Summary
The synthesis of selenoproteins requires the translational recoding of the UGA stop codon as selenocysteine. During selenium deficiency, there is a hierarchy of selenoprotein expression, with certain selenoproteins synthesized at the expense of others. The mechanism by which the limiting selenocysteine incorporation machinery is preferentially utilized to maintain the expression of essential selenoproteins has not been elucidated. Here, we demonstrate that eukaryotic initiation factor 4a3 (eIF4a3) is involved in the translational control of a subset of selenoproteins. The interaction of eIF4a3 with the selenoprotein mRNA prevents the binding of SECIS binding Protein 2, which is required for selenocysteine insertion, thereby inhibiting the synthesis of the selenoprotein. Furthermore, the expression of eIF4a3 is regulated in response to selenium. Based on knockdown and overexpression studies, eIF4a3 is necessary and sufficient to mediate selective translational repression in cells. Our results support a model in which eIF4a3 links selenium status with differential selenoprotein expression.
INTRODUCTION
Selenium is an essential micronutrient in many organisms including humans. This element is incorporated into selenoproteins as selenocysteine (Sec) through a unique mechanism whereby the UGA stop codon is recoded as Sec. In eukaryotes, this process is a cotranslational event that requires a highly structured stem loop, termed the SECIS (Sec insertion sequence) element, in the 3′-untranslated region (3′-UTR) of the selenoprotein mRNA (Berry et al., 1991). Sec incorporation also depends on a number of trans-acting proteins, including SECIS Binding Protein 2 (SBP2) (Copeland et al., 2000), a specialized elongation factor (EFsec) (Fagegaltier et al., 2000; Tujebajeva et al., 2000), and ribosomal protein L30 (Chavatte et al., 2005).
Although much is known regarding the mechanism of selenoprotein biosynthesis, the regulation of selenoprotein expression remains an elusive aspect of selenoprotein biology. The prioritization of selenium utilization for the synthesis of a subset of selenoproteins when dietary selenium is reduced has been well documented (Hill et al., 1992). The classic example for this hierarchy is the preservation of phospholipid hydroperoxide glutathione peroxidase (PHGPx) expression while the synthesis of glutathione peroxidase 1 (GPx1) is greatly reduced when selenium becomes limiting (Bermano et al., 1996; Lei et al., 1995). The reduction of GPx1 expression in selenium deficiency has been shown to take place not at the level of GPx1 mRNA transcription, but instead at the post-transcriptional level (reviewed in Sunde, 2006). When selenium is not available, there is a dramatic decrease in GPx1 mRNA levels, caused by an alteration in GPx1 mRNA stability (Bermano et al., 1996; Muller et al., 2003; Wingler et al., 1999). The decreased stability is proposed to occur via nonsense-mediated decay (NMD), a pathway that targets mRNAs containing a premature termination codon for degradation (Moriarty et al., 1998; Weiss and Sunde, 1998). Studies in selenium deficient rats suggested that GPx1 expression is also regulated at the translational level. When dietary selenium was low, GPx1 mRNA was barely detected in the liver whereas at high selenium status, both GPx1 mRNA and protein were expressed at the maximum level (Lei et al., 1995). However, when dietary selenium was half of the concentration required for maximal GPx1 expression, there was an 80% decrease in GPx1 protein but only a 20% decrease in the mRNA (Weiss-Sachdev and Sunde, 2001). Additionally, GPx1 belongs to a subset of stress-related selenoproteins (Carlson et al., 2005) whose synthesis depends on the Um34 isoform of the Sec-tRNASec, which contains a 2′-O-hydroxymethyl group on the ribosyl moiety at position 34 (Diamond et al., 1993). GPx1 protein is not detected in the livers of mice lacking the Um34 isoform despite having normal GPx1 mRNA levels (Carlson et al., 2005). Taken together, these observations suggest that the biosynthesis of GPx1 is a complex process involving multiple levels of regulation.
One possible control point for the translational regulation of GPx1 expression is the SBP2-SECIS interaction. This interaction, which is essential for the recoding of the UGA stop codon as Sec, has been shown to be the rate determining step in selenoprotein biosynthesis (Low et al., 2000). A naturally occurring point mutation within the RNA binding domain of SBP2 resulted in differential expression of a subset of selenoproteins in humans (Bubenik and Driscoll, 2007; Dumitrescu et al., 2005; Squires et al., 2007). In addition, mutations within the SECIS element, which alter SBP2 binding, also result in dramatic decreases or loss of selenoprotein expression (Allamand et al., 2006). Thus, SBP2 is a potential candidate for establishing the regulation of selenoprotein synthesis upon selenium deprivation. However, neither the activity nor expression level of SBP2 is modulated by selenium (Copeland et al., 2000; Driscoll and Copeland, 2003), suggesting that other factors might be involved in dictating the hierarchy of selenoprotein expression.
