4 virus ≤2 years after seroconversion; all 4 were rapid progressors, with development of an AIDS-defining illness and/or a CD4+ T cell count of <200 cells/μL by 3.5 years after seroconversion.
Figure 3.
Prevalence of X4 viruses, by time after HIV-1 seroconversion, among specimens obtained from a population of men from the Multicenter AIDS Cohort Study.
Risk of X4 virus emergence, by rate of disease progression
Of the 67 men studied, 9 started HAART ≤9 years after seroconverting without having developed an AIDS-defining illness or a CD4+ T cell count of <200 cells/μL. These men were excluded from the analysis of HIV-1 disease progression. Fourteen (24.1%) of the remaining 58 subjects were rapid progressors, 23 (39.7%) had moderate progression of disease (hereafter, “moderate progressors”), 7 (12.1%) were slow progressors, and 14 (24.1%) were very slow progressors. A total of 55 (95%) developed an AIDS-defining illness or a CD4+ T cell count of <200 cells/μL before HAART initiation. Three moderate progressors who developed AIDS shortly after initiating HAART were retained in the analysis because HAART initiation and AIDS onset both occurred 5–9 years after seroconversion. Compared with very slow progressors, all other groups were more likely to have X4 virus detected at any time point before development of an AIDS-defining illness or a CD4+ T cell count of <200 cells/μL, with odds ratios (ORs) of 4.9, 6.9, and 1.5 for the rapid, moderate, and slow progressors, respectively (table 2). When progression to AIDS was defined separately as the development of a CD4+ T cell count of <200 cells/μL or as the development of an AIDS-defining illness, the corresponding ORs were somewhat higher, ranging from 4.0 to 15.9 (table 2). Among men who developed either a CD4+ T cell count of <200 cells/μL or an AIDS-defining illness, X4 virus emerged before the AIDS-defining event in 52% of subjects (25 of 48) and at the same time as the event or shortly thereafter in 13% (6 of 48). In contrast, X4 virus emerged in only 30% of men (3 of 10) for whom neither event occurred before HAART initiation (P = .04). By nonparametric survival analysis of the 28 men with X4 emergence before an AIDS-defining event, the median time from emergence of detectable X4 virus to development of either a CD4+ T cell count of <200 cells/μL or an AIDS-defining illness was 13.2 months (interquartile range [IQR], 11.8–28.2 months). X4 HIV-1 was detected more frequently among men who developed AIDS within 11 years, compared with men who did not (P = .005).
Table 2.
Progression to AIDS-defining events in a population of seroconverting men from the Multicenter AIDS Cohort Study, by R5 virus persistence and X4 virus emergence.
| Progression rate |
|||||
|---|---|---|---|---|---|
| AIDS-defining event | Rapid | Moderate | Slow | Very slow | Total |
| A. CD4+ T cell count <200 cells/μL or AIDS-defining illness | |||||
| R5 persistence at all visits | 4 | 6 | 3 | 11 | 24 |
| X4 emergence at any visit | |||||
| Before AIDS onset | 8 | 15 | 2 | 3 | 28 |
| At or after AIDS onset | 2 | 2 | 2 | 0 | 6 |
| Total | 14 | 23 | 7 | 14 | 58a |
| OR (P) for X4 emergence | |||||
| Before AIDS onsetb | 4.9 (.061) | 6.9 (.014) | 1.5 (.718) | … | … |
| At any timec | 9.2 (.012) | 10.4 (.004) | 4.9 (.114) | … | … |
| B. CD4+ T cell count <200 cells/μL | |||||
| R5 persistence at all visits | 4 | 1 | 2 | 13 | 20 |
| X4 emergence at any visit | |||||
| Before AIDS onset | 7 | 11 | 4 | 3 | 25 |
| At or after AIDS onset | 2 | 2 | 2 | 0 | 6 |
| Total | 13 | 14 | 8 | 16 | 51d |
| OR (P) for X4 emergence | |||||
| Before AIDS onsetb | 5.1 (.056) | 15.9 (.003) | 4.3 (.124) | … | … |
| At any timec | 9.8 (.01) | 56.3 (<.001) | 13.0 (.013) | … | … |
| C. AIDS-defining illness | |||||
| R5 persistence at all visits | 3 | 5 | 3 | 12 | 23 |
| X4 emergence at any visit | |||||
| Before AIDS onset | 4 | 17 | 4 | 4 | 29 |
| At or after AIDS onset | 0 | 0 | 0 | 0 | 0 |
| Total | 7 | 22 | 7 | 16 | 52e |
| OR (P) for X4 emergence before AIDS onsetf | 4.0 (.148) | 10.2 (.003) | 4.0 (.148) | … | … |
NOTE. Data are no. of subjects, unless otherwise indicated. Progression to AIDS was defined as a CD4+ cell count of <200 cells/μL on 2 consecutive study visits and/or onset of an AIDS-defining illness and was considered rapid if the time since seroconversion was ≤5 years, moderate if the time was >5 to 9 years, slow if the time was >9 to 11 years, and very slow if the time was >11 years or if no event was recorded during >11 years of follow-up.
