Abstract
The endosomal sorting complex required for transport (ESCRT) machinery controls the incorporation of cargo into intraluminal vesicles of multivesicular bodies. This machinery is used during envelopment of many RNA viruses and some DNA viruses, including herpes simplex virus type 1. Other viruses mature independent of ESCRT components, instead relying on the intrinsic behavior of viral matrix and envelope proteins to drive envelopment. Human cytomegalovirus (HCMV) maturation has been reported to proceed independent of ESCRT components (A. Fraile-Ramos et al. Cell. Microbiol. 9:2955-2967, 2007). A virus complementation assay was used to evaluate the role of dominant-negative (DN) form of a key ESCRT ATPase, vacuolar protein sorting-4 (Vps4DN) in HCMV replication. Vps4DN specifically inhibited viral replication, whereas wild-type-Vps4 had no effect. In addition, a DN form of charged multivesicular body protein 1 (CHMP1DN) was found to inhibit HCMV. In contrast, DN tumor susceptibility gene-101 (Tsg101DN) did not impact viral replication despite the presence of a PTAP motif within pp150/ppUL32, an essential tegument protein involved in the last steps of viral maturation and release. Either Vps4DN or CHMP1DN blocked viral replication at a step after the accumulation of late viral proteins, suggesting that both are involved in maturation. Both Vps4A and CHMP1A localized in the vicinity of viral cytoplasmic assembly compartments, sites of viral maturation that develop in CMV-infected cells. Thus, ESCRT machinery is involved in the final steps of HCMV replication.
Cellular endosomal sorting complex required for transport (ESCRT) machinery controls the evolutionarily conserved process (33) of membrane budding that is normally a component of cytokinesis (6, 46), endosome sorting and multivesicular body (MVB) formation (28). After the initial characterization in retroviruses, many enveloped viruses have been shown to rely on this machinery during envelopment and release from cells (1, 18, 35, 40, 47, 69). Other viruses, such as influenza virus, mature independent of ESCRT machinery and are believed to use an alternative virus-intrinsic pathway (7). The core of the ESCRT machinery consists of five multiprotein complexes (ESCRT-0, -I, -II, and -III and Vps4-Vta1) (27). Vacuolar protein sorting-4 (Vps4) is a critical ATPase that functions downstream of most ESCRT components. Based on sensitivity to dominant-negative (DN) inhibitors of protein function, replication of several RNA viruses, as well as of the DNA virus herpes simplex virus type 1 (HSV-1) (5, 10), have been shown to rely on Vps4 in a manner that is analogous to the formation of MVBs (endosomal compartments containing intraluminal vesicles) (10, 45). Evidence based exclusively on small interfering RNA (siRNA) methods suggested cytomegalovirus (CMV) maturation was independent of ESCRT components, although the maturation of this virus remained MVB associated (16).
ESCRT machinery facilitates envelopment and release at cytoplasmic membranes and recruits cargo for sorting via any of three alternative pathways that converge on a Vps4-dependent downstream step: (i) a tumor susceptibility gene-101 (Tsg101)-dependent pathway, (ii) an apoptosis linked gene-2 interacting protein X (ALIX)-dependent pathway, and (iii) a pathway that relies on a subset of Nedd4-like HECT E3 ubiquitin ligases (35). The involvement of ESCRT in viral envelopment and egress was first observed in human immunodeficiency virus (HIV) (18, 19, 40, 60) and has been extended to equine infectious anemia virus (34, 40, 52, 60), Rous sarcoma virus (29, 70, 71), Mason-Pfizer monkey virus (20, 72), rabies virus (24), Ebola virus (23), hepatitis B virus (68), vaccinia virus (25), HSV-1 (5, 10), and several other RNA and DNA viruses (7). Structural proteins in most of these viruses carry late (L) domains characterized by conserved amino acid motifs (PTAP, PPXY, and YXXL) that mediate protein-protein interactions and facilitate recruitment of ESCRT components to facilitate virus budding. The introduction of mutations in these motifs leads to defects in viral maturation and release from cells (40).
Vps4 controls the release of ESCRT complexes from membranes (18, 40). Inhibition of Vps4A and Vps4B using Vps4ADN reduces levels of viral maturation mediated by L domains (47). For this reason, inhibition by a Vps4DN is considered the gold standard test to establish the role of ESCRT machinery in maturation of any virus (7). Tsg101, a component of ESCRT-I, normally functions to deliver ubiquitinated transmembrane proteins to MVBs (35). HIV-1 p6 Gag PTAP domain interacts with Tsg101 (18) and directs viral cores (capsids) to sites of viral envelopment (39). Upon disruption of HIV-1 PTAP domain, particle release becomes dependent on auxiliary factors, including an ALIX-binding YXXL domain within p6 Gag (60). A minimal amino-terminal L domain of Tsg101 functions as a DN inhibitor of PTAP-mediated viral budding without inhibiting Tsg101-independent PPXY- or YXXL-dependent pathways (40). The murine leukemia virus PPXY domain recruits a subset of Nedd4-like HECT E3 ubiquitin ligases (WWP1, WWP2, and Itch) (36) that in turn recruit ESCRT-III components (35). The YXXL L domain binds to the cellular protein ALIX (60). ALIX binds to Tsg101 (38) and also with ESCRT-III protein CHMP-4B (60), thus linking ESCRT-I and ESCRT-III. Green fluorescent protein (GFP)-, red fluorescent protein, or yellow fluorescent protein (YFP)-fused CHMPs are general DN inhibitors of all natural CHMP-associated activities and cause the formation of aberrant endosomal compartments that sequester ESCRT complexes (26, 31, 60). Through the use of these DN constructs, the recruitment and assembly of ESCRT components can be inhibited to specifically disrupt different steps of the ESCRT pathway.
The best evidence supporting involvement of ESCRT machinery in the life cycle of herpesviruses comes from the inhibition of HSV-1 envelopment by Vps4DN (10), as well as by CHMP3DN (5), together with the association of HSV-1 maturation with MVB. It was recently reported that HHV-6 also induces MVB formation that controls viral egress via an exosomal release pathway (45). After losing primary envelope acquired at the nuclear membrane, Human CMV (HCMV) undergoes a secondary, or final, envelopment step within a cytoplasmic assembly compartments (AC) (59). Secondary envelopment is thought to occur within early endosomal compartments based on diverse observations: (i) purified virions and dense bodies have a lipid composition that is similar to this compartment (64); (ii) the AC of HCMV-infected fibroblasts contain endosomal markers (11); and (iii) a number of HCMV envelope proteins, including US28 (14), UL33, US27 (15), and gB (9), colocalize with endosomal markers in infected cells. A model of HCMV egress via early endosomes has been proposed (11).