Nine men were excluded because they initiated highly active antiretroviral therapy (HAART) ≤9 years before development of a CD4+ T cell count of <200 cells/μL or an AIDS-defining illness.
Odds ratios (ORs) and P values for X4 emergence before event versus R5 persistence or X4 emergence at or after the event, using the very slow progressors as the reference group.
ORs and P values for X4 emergence at any time (before or at/after event) versus R5 persistence, using the very slow progressors as the reference group.
Sixteen men were excluded because they initiated HAART before development of a CD4+ T cell count of <200 cells/μL.
Fifteen men were excluded because they initiated HAART before development of an AIDS-defining illness.
ORs and P values for X4 emergence before AIDS onset versus R5 persistence, using the very slow progressors as the reference group.
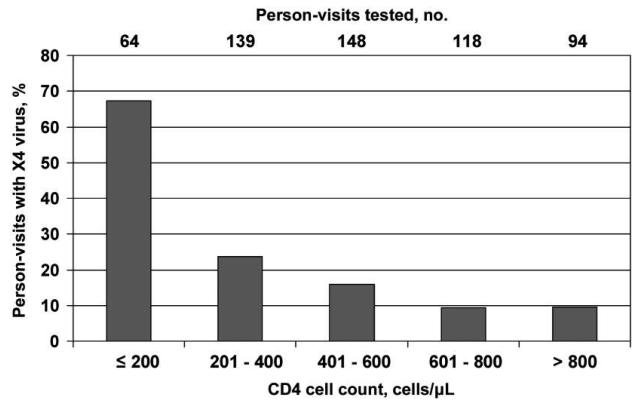
Among 29 men in whom X4 virus was detected before onset of an AIDS-defining illness, the median CD4+ T cell count at the visit when X4 virus was first detected was 475 cells/μL (IQR, 265–718 cells/μL; mean, 505 cells/μL). The median CD4+ T cell counts at the time of first X4 emergence before AIDS onset were 308 cells/μL among 4 rapid progressors, 481 cells/μL among 17 moderate progressors, 189 cells/μL among 4 moderately slow progressors, and 707 cells/μL among 4 slow progressors. Neither CD4+ T cell count nor viral load was associated with the likelihood of having detectable X4 virus at the next visit (within 1 year). The proportion of person-visits in which X4 virus was detected increased from ~10% among person-visits where the CD4+ T cell count was >600 cells/μL to >67% among person-visits where the CD4+ T cell count was ≤200 cells/μL (figure 4). Of the 502 person-visits at which CD4+ T cell counts were >200 cells/μL, X4 virus was detected at 77 (15.3%).
Figure 4.
Prevalence of X4 viruses, by CD4+ T cell count, among specimens obtained from a population of men from the Multicenter AIDS Cohort Study.
Temporal association between emergence of X4 virus and loss of T cell homeostasis
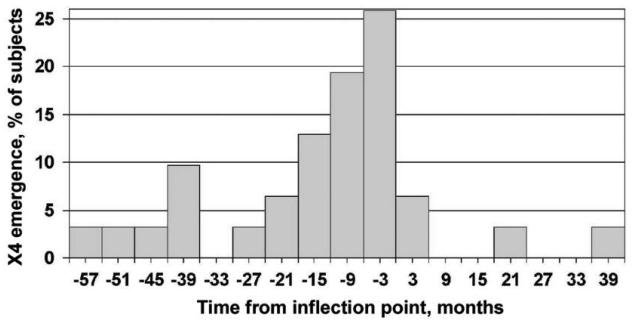
Onset of circulating CD3+ T lymphocyte decline has been described as a marker for impending immunological decline [4, 11, 17, 20] and is referred to as the CD3+ T cell inflection point. Fifty (75%) of 67 men in this study had a clearly defined CD3+ inflection point, which is comparable to the prevalence (77%) among men with progressive HIV-1 infection described in the study by Gange et al. [17]. Twenty-seven men (54%) with a CD3+ inflection point had X4 virus detected before inflection, compared with only 4 (24%) with no inflection point (P = .03). Nonparametric survival analysis revealed that the median time from X4 emergence to the inflection point was 1.23 years (IQR, 0.33–3.22 years). For 31 men with an inflection point in whom X4 virus emerged, the median time from X4 virus detection to the inflection point was 0.83 years (IQR, 0.28–1.99 years), with X4 virus emerging in 21 (66%) ≤2 years before the inflection point (figure 5) and in 27 (87%) before the inflection point. There were 28 men who developed CD4+ T cell counts of <200 cells/μL and had X4 virus emergence; 22 (79%) of these emergences preceded the low CD4+ T cell count. For those who developed AIDS, X4 virus emergence, if it occurred, always preceded AIDS onset. For the 6 men in whom X4 virus emerged after the first CD4+ T cell count of <200 cells/μL, the X4 virus usually emerged at the same time or in the first sample after this event.
Figure 5.
Time of X4 virus emergence in relation to CD3+ inflection point among 31 men from the Multicenter AIDS Cohort Study who exhibited both X4 emergence and a CD3+ inflection point. See Subjects, Materials, and Methods for a description of the inflection point.
DISCUSSION
In this nonrandom selection of individuals, approximately two-thirds of the men who developed AIDS also developed X4-tropic or dual (i.e., X4R5)–tropic virus. This proportion was significantly higher among men with more rapid progression than among those with slower progression. Men who developed X4 viruses had a significant acceleration in the decline of CD3+ T lymphocytes, which commenced, on average, ~1 year after the first appearance of these viruses, similar to previous reports. These data are consistent with a causal relationship between emergence of X4 virus and onset of decreases in the T cell count, although the presence of X4 variants at a time before they were detected by the assay used cannot be excluded. Approximately 37% of individuals developed a CD4+ T cell count of <200 cells/μL and/or an AIDS-defining illness without detectable X4 virus; this was more common among slow and very slow progressors but was also seen in rapid and moderate progressors. It is possible that more frequent sampling might reveal a higher prevalence of X4 viruses among persons progressing to AIDS, especially considering the observation in this and other studies [2, 21] that X4 viruses may decrease in frequency or disappear during HIV-1 infection.
We found that men who did not have AIDS first exhibited X4 virus at relatively high CD4+ T cell counts, with a median value of 475 cells/μL. This closely resembles the corresponding value of 0.48 × 109 cells/L found by Koot et al. [3] in a prospective study that used the MT-2 assay for detection of syncytium-inducing viruses. Prospective studies would be expected to yield earlier times of detection and, thus, higher CD4+ T cell counts at detection, compared with cross-sectional studies. In the present study, median CD4+ T cell counts at first detection of X4 viruses were relatively high in all progression groups, indicating that X4 expression at high CD4+ T cell counts is quite common. In cross-sectional studies, the presence of X4 virus was strongly associated with a low CD4+ T cell count [22-25], but even in such studies it has been noted that a substantial proportion of subjects with X4 viruses can have high CD4+ T cell counts [25]. Taken together, the results of both longitudinal and cross-sectional studies suggest that restriction of tropism testing to HIV-infected persons with low CD4+ T cell counts will lead to substantial underdetection of X4 viruses in the overall population. In particular, on the basis of current treatment guidelines, many patients will have developed X4 viruses before they are considered for antiretroviral therapy. Among newly infected men, those in whom X4 virus emerged at some later date tended to have higher plasma HIV RNA values than those who expressed only R5 viruses (P = .08). Plasma HIV RNA load was also found to be a strong predictor in another study [23]. Finally, the finding of relatively high CD4+ T cell counts at the first appearance of X4 viruses supports an etiologic role for these viruses in the decrease of T cell counts.