The approach that we have used here employed human foreskin fibroblasts (HFs) and restricted viral replication to cells that expressed the DN or wild-type (WT) component of the ESCRT pathway by including a requirement that transfected cells complement replication of virus. Confirming expression of both DN and complementing protein in transfected cells by epifluorescence microscopy ensured that an overwhelming majority of cells coexpressed these proteins. The results were scored as inhibition of viral spread to adjacent cells as well as demonstration of late gene expression in the transfected and/or infected cell. Viral progeny is released within 48 to 72 h from CMV-infected cells (44), reducing the likelihood that nonspecific or long-term toxicity of DN-ESCRT proteins would impact our analysis. This assay has been effectively used earlier for both immediate-early gene (54) and late gene (2, 62) mutants, and similar complementation assay results have been reported in diverse systems (8, 49, 73). This assay further provided an opportunity to determine when inhibition occurred relative to the viral replication cycle. Our data implicate ESCRT machinery late during HCMV maturation, which is consistent with a role in secondary envelopment and release.
MATERIALS AND METHODS
Cells.
Primary HFs were cultured in Dulbecco modified Eagle medium (DMEM; Invitrogen Corp., Carlsbad, CA) supplemented with 10% FetalClone III (HyClone, Logan, UT) at 37°C with 5% CO2. HFs between passages 5 and 15 were used for transfections and infections. Medium was changed every other day in cell culture. HFs that stably express IE1 protein (ihfie1.3) (21) were used for viral plaque assays of the IE1-deficient Towne virus (CR208) (54).
Viruses, DNA constructs, and complementation assays.
HCMV parental Towne strain (51) and IE1-deficient CR208 mutant (54) have been described previously. Plasmids expressing WT and DN forms of Vps4A (pBJ-Vps4A and pBJ-Vps4AE228Q, respectively) (60) were provided by Arianna Calistri, University of Padua, Padua, Italy. Plasmid expressing CHMP1AWT protein (pCDNA3-myc-CHMP1A) (55) was obtained from Sylvie Urbe, University of Liverpool, Liverpool, United Kingdom, and the plasmids expressing Tsg101DN (pCR3.1-GFP-Tsg1011-157) (37), ALIXDN (pCR3.1-YFP-ALIX176-869) (38), and WWP1DN (pCR3.1-YFP-WWP1341-547) (36) were obtained from Juan Martin-Serrano, King's College, London, England. Plasmid expressing CHMP1ADN protein (pEYFPN1-CHMP1A) was kindly provided by Colin Crump, Cambridge University, United Kingdom. Glycoprotein M (gM) and gN amplified from HCMV-TowneBAC were cloned into pDsRed-Monomer-N1 vector (Clontech Laboratories, Inc., Mountain View, CA) to generate DsRed-tagged gM (pON2995) and gN (pON2996).
CR208 complementation assays were performed as described earlier (54). Briefly, primary HFs in six-well tissue culture plates were cotransfected 24 h postseeding with pCMV-IE1491-GFP plasmid (pON2994) (54) and either vector control or one of the plasmids expressing WT or a DN ESCRT protein. Cells were analyzed by epifluorescence microscopy for expression and evaluation of IE1491-GFP (localized to the nucleus) and ESCRT proteins (expressed predominantly in cytoplasm) in all wells at 24 h posttransfection (hpt). Immunofluorescence was used to optimize plasmid concentrations providing equivalent number of cells coexpressing these proteins. This optimum concentration of plasmids also resulted in maximum inhibition. These cells were infected at 48 hpt with CR208 at a multiplicity of infection (MOI) of 0.002. At such low MOIs, CR208 replicates only in the cells that express IE1 (54). DMEM containing 0.16 mg of pooled human γ-immunoglobulin/ml (to restrict viral spread through medium) was added to the cells after adsorption, and medium was changed every 2 days until day 10 postinfection when the cells were fixed in methanol for 5 min and stained with Giemsa stain (standard Fluka) (Sigma-Aldrich Corp., St. Louis, MO) 1:10 in phosphate-buffered saline (PBS) for 1 h. After staining, plates were washed with distilled water and dried in air, and plaques were counted under optical microscope. Alternatively, antigen (ppUL44) spread was assessed in this assay after fixing cells in 3.7% formaldehyde for 10 min in 1× PBS at day 6 postinfection. Protocols for immunofluorescence microscopy (62) were followed to detect viral antigens. During WT or a complemented virus infection, ppUL44 was expressed in the nucleus with a delayed early kinetics; staining the nucleus diffusely during early infection and localized to nuclear replication compartments later in the infection (21). Noncomplemented CR208 either failed to express ppUL44 or expressed it very poorly, making it easier to identify cells that had expressed IE1 from complementing plasmid before infection. Spread of ppUL44 to cells around the primary infected cell depends on successful maturation and release of virus with spread to adjacent cells where replication ensued. Therefore, restriction of ppUL44 expression to nuclear replication compartments of a single cell was used to indicate the inhibition of viral spread from a primary infected cell in this experiment. A significant decrease in the number of ppUL44 foci (three or more adjacent ppUL44+ cells) or plaque formation due to coexpression of a particular DN protein compared to WT-ESCRT protein was interpreted as a viral replication defect. To examine whether infected cell lysates from CR208 complementation assays form plaques on IE1 expressing ihfie1.3 cells, medium was discarded after 3 or 6 days postinfection (dpi), and cells were harvested by brief trypsinization and pipetting in 750 μl of fresh DMEM. These harvested cells were mixed with equal amounts of autoclaved skim milk, sonicated for 10 s on ice, and diluted 10- to 100-fold in serum-free medium for plaque assay on ihfie1.3 cells (21). Medium (DMEM containing 10% serum and 0.16 mg of pooled human γ-immunoglobulin/ml) was changed after 5 days and, at 10 days postinoculation, the cells were washed twice with 1× PBS, fixed in methanol for 5 min, and stained with Giemsa stain as described above. After staining, plates were washed with distilled water and dried in air, and the plaques were counted under a dissecting microscope.