A limitation of the present study is the possibility that minority variants of HIV-1 were missed. This may have happened if they were present at less than 5%–10% of the viral population at a given time point. However, the assay used has been validated in laboratory studies [19] and in several large clinical trials, including ACTG 5211, MOTIVATE 1, MOTIVATE 2, and Maraviroc Study A4001029. In the latter study, treatment-experienced patients with dual/mixed-tropic HIV-1 demonstrated no significant reduction in HIV-1 RNA, as opposed to the previous studies involving patients with CCR5-tropic virus [26]. Therefore, it is likely that the vast majority of the results reported here reflect the true HIV-1 coreceptor phenotype in these men, at least to the extent that we can measure it today. Reproducibility of the test could also be a limitation, because a small percentage of subjects entering clinical trials have exhibited changes from R5 to X4 virus during the first few weeks after study entry without concomitant changes in antiretroviral therapy [25, 26]. However, these changes are believed to reflect variability of the test at low or borderline levels of X4 virus, which is probably due to the biologic variability of viral populations in the host, because the reproducibility of the test on replicate samples has been excellent [19]. In most of the samples tested in the present study, X4 virus levels were high and unequivocal.
The factors responsible for the emergence of X4 viruses are not known. In this study, neither CD4+ T cell count nor plasma HIV-1 RNA level at a given visit was associated with the presence of X4 virus at the following visit. It has been suggested that an increase in natural ligands of CCR5 or a down-regulation of CCR5 itself may confer a selective advantage on viruses that use other coreceptors [27]. Alternatively, a change in the relative abundance of memory and naive T cells may favor emergence of X4 viruses later during the course of disease [28].
The results of this study are consistent with the hypothesis that it may be prudent to initiate HAART as soon as X4 viruses are detected, because such treatment can prevent X4 virus–associated disease progression and thereby improve treatment outcome. A recent report that compared viral tropism between treatment-experienced patients and treatment-naive patients by means of the same assay used here found a significant increase in the prevalence of X4 virus in the treated group [29]. This finding was somewhat confounded by the more advanced stage of disease in the treated group, but not completely so. An early report suggested that HAART initiation before genotypic evidence of CXCR4-tropism development was associated with a better long-term outcome than HAART initiation after X4 virus had been detected [22]. However, a more recent report from the same group, classifying viruses as R5 or X4 by means of the same assay used in this study, did not find that the presence of X4 virus before treatment initiation was correlated with poorer response to treatment [23]. Further complicating this question is the possibility that X4 virus may be more susceptible to antiviral drugs other than entry inhibitors [30]. Data also suggest that cellular reservoirs of HIV-1 may become more CXCR4 tropic in the presence of HAART [31]. Thus, caution in the use of CCR5-entry inhibitors and close monitoring of persons who receive them are warranted. However, emergence of X4 virus during therapy with maraviroc may not be associated with immunologic decline and may be reversible on discontinuation of maraviroc [26].
The results from this study were derived from men with subtype B HIV-1 infection. Emergence of X4 viruses has been reported in persons infected with other subtypes of HIV-1, but it should be kept in mind that relationships between X4 viruses associated with non-B subtypes and HIV-1 disease progression may be different from the relationships described in this study.
STUDY GROUP MEMBERS
The Multicenter AIDS Cohort Study (MACS) includes the following: Baltimore: The Johns Hopkins Bloomberg School of Public Health: Joseph B. Margolick (Principal Investigator), Haroutune Armenian, Barbara Crain, Adrian Dobs, Homayoon Farzadegan, Joel Gallant, John Hylton, Lisette Johnson, Shenghan Lai, Ned Sacktor, Ola Selnes, James Shepherd, Chloe Thio. Chicago: Howard Brown Health Center, Feinberg School of Medicine, Northwestern University, and Cook County Bureau of Health Services: John P. Phair (Principal Investigator), Joan S. Chmiel (Co-Principal Investigator), Sheila Badri, Bruce Cohen, Craig Conover, Maurice O'Gorman, David Ostrow, Frank Palella, Daina Variakojis, Steven M. Wolinsky. Los Angeles: University of California, UCLA Schools of Public Health and Medicine: Roger Detels (Principal Investigator), Barbara R. Visscher (Co-Principal Investigator), Aaron Aronow, Robert Bolan, Elizabeth Breen, Anthony Butch, Thomas Coates, Rita Effros, John Fahey, Beth Jamieson, Otoniel Martínez-Maza, Eric N. Miller, John Oishi, Paul Satz, Harry Vinters, Dorothy Wiley, Mallory Witt, Otto Yang, Stephen Young, Zuo Feng Zhang. Pittsburgh: University of Pittsburgh, Graduate School of Public Health: Charles R. Rinaldo (Principal Investigator), Lawrence Kingsley (Co-Principal Investigator), James T. Becker, Robert L. Cook, Robert W. Evans, John Mellors, Sharon Riddler, Anthony Silvestre. Data Coordinating Center: The Johns Hopkins Bloomberg School of Public Health: Lisa P. Jacobson (Principal Investigator), Alvaro Muñoz (Co-Principal Investigator), Haitao Chu, Stephen R. Cole, Christopher Cox, Gypsyamber D'Souza, Stephen J. Gange, Janet Schollenberger, Eric C. Seaberg, Sol Su. National Institutes of Health: National Institute of Allergy and Infectious Diseases: Robin E. Huebner; National Cancer Institute: Geraldina Dominguez; National Heart, Lung and Blood Institute: Cheryl McDonald.