Mutagenesis and secondary spread assays.
HCMV UL32 open reading frame (encoding tegument phosphoprotein pp150) cloned in pLNCX vector (pON2780) (2) was used as a template for QuikChange mutagenesis (Qiagen, Inc., Valencia, CA). Proline and tyrosine in PTAP domain of pp150 were mutated simultaneously to glutamate and phenylalanine, respectively. Inserted mutations were confirmed by diagnosis of new silent restriction sites that were introduced and by sequencing in this region to confirm insertion of desired mutations without undesirable changes. Secondary spread assays were performed as described previously (2, 62). Briefly, primary HFs in six-well tissue culture dishes were cotransfected 24 h after seeding with either Towne-BAC-GFP (13) or ΔUL32-BAC-GFP (2, 62) with pp71 expression plasmid (pON2788) (41) and either WT or mutant UL32 LNCX constructs. Medium (DMEM containing 10% serum and 0.16 mg of pooled human γ-immunoglobulin/ml) was changed every other day until day 10 posttransfection when the cells were fixed in 3.7% formaldehyde-PBS for 10 min prior to evaluation by epifluorescence microscopy. Viral spread was defined as three or more adjacent eGFP-positive cells.
Antibodies.
A mouse monoclonal antibody to ppUL44 (ICP36, cell clone CH16) was purchased from Virusys Corporation, Sykesville, MD. Monoclonal anti-c-myc antibody (9E10; Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was used to detect myc-tagged CHMP1A and monoclonal anti-FLAG antibody (F3165; Sigma-Aldrich Corporation, St. Louis, MO) was used to detect FLAG-tagged VPS proteins. Hoechst 33258 (AnaSpec Corporation, San Jose, CA) staining identified the nuclei. Secondary antibodies Alexa-594 anti-mouse immunoglobulin G (IgG; H+L) or Alexa-488 anti-mouse IgG (H+L) were purchased from Molecular Probes/Invitrogen, Inc., (Carlsbad, CA).
Microscopy.
Samples were prepared by using established protocols for immunofluorescence assay and confocal fluorescence microscopy. Briefly, HFs were grown on coverslip inserts in 24-well tissue culture dishes. After transfections and infections, at the endpoint, the cells were fixed in 3.7% formaldehyde for 10 min and incubated in 50 mM NH4Cl in 1× PBS for 10 min to reduce the autofluorescence. This followed washing in 1× PBS, incubation in 0.5% Triton X-100 for 20 min to permeabilize the cells, and finally washing and incubation with primary and secondary antibodies at a 1:1,000 dilution in 0.1% bovine serum albumin in 1× PBS. Coverslips were retrieved from the wells and were mounted on glass slides with a drop of mounting medium (Gel/Mount; Biomeda, Foster City, CA) and dried overnight before imaging. Images were acquired on a Carl-Zeiss LSM 510 META confocal fluorescence microscope or Zeiss Axio Imager A1 epifluorescence microscope.
RESULTS
Employment of ESCRT machinery by HCMV in a Vps4- and CHMP1-dependent manner.
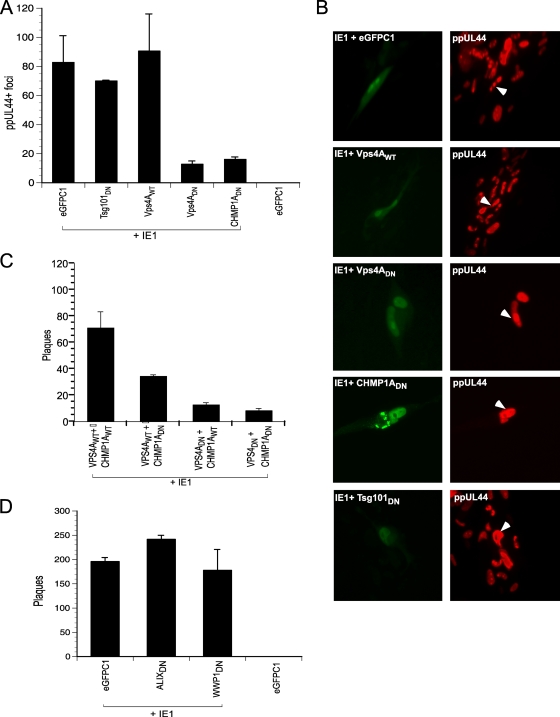
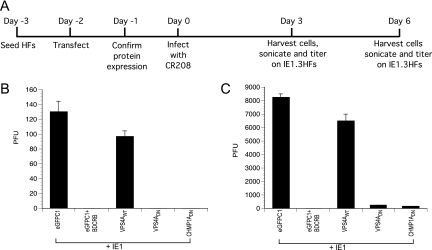
We inhibited the function of individual ESCRT proteins to evaluate impact on HCMV replication in an assay where an HCMV-Towne IE1 mutant virus (CR208) was complemented in trans (54). HFs were infected with CR208 under conditions where viral replication was restricted to cells that transiently expressed the IE1 protein, as well as a WT or DN ESCRT protein. This assay allowed disruption of ESCRT components specifically in virus-infected cells where replication proceeds. A vector (eGFPC1) lacking IE1 was used as a negative control in this assay. The transfection efficiency of HFs is low (<10%), such that only the infected subpopulation of cells were studied. Equivalent coexpression of IE1 and ESCRT proteins was confirmed by fluorescent protein detection (IE1, Tsg101, and CHMP1A) or immunofluorescence (Vps4) at 48 hpt (data not shown). IE1 was consistently nuclear, and ESCRT components were localized to the cytoplasm. At 48 hpt, monolayers were exposed to CR208 virus at an MOI of 0.002, conditions where this mutant fails to replicate unless complemented. At 72 h postinfection, HCMV-infected cells contain nuclear antigen ppUL44 in a replication compartment pattern. The number of ppUL44-positive cells was not influenced by expression of WT or DN ESCRT components (data not shown). Cells were fixed at endpoint (6 dpi) and stained for ppUL44 expression to evaluate virus production and spread (Fig. 1). Coexpression of IE1 with eGFPC1, Vps4AWT, or Tsg101DN yielded similar numbers of ppUL44-positive foci, whereas Vps4ADN or CHMP1ADN expression resulted in a seven- to eightfold reduction in numbers of infected foci (Fig. 1A). At this time, ppUL44 was localized to replication compartments in primary transfected cells independent of viral spread (Fig. 1B) confirming that DN ESCRT components inhibit viral replication at late times (after viral DNA replication and during maturation). A combination of CHMP1ADN and Vps4ADN resulted in a marginal further reduction in the levels of viral spread compared to Vps4ADN alone (at 10 dpi), suggesting these proteins function within the same pathway (Fig. 1C), a finding consistent with their roles in MVB formation (27). ALIXDN and WWP1DN did not inhibit CMV replication in a similar complementation assay (Fig. 1D). Thus, HCMV replication was inhibited by disruption of either Vps4A (ATPase) or CHMP1A (ESCRT-III component) function. These proteins typically influence the outcome of ESCRT recruitment via upstream parallel pathways (using Tsg101, ALIX, or HECT-Ub ligases) utilized by other viruses (35).
FIG. 1.
DN ESCRT proteins inhibit CMV replication. HCMV IE1 protein was coexpressed with WT or DN proteins of cellular ESCRT machinery in HFs, and cells were infected with IE1-deficient HCMV (CR208). (A) At the endpoint (6 dpi), ppUL44+ foci were quantified. (B) ESCRT inhibition allowed expression of delayed early viral antigen (ppUL44). The native cytoplasmic fluorescence of GFP-tagged Tsg101DN, eYFP tagged-CHMP1ADN, and nuclear fluorescence of eGFP-tagged IE1 is shown in the left panels. Vps4A was detected by anti-FLAG primary antibody and Alexa-488 secondary antibody. ppUL44 concentrated in the nuclear replication compartments in the primary transfected cell (arrowheads) in all transfections suggesting progression to a late stage of viral replication. (C and D) Giemsa-stained viral plaques were quantified in a similar CR208 complementation assay at 10 dpi.
Tsg101-independent nature of ESCRT engagement by HCMV.
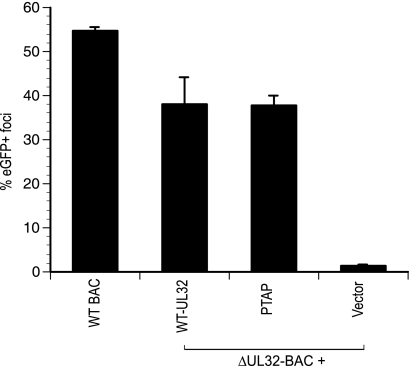
Given our observations that Vps4A and CHMP1A are important for HCMV replication, opposite to those previously reported (16), we also evaluated the role of Tsg101 and PTAP recognition in CMV replication. There are only two HCMV proteins (pUL56 and pp150) with putative PTAP motifs that characterize a subset of L domains. These motifs are conserved in all characterized strains of HCMV. pUL56 is a nonstructural protein involved in packaging of viral DNA in the nucleus (4), whereas pp150 (ppUL32) is a major tegument protein required in cytoplasmic maturation steps (2, 62), making this protein an attractive candidate for interaction with ESCRT components. To further examine any role of Tsg101DN in viral replication, we mutated the pp150 PTAP motif to EFAP. When this mutant was assessed for its ability to complement viral replication in a standard assay (2, 62), the mutant protein retained activity similar to WT-pp150 (Fig. 2). Thus, pp150 PTAP was completely dispensable for virus replication in HFs, a result that was consistent with the lack of any impact of Tsg101DN in the present study. HCMV thus appears to exploit ESCRT machinery in a Tsg101-independent manner.
FIG. 2.
UL32-PTAP mutant complements UL32 defect similar to WT protein in a secondary spread assay. WT (Towne) or ΔUL32-BAC strains were transfected in HFs, along with indicated expression constructs, and the eGFP+ foci were counted at 10 days posttransfection.
Localization of ESCRT proteins in the vicinity of cytoplasmic viral AC.
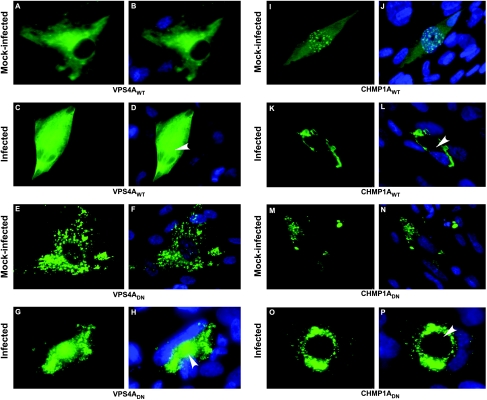
It has been reported that HCMV derives its final envelope from endocytic membranes (64) and that viral envelope carries the MVB marker CD63 (16) in a pattern that is reminiscent of the role ESCRT machinery plays in the generation of MVBs in uninfected cells (50). To gain insights into the recruitment of ESCRT machinery by HCMV during infection, we studied the localization of ESCRT proteins in HCMV-infected and mock-infected HFs. HCMV-infected cells develop morphologically distinct nuclear (replication compartment) and cytoplasmic (AC) inclusions, where virus replication and maturation, respectively, take place (43). Here, AC was recognized based on the hallmark morphology of CMV-infected cells with nucleus transforming into a kidney bean shape and the AC pressing against this newly formed depression in the nucleus (11, 62). We determined the localization pattern of ESCRT proteins relative to the AC. Both Vps4AWT (Fig. 3A and B) and Vps4ADN (Fig. 3E and F) proteins were distributed in the cytoplasm of mock-infected HFs, with Vps4ADN also accumulating in aggregates at the periphery of the nucleus. During the late phase (5 dpi) of WT-HCMV infection, both of these proteins were found in cytoplasm, as well as in the AC region (Fig. 3C, D, G, and H). Cells containing either Vps4AWT or Vps4ADN expressed both early (ppUL44) (Fig. 1B) and late (MCP, gB, gM-gN, and pp150) viral proteins (data not shown), confirming a late replication defect. This was further supported by localization of ppUL44 in nuclear replication compartments in these cells, an expression pattern associated with late infection. CHMP1AWT protein (Fig. 3I and J) was expressed diffusely in the cytoplasm, as well as in punctate form in the nucleus of mock-infected HFs, but the CHMP1ADN (Fig. 3M and N) expressed only in cytoplasm of mock-infected HFs in punctate form. In infected cells, CHMP1AWT showed a predominantly cytoplasmic expression pattern, adjacent to the AC region in infected HFs (Fig. 3K and L), and a similar expression pattern was observed for CHMP1ADN (Fig. 3O and P) during late stages of infection. Cells expressing either CHMP1AWT or CHMP1ADN proteins expressed early (ppUL44) and late (MCP, gB, gM-gN, and pp150) (Fig. 4F and I and data not shown) viral proteins. Appearance of distinct aberrant endosomal vesicles (class E compartments) has been used as a criterion for impairment of ESCRT function in yeast (3, 53) and mammalian cells (12, 17, 26). Aberrant vesicles in the present study were observed only on expression of DN-ESCRT proteins (Fig. 3E to H and M to P) and not on WT-ESCRT proteins expression (Fig. 3A to D and I to L), confirming inhibition of ESCRT pathway by DN-ESCRT proteins.