Acknowledgments
We thank Pfizer, particularly Howard Mayer, Ann Kolokathis, and Kay Mitzel, for funding the tropism determinations and providing support to J.C.S. during the study; Kathy Valiasek, Jeanette Whitcomb, and Judy Jang Jan of Monogram Biosciences, for organizing samples for the tropism determinations; Janet Schollenberger and staff, for sample management, seletion, and shipping; Judy Konig, for preparing some of the figures; and the men who have participated in the Multicenter AIDS Cohort Study, whose dedication to the study made this report possible.
Financial support: Multicenter AIDS Cohort Study (grants UO1-AI-35042, 5-MO1-RR-00722 [GCRC], UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613, and UO1-AI-35041); Pfizer.
Footnotes
Potential conflicts of interest: H.M. is an employee of Pfizer. All other authors: none reported.
Presented in part: XIVth International Conference on AIDS, Toronto, Canada, August 2006 (abstract TUPE0001).
References
- 1.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koot M, Keet IP, Vos AH, et al. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann Intern Med. 1993;118:681–8. doi: 10.7326/0003-4819-118-9-199305010-00004. [DOI] [PubMed] [Google Scholar]
- 4.Maas JJ, Gange SJ, Schuitemaker H, Coutinho RA, van Leeuwen R, Margolick JB. Strong association between failure of T cell homeostasis and the syncytium-inducing phenotype among HIV-1-infected men in the Amsterdam Cohort Study. AIDS. 2000;14:1155–61. doi: 10.1097/00002030-200006160-00012. [DOI] [PubMed] [Google Scholar]
- 5.Decrion AZ, Varin A, Estavoyer JM, Herbein G. CXCR4-mediated T cell apoptosis in human immunodeficiency virus infection. J Gen Virol. 2004;85:1471–8. doi: 10.1099/vir.0.79933-0. [DOI] [PubMed] [Google Scholar]
- 6.Jekle A, Keppler OT, De Clercq E, Schols D, Weinstein M, Goldsmith MA. In vivo evolution of human immunodeficiency virus type 1 toward increased pathogenicity through CXCR4-mediated killing of uninfected CD4 T cells. J Virol. 2003;77:5846–54. doi: 10.1128/JVI.77.10.5846-5854.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kreisberg JF, Kwa D, Schramm B, et al. Cytopathicity of human immunodeficiency virus type 1 primary isolates depends on coreceptor usage and not patient disease status. J Virol. 2001;75:8842–7. doi: 10.1128/JVI.75.18.8842-8847.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berkowitz RD, Alexander S, McCune JM. Causal relationships between HIV-1 coreceptor utilization, tropism, and pathogenesis in human thymus. AIDS Res Hum Retroviruses. 2000;16:1039–45. doi: 10.1089/08892220050075291. [DOI] [PubMed] [Google Scholar]
- 9.Grivel JC, Margolis LB. CCR5- and CXCR4-tropic HIV-1 are equally cytopathic for their T-cell targets in human lymphoid tissue. Nat Med. 1999;5:344–6. doi: 10.1038/6565. [DOI] [PubMed] [Google Scholar]
- 10.Shankarappa R, Gupta P, Learn GH, Jr, et al. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology. 1998;241:251–9. doi: 10.1006/viro.1997.8996. [DOI] [PubMed] [Google Scholar]
- 11.Margolick JB, Munoz A, Donnenberg AD, et al. Failure of T-cell homeostasis preceding AIDS in HIV-1 infection. The Multicenter AIDS Cohort Study. Nat Med. 1995;1:674–80. doi: 10.1038/nm0795-674. [DOI] [PubMed] [Google Scholar]
- 12.Shepherd JC, Quinn TC. Entry inhibitors: a primer on the class and on its most promising agents. Hopkins HIV Rep. 2004;16:1–4, 11. [PubMed] [Google Scholar]
- 13.Westby M, van der Ryst RE. CCR5 antagonists: host-targeted antivirals for the treatment of HIV infection. Antivir Chem Chemother. 2005;16:339–54. doi: 10.1177/095632020501600601. [DOI] [PubMed] [Google Scholar]