FIG. 3.
Cellular rearrangement of ESCRT proteins during HCMV infection. HFs transfected with plasmids expressing Vps4AWT, Vps4ADN, CHMP1A WT, or CHMP1A DN proteins were infected with HCMV Towne virus at 24 hpt. Cells were fixed and photographed for immunofluorescence microscopy at day 5 postinfection. Arrowheads point to the AC in infected cells. The native fluorescence of eYFP-tagged CHMP1ADN protein (M, N, O, and P) and immunofluorescence localization of CHMP1AWT protein using anti-myc primary antibody and Alexa-488 secondary antibody (I, J, K, and L) is shown. Vps4A was detected by anti-FLAG primary antibody and Alexa-488 secondary antibody (A, B, C, D, E, F, G, and H). Hoechst 33342 staining (blue) marks the nucleus in merged images (B, D, F, H, J, L, N, and P).
FIG. 4.
HCMV-infected HFs express gM-gN glycoproteins in the presence of DN or WT proteins of the ESCRT machinery. Epifluorescence micrograph of cells coexpressing control eGFPC1 protein (A to C), myc-tagged CHMP1AWT protein (D to F), or eYFP-tagged CHMP1ADN protein (G to I) and DsRed-tagged viral proteins. CHMP1AWT protein was detected using anti-myc primary antibody and Alexa 488-conjugated secondary antibody (E and F). Transfected cells were infected at 24 hpt and were fixed and photographed at day 5 postinfection. Hoechst 33342 (blue) stains for the nucleus (C, F, and I). Arrowheads point to the AC in infected cells (C, F, and I).
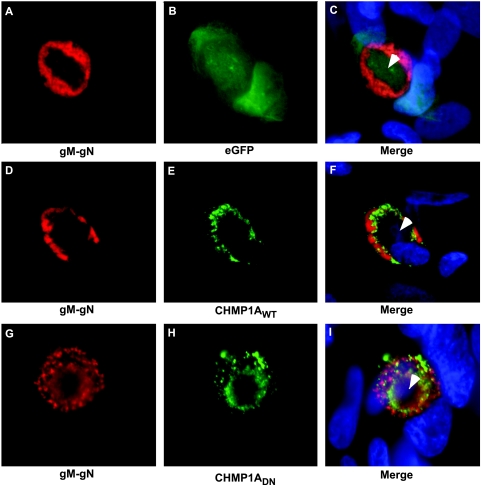
In order to determine the localization of CHMP1AWT and CHMP1ADN relative to viral markers of maturation, both were evaluated relative to viral glycoproteins (gM-gN). Localization of gM-gN proteins has been correlated with the site of viral envelopment in CMV-infected cells (30). Here, gM-gN proteins were expressed in a ring pattern in the vicinity of AC (Fig. 4A and C) and eGFP control protein expressed in diffused form in the whole cell (Fig. 4B and C). WT CHMP1A localized to the periphery of AC during late infection (5 dpi) and appeared to form a concentric ring inside of the gM-gN region (Fig. 4D to F). Similar expression pattern was observed for DN CHMP1A (Fig. 4G to I). Thus, both WT and DN CHMP1A proteins showed similar expression patterns, although DN CHMP1A blocked CMV replication and WT CHMP1A did not (Fig. 1A). The presence of late viral proteins in cells that express DN ESCRT proteins suggests a block in viral life cycle at late stages of infection. Localization of CHMP1A and Vps4A proteins in the vicinity of AC, where the final steps in virus maturation and envelopment occur, indicates an important role of ESCRT pathway during these processes in HCMV life cycle. AC is formed during late stages of HCMV infection (11) and has a concentration of several viral (pp28, pp150, gM-gN, and gB) and cellular (Golgin-97, EEA-1, mannosidase-II, GM130, p230, and Bip/GRP78) markers (11, 30, 62). The AC is a highly organized structure (11), and a number of HCMV mutants with defects in maturation accumulate in AC (57, 58, 62). It may be hypothesized that viral proteins recruit ESCRT components to these sites and ESCRT machinery plays an important role in CMV maturation steps that are associated with this distinct cellular compartment.
Inhibition of ESCRT components and CMV maturation.
Because maturation and envelopment occurs in AC (43) and these compartments incorporate endosomal markers (11) corresponding to MVBs (16), we examined cells that expressed DN-ESCRT components VPS4DN or CHMP1ADN to determine whether virus particles that were detected in these cells were replication competent (mature). For this, we evaluated the ability of infected cell lysates from CR208 complementation experiments to form plaques on complementing ihfie1.3 cells. At day 3 postinfection, more than 100 viral plaques were produced from lysates of cells expressing WT-ESCRT or eGFP control proteins, whereas viral plaques were not detected when the lysates of ESCRT inhibited cells were tested (Fig. 5B). BDCRB [2-bromo-5,6-dichloro-1-(-d-ribofuranosyl) benzimidizole] blocks encapsidation of CMV DNA in the nucleus (65); therefore, BDCRB treated cells served as a negative control in this assay. At day 6 postinfection, lysates from ESCRT inhibited cells produced ∼100-fold fewer viral plaques compared to lysates from cells with WT-ESCRT or control plasmids (Fig. 5C). These experiments demonstrated that inhibition of ESCRT machinery blocked HCMV replication prior to virus release. We believe that these data, together with the localization patterns, suggest a block to secondary envelopment within the AC, a step analogous to HSV maturation, as well as to RNA virus envelopment.
FIG. 5.
Viral plaque assays to analyze ability of cytoplasmic viral particles in lysates from ESCRT inhibited or uninhibited cells to form plaques on IE1-complementing cells. CR208 complementation assays were set up as described previously (A) and at day 3 (B) or day 6 (C) postinfection, and the cells were harvested, sonicated, and titered on ihfie1.3 cells. Plaques were counted at day 10 postinoculation.
DISCUSSION
ESCRT machinery is necessary for envelopment of RNA viruses and controls virus release from cells. This is a burgeoning area of interest in DNA viruses. Although HCMV particles were detected in MVBs, prior investigation using siRNA- mediated inhibition of VPS4, Tsg101, and ALIX expression did not reduce HCMV replication levels (16). These results were surprising considering the evidence supporting a role for the ESCRT pathway in HSV-1 (10) and the close association of both HCMV (16) and HHV-6 (45) envelopment with MVB. Despite controls showing that siRNAs to VPS4A and VPS4B inhibited maturation of HIV, these approaches may not be as complete as those that use DN ESCRT components. DN proteins work in a competitive manner and would inhibit all functional members of a family, whereas siRNA would knock down only those specific transcripts with significant nucleotide sequence homology. In fact, depletion of CHMP5 and CHMP6 using siRNA has a negligible effect on HIV release (67), whereas DN-CHMP5 or DN-CHMP6 significantly inhibit HIV release (31, 38, 60, 61). Assembly of ESCRT machinery is highly complex, especially in metazoan cells, where more than 25 individual proteins play more or less critical roles in ESCRT function (69). There is evidence of redundancy in functional ESCRT components in human cells (32), where different isoforms of the same protein have been identified (e.g., CHMP4-A, -B, and -C) (63). Therefore, it is possible that depletion of some components of ESCRT by siRNA would have slight effect on ESCRT function, whereas DN knockdown of the same protein would lead to significant inhibition of ESCRT function because of the sequestration of other ESCRT components in the aberrant endosomal compartments formed when DN proteins are expressed (31). Also, there can be pathway or pathogen specific requirements for particular ESCRT proteins or their levels that would explain the results obtained in Fraile-Ramos et al. (16) study where HIV release was impaired but HCMV maturation was unaffected upon siRNA depletion of several ESCRT proteins. The studies by Fraile-Ramos et al. were further confounded by the choice to evaluate the replication of a laboratory-propagated strain of HCMV (AD169 strain derivative RCMV288) in a cell type that is not considered to be susceptible to this strain (42). The AD169 strain is defective in the expression of UL131, an envelope protein that contributes to efficient entry and replication in RPE-1 (epithelial) cells (22, 48, 56, 66) used in that study. Thus, the redundancy in functional ESCRT components, the complexity of the ESCRT system, the differences in the way DN proteins work compared to siRNA, and the cell system used for propagation of AD169 virus by Fraile-Ramos et al. would explain the strikingly different results reported earlier (16). In the present study, we evaluated the importance of ESCRT pathway in HCMV replication using more conventional DN inhibitors that have been effective where siRNA has failed (31). In the complementation assay used here, a very small number of transfected cells lead to viral spread and plaque formation (54). This, however, does not compromise the quality of the results or the interpretation of the data because no spreads are seen in the absence of complementing protein (Fig. 1A) and, once complemented, CR208 virus replicates as a WT virus, leading to very distinct viral spreads and plaques (Fig. 1) and significant virus titers (Fig. 5).
Vps4A contributes to all three known primary mechanisms of ESCRT recruitment by viruses (47); therefore, we first examined the effects of inhibiting Vps4A on HCMV replication and observed a significant reduction in viral plaque formation. We evaluated the impact of inhibition of ESCRT-III component CHMP1A using CHMP1ADN, which also significantly reduced HCMV plaque formation. Moreover, Vps4A and CHMP1A localized in the vicinity of the AC, where final envelopment occurs in infected fibroblasts, suggesting recruitment of ESCRT machinery to the site of cytoplasmic virus assembly and maturation. Localization of ppUL44 to nuclear replication compartments in cells where ESCRT components were inhibited is consistent with a role of ESCRT machinery in maturation rather than at early times postinfection. Thus, viral infection proceeds through viral DNA replication and formation of the AC appears to be independent of the ESCRT machinery. The results from Vps4ADN and CHMP1ADN experiments suggested that ESCRT machinery is directly involved in CMV maturation, assembly, or egress. To investigate whether ESCRT inhibition in CMV is specific to virus release as shown for RNA viruses, or other parts of the virus life cycle prior to egress are affected, we tested the ability of lysates of HCMV-infected cells to form plaques. The lysates from WT or control plasmid transfected cells formed quantifiable plaques, whereas lysates from DN plasmid transfected cells failed to do so or formed very few plaques, suggesting that the defects are not specific to virus release but most probably impact secondary envelopment in AC, a process analogous to RNA virus budding.