- 14.Westby M, Lewis M, Whitcomb J, et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol. 2006;80:4909–20. doi: 10.1128/JVI.80.10.4909-4920.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR., Jr The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126:310–8. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 16.Lyles RH, Munoz A, Yamashita TE, et al. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. J Infect Dis. 2000;181:872–80. doi: 10.1086/315339. [DOI] [PubMed] [Google Scholar]
- 17.Gange SJ, Munoz A, Chmiel JS, et al. Identification of inflections in T-cell counts among HIV-1-infected individuals and relationship with progression to clinical AIDS. Proc Natl Acad Sci U S A. 1998;95:10848–53. doi: 10.1073/pnas.95.18.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coakley E, Petropoulos CJ, Whitcomb JM. Assessing chemokine co-receptor usage in HIV. Curr Opin Infect Dis. 2005;18:9–15. doi: 10.1097/00001432-200502000-00003. [DOI] [PubMed] [Google Scholar]
- 19.Whitcomb JM, Huang W, Fransen S, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother. 2007;51:566–75. doi: 10.1128/AAC.00853-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margolick JB, Donnenberg AD, Chu C, O'Gorman MR, Giorgi JV, Munoz A. Decline in total T cell count is associated with onset of AIDS, independent of CD4+ lymphocyte count: implications for AIDS pathogenesis. Clin Immunol Immunopathol. 1998;88:256–63. doi: 10.1006/clin.1998.4577. [DOI] [PubMed] [Google Scholar]
- 21.Kitrinos K, Irlbeck D, Bonny T, et al. Program and abstracts of the International AIDS Society (abstract MOPE0006) Toronto, Canada: 2006. HIV-1 coreceptor tropism changes over time in drug naive patients in the absence of ART. [Google Scholar]
- 22.Brumme ZL, Dong WW, Yip B, et al. Clinical and immunological impact of HIV envelope V3 sequence variation after starting initial triple antiretroviral therapy. AIDS. 2004;18:F1–9. doi: 10.1097/00002030-200403050-00001. [DOI] [PubMed] [Google Scholar]
- 23.Brumme ZL, Goodrich J, Mayer HB, et al. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J Infect Dis. 2005;192:466–74. doi: 10.1086/431519. [DOI] [PubMed] [Google Scholar]
- 24.Moyle GJ, Wildfire A, Mandalia S, et al. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J Infect Dis. 2005;191:866–72. doi: 10.1086/428096. [DOI] [PubMed] [Google Scholar]
- 25.Wilkin TJ, Su Z, Kuritzkes DR, et al. HIV type 1 chemokine coreceptor use among antiretroviral-experienced patients screened for a clinical trial of a CCR5 inhibitor: AIDS Clinical Trial Group A5211. Clin Infect Dis. 2007;44:591–5. doi: 10.1086/511035. [DOI] [PubMed] [Google Scholar]
- 26.van der Ryst RE, Westby M. Changes in HIV co-receptor tropism for patients participating in the maraviroc MOTIVATE 1 and 2 clinical trials. Program and abstracts of the 47th Interscience Conference on Antimicrobial Agents and Chemotherapy (abstract H-715) 2007 [Google Scholar]
- 27.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 28.Ribeiro RM, Hazenberg MD, Perelson AS, Davenport MP. Naive and memory cell turnover as drivers of CCR5-to-CXCR4 tropism switch in human immunodeficiency virus type 1: implications for therapy. J Virol. 2006;80:802–9. doi: 10.1128/JVI.80.2.802-809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt PW, Harrigan PR, Huang W, et al. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J Infect Dis. 2006;194:926–30. doi: 10.1086/507312. [DOI] [PubMed] [Google Scholar]
- 30.Skrabal K, Trouplin V, Labrosse B, et al. Impact of antiretroviral treatment on the tropism of HIV-1 plasma virus populations. AIDS. 2003;17:809–14. doi: 10.1097/00002030-200304110-00005. [DOI] [PubMed] [Google Scholar]
- 31.Delobel P, Sandres-Saune K, Cazabat M, et al. R5 to X4 switch of the predominant HIV-1 population in cellular reservoirs during effective highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2005;38:382–92. doi: 10.1097/01.qai.0000152835.17747.47. [DOI] [PubMed] [Google Scholar]