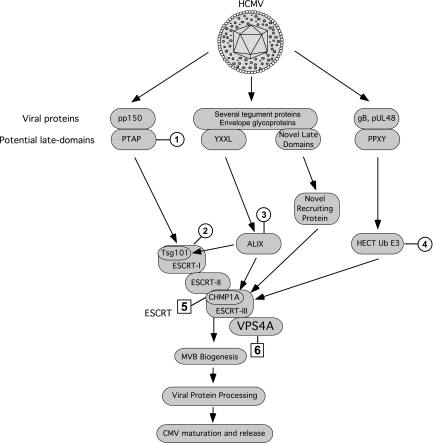
In many RNA viruses, late domains have been identified within viral proteins that interact with ESCRT machinery, with different pathways important in different viruses. Searching the HCMV tegument and envelope protein sequences for homology with L domains revealed one PTAP motif (in pp150), two PPXY motifs (gB and UL48), and more than 30 YXXL motifs, as reported earlier (16). Neither Tsg101DN nor PTAP mutants in pp150 influenced viral maturation, suggesting that HCMV uses PTAP-Tsg101 independent pathways. Comprehensive functional screening of mutations in PPXY and YXXL domains in CMV structural proteins has not been completed, and it may or may not reveal L domains responsible for HCMV-ESCRT interactions. A model of exploitation of ESCRT machinery by HCMV is proposed (Fig. 6) wherein Tsg101-, ALIX-, and WWP1-independent novel L domains in HCMV structural proteins may play a role in ESCRT recruitment. Upstream ESCRT complexes (ESCRT-I and ESCRT-II) are believed to work in parallel to recruit the cargo, whereas Vps4 and ESCRT-III work in series (27). This redundancy in cargo recruitment may explain why CHMP1A (ESCRT-III component) and Vps4A disruption leads to significant inhibition of CMV maturation, whereas Tsg101 (ESCRT-I component) disruption has no effect. It is also possible that HCMV does not engage ESCRT directly, but disruption of ESCRT leads to maturation defects in HCMV due to disruption of the biogenesis of MVBs that are known to be important for herpesvirus maturation (5, 10, 16). This can also explain why disruption of ESCRT cargo recruitment (by Tsg101DN, ALIXDN, and WWP1DN) did not have an effect, whereas disruption of ESCRT assembly (by CHMP1DN) or recycling (Vps4DN) had pronounced effects on HCMV maturation.
FIG. 6.
Proposed model for HCMV exploitation of host-cell ESCRT machinery incorporating the results obtained in the present study. Mutation of PTAP domain in pUL32 (pp150) (circled 1) and DN inhibition of Tsg101 (circled 2), ALIX (circled 3), or WWP1 (HECT Ub E3) (circled 4) did not affect HCMV replication but inhibition of CHMP1A (ESCRT-III component) (boxed 5) or Vps4A (ATPase) (boxed 6) impaired HCMV replication. HCMV is proposed to either utilize novel late domains and novel ESCRT recruiting proteins for engaging ESCRT, or it depends on ESCRT machinery for the biogenesis of MVBs for viral protein processing.
HCMV particles are believed to mature as they proceed into the core of AC and pass through concentric layers of cellular organelles. Virus envelopment occurs by budding into endosomal vesicles, which fuse with the plasma membrane to release virions to the extracellular space (11). Accumulation of viral glycoproteins (gM-gN) and ESCRT proteins at the periphery of AC in our studies support this model of viral egress, where these proteins would provide essential functions (envelopment and endosomal budding) before viral exit. Formation of intact AC and defects in viral maturation rather than an early or very late replication defect in ESCRT-inhibited cells further support viral maturation and exit through this model. Thus, our studies support an existing model of CMV maturation and implicate the ESCRT system as an important component of this process. Targeted disruption of all motifs in HCMV structural proteins with similarity to L domains and elaborate electron microscopic studies of defects in CMV maturation on ESCRT inhibition will help finely define the mechanism of ESCRT engagement by HCMV.
Acknowledgments
We appreciate the help of William J. Kaiser in cell culture. We thank A. Louise McCormick and Colin M. Crump for critical reading of the manuscript; Colin M. Crump, Arianna Calistri, Sylvie Urbe, and Juan Martin-Serrano for DNA expression constructs; and Daniel Kalman and Patrick Reeves for assistance with the confocal microscopy. We acknowledge Davide A. Abate for early investigations on the PTAP domain of CMV that were pursued with the guidance of Stanley Cohen at Stanford University School of Medicine.
PHS grant RO1 AI20211 funded this research.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., G. B. Smith, C. D. Meiering, and E. S. Mocarski. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 80:8199-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilodeau, P. S., S. C. Winistorfer, W. R. Kearney, A. D. Robertson, and R. C. Piper. 2003. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 163:237-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calistri, A., P. Sette, C. Salata, E. Cancellotti, C. Forghieri, A. Comin, H. Gottlinger, G. Campadelli-Fiume, G. Palu, and C. Parolin. 2007. Intracellular trafficking and maturation of herpes simplex virus type 1 gB and virus egress require functional biogenesis of multivesicular bodies. J. Virol. 81:11468-11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316:1908-1912. [DOI] [PubMed] [Google Scholar]
- 7.Chen, B. J., and R. A. Lamb. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Y. L., P. W. Ts'ai, C. C. Yang, and C. T. Wang. 1997. Generation of infectious virus particles by transient coexpression of human immunodeficiency virus type 1 gag mutants. J. Gen. Virol. 78(Pt. 10):2497-2501. [DOI] [PubMed] [Google Scholar]
- 9.Crump, C. M., C. H. Hung, L. Thomas, L. Wan, and G. Thomas. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105-11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crump, C. M., C. Yates, and T. Minson. 2007. Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81:7380-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das, S., A. Vasanji, and P. E. Pellett. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81:11861-11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doyotte, A., M. R. Russell, C. R. Hopkins, and P. G. Woodman. 2005. Depletion of TSG101 forms a mammalian “class E” compartment: a multicisternal early endosome with multiple sorting defects. J. Cell Sci. 118:3003-3017. [DOI] [PubMed] [Google Scholar]
- 13.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraile-Ramos, A., T. N. Kledal, A. Pelchen-Matthews, K. Bowers, T. W. Schwartz, and M. Marsh. 2001. The human cytomegalovirus US28 protein is located in endocytic vesicles and undergoes constitutive endocytosis and recycling. Mol. Biol. Cell 12:1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraile-Ramos, A., A. Pelchen-Matthews, T. N. Kledal, H. Browne, T. W. Schwartz, and M. Marsh. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218-232. [DOI] [PubMed] [Google Scholar]
- 16.Fraile-Ramos, A., A. Pelchen-Matthews, C. Risco, M. T. Rejas, V. C. Emery, A. F. Hassan-Walker, M. Esteban, and M. Marsh. 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol. 9:2955-2967. [DOI] [PubMed] [Google Scholar]
- 17.Fujita, H., M. Yamanaka, K. Imamura, Y. Tanaka, A. Nara, T. Yoshimori, S. Yokota, and M. Himeno. 2003. A dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane trafficking out of endosomal/lysosomal compartments: class E Vps phenotype in mammalian cells. J. Cell Sci. 116:401-414. [DOI] [PubMed] [Google Scholar]
- 18.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 19.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Honeychurch, K. M., G. Yang, R. Jordan, and D. E. Hruby. 2007. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J. Virol. 81:7310-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard, T. L., D. R. Stauffer, C. R. Degnin, and S. M. Hollenberg. 2001. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 114:2395-2404. [DOI] [PubMed] [Google Scholar]
- 27.Hurley, J. H., and X. Ren. 2009. The circuitry of cargo flux in the ESCRT pathway. J. Cell Biol. 185:185-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 29.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krzyzaniak, M., M. Mach, and W. J. Britt. 2007. The cytoplasmic tail of glycoprotein M (gpUL100) expresses trafficking signals required for human cytomegalovirus assembly and replication. J. Virol. 81:10316-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langelier, C., U. K. von Schwedler, R. D. Fisher, I. De Domenico, P. L. White, C. P. Hill, J. Kaplan, D. Ward, and W. I. Sundquist. 2006. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol. 80:9465-9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lata, S., G. Schoehn, J. Solomons, R. Pires, H. G. Gottlinger, and W. Weissenhorn. 2009. Structure and function of ESCRT-III. Biochem. Soc. Trans. 37:156-160. [DOI] [PubMed] [Google Scholar]
- 33.Leung, K. F., J. B. Dacks, and M. C. Field. 2008. Evolution of the multivesicular body ESCRT machinery; retention across the eukaryotic lineage. Traffic 9:1698-1716. [DOI] [PubMed] [Google Scholar]
- 34.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Serrano, J. 2007. The role of ubiquitin in retroviral egress. Traffic 8:1297-1303. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Serrano, J., S. W. Eastman, W. Chung, and P. D. Bieniasz. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 40.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCormick, A. L., C. D. Meiering, G. B. Smith, and E. S. Mocarski. 2005. Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J. Virol. 79:12205-12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McSharry, B. P., C. J. Jones, J. W. Skinner, D. Kipling, and G. W. Wilkinson. 2001. Human telomerase reverse transcriptase-immortalized MRC-5 and HCA2 human fibroblasts are fully permissive for human cytomegalovirus. J. Gen. Virol. 82:855-863. [DOI] [PubMed] [Google Scholar]
- 43.Mocarski, E. S. 2007. Betaherpesvirus genes and their functions, p. 202-228. In A. M. Arvin, E. S. Mocarski, P. Moore, R. Whitley, K. Yamanishi, G. Campadelli-Fiume, and B. Roizman (ed.), Human herpesviruses: biology, therapy and immunoprophylaxis. Cambridge Press, Cambridge, England. [PubMed]
- 44.Mocarski, E. S., Jr., T. Shenk, and R. F. Pass. 2006. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields Virology, 5th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 45.Mori, Y., M. Koike, E. Moriishi, A. Kawabata, H. Tang, H. Oyaizu, Y. Uchiyama, and K. Yamanishi. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morita, E., V. Sandrin, H. Y. Chung, S. G. Morham, S. P. Gygi, C. K. Rodesch, and W. I. Sundquist. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morita, E., and W. I. Sundquist. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395-425. [DOI] [PubMed] [Google Scholar]
- 48.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nichols, R. J., E. Stanitsa, B. Unger, and P. Traktman. 2008. The vaccinia virus gene I2L encodes a membrane protein with an essential role in virion entry. J. Virol. 82:10247-10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nickerson, D. P., M. R. Russell, and G. Odorizzi. 2007. A concentric circle model of multivesicular body cargo sorting. EMBO Rep. 8:644-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plotkin, S. A., S. E. Starr, H. M. Friedman, E. Gonczol, and R. E. Weibel. 1989. Protective effects of Towne cytomegalovirus vaccine against low-passage cytomegalovirus administered as a challenge. J. Infect. Dis. 159:860-865. [DOI] [PubMed] [Google Scholar]
- 52.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reinhardt, J., G. B. Smith, C. T. Himmelheber, J. Azizkhan-Clifford, and E. S. Mocarski. 2005. The carboxyl-terminal region of human cytomegalovirus IE1491aa contains an acidic domain that plays a regulatory role and a chromatin-tethering domain that is dispensable during viral replication. J. Virol. 79:225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Row, P. E., H. Liu, S. Hayes, R. Welchman, P. Charalabous, K. Hofmann, M. J. Clague, C. M. Sanderson, and S. Urbe. 2007. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 282:30929-30937. [DOI] [PubMed] [Google Scholar]
- 56.Ryckman, B. J., B. L. Rainish, M. C. Chase, J. A. Borton, J. A. Nelson, M. A. Jarvis, and D. C. Johnson. 2008. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 82:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Severi, B., M. P. Landini, and E. Govoni. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51-64. [DOI] [PubMed] [Google Scholar]
- 60.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 61.Stuchell, M. D., J. E. Garrus, B. Muller, K. M. Stray, S. Ghaffarian, R. McKinnon, H. G. Krausslich, S. G. Morham, and W. I. Sundquist. 2004. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 279:36059-36071. [DOI] [PubMed] [Google Scholar]
- 62.Tandon, R., and E. S. Mocarski. 2008. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J. Virol. 82:9433-9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teis, D., S. Saksena, and S. D. Emr. 2008. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev. Cell 15:578-589. [DOI] [PubMed] [Google Scholar]
- 64.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 65.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, D., and T. Shenk. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 79:10330-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ward, D. M., M. B. Vaughn, S. L. Shiflett, P. L. White, A. L. Pollock, J. Hill, R. Schnegelberger, W. I. Sundquist, and J. Kaplan. 2005. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 280:10548-10555. [DOI] [PubMed] [Google Scholar]
- 68.Watanabe, T., E. M. Sorensen, A. Naito, M. Schott, S. Kim, and P. Ahlquist. 2007. Involvement of host cellular multivesicular body functions in hepatitis B virus budding. Proc. Natl. Acad. Sci. USA 104:10205-10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams, R. L., and S. Urbe. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell. Biol. 8:355-368. [DOI] [PubMed] [Google Scholar]
- 70.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu, L. A., and S. K. Weller. 1992. The six conserved helicase motifs of the UL5 gene product, a component of the herpes simplex virus type 1 helicase-primase, are essential for its function. J. Virol. 66:469-479. [DOI] [PMC free article] [PubMed] [Google Scholar]