In this study, we show that eukaryotic initiation factor 4a isoform 3 (eIF4a3), a DEAD-box family helicase with no defined function in translation, plays a key role in regulating the hierarchy of selenoprotein expression at the translational level.
RESULTS
Identification of a 48 kDa SECIS Binding Protein
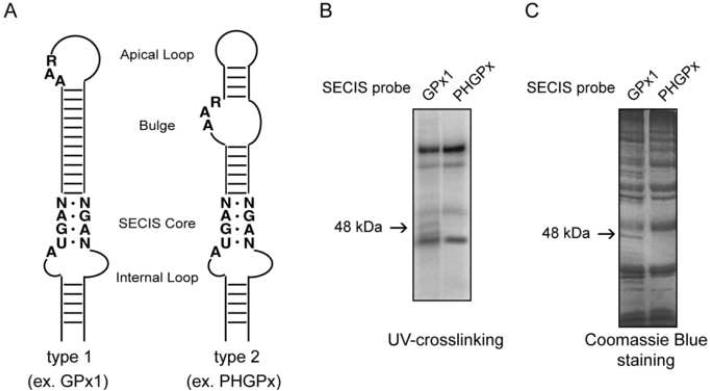
In eukaryotes, gene expression relies on many aspects of cytoplasmic mRNA metabolism such as its export, stability and translation efficiency. These processes are highly regulated and commonly mediated by various ribonucleoproteins (RNP), which are mainly assembled and formed in the nucleus. Therefore, the fate of an mRNA in the cytoplasm is strongly dependent on the nuclear history of its regulatory events (Giorgi and Moore, 2007). Thus, we hypothesized that selenoprotein mRNAs may acquire SECIS binding proteins, which subsequently regulate their metabolism, before their export to the cytoplasm. To test this hypothesis, we performed UV-crosslinking experiments with nuclear extracts from McArdle 7777 cells, a rat hepatoma cell line that expresses multiple selenoproteins including PHGPx and GPx1. The SECIS elements from these two selenoproteins were used as probes to identify protein factors that differentially interact with the PHGPx and GPx1 mRNAs. Within a SECIS element, there are two highly conserved regions, the SECIS core and the AAR motif (Fig. 1A). The SECIS core contains two sheared tandem G•A pairs (G•A/A•G) that are required for SBP2 binding. The structural context of the AAR motif is used to classify the SECIS element into two subgroups, type 1 and type 2. In the GPx1 SECIS (type 1), the AAR motif is located in the apical loop, whereas the AAR motif in the PHGPx SECIS (type 2) is found in the apical bulge (Fig. 1A). 32P-labeled SECIS elements were incubated with nuclear extract. After incubation, the samples were crosslinked with UV light, treated with RNase A to digest unbound RNA, resolved by SDS-PAGE and analyzed by autoradiography. Nuclear extracts contained several proteins that crosslinked to both probes as well as a prominent 48 kDa protein, which crosslinked to the GPx1 SECIS, but not to the PHGPx SECIS (Fig. 1B).
Figure 1. Identification of a 48 kDa SECIS binding protein.
(A) Schematic representation of the GPx1 and PHGPx SECIS elements. The SECIS Core, AAR motif and Internal Loop region are indicated. The location of the AAR motif determines whether the element is a type 1 SECIS (Apical Loop) or type 2 SECIS (Bulge). (B) Radiolabeled GPx1 or PHGPx SECIS probes (1 nM) were used with McArdle 7777 nuclear extract (50 μg) in UV-crosslinking assays. The 48 kDa protein that binds only to the GPx1 probe is indicated by the arrow. (C) Biotinylated GPx1 or PHGPx SECIS RNAs were incubated with McArdle 7777 nuclear extract. The bound proteins were eluted and analyzed by SDS-PAGE and Coomassie Blue-staining as described in Experimental Procedures. The arrow indicates the 48 kDa protein identified by LCMS.
To isolate and identify the 48 kDa protein, we carried out a single-step biotin/streptavidin RNA affinity purification scheme followed by mass spectrometry analysis. Biotinylated GPx1 and PHGPx SECIS RNAs were generated by in vitro transcription and incubated with McArdle 7777 nuclear extract. The SECIS-protein complexes were captured using streptavidin-coated magnetic beads. The bound proteins were eluted from the beads, analyzed by SDS-PAGE, and stained with Coomassie Blue. Multiple proteins eluted from both the GPx1 and PHGPx SECIS RNA beads (Fig. 1C). However, several proteins were specific for the GPx1 SECIS, including a 48 kDa protein similar to the band observed by UV-crosslinking. The 48 kDa band was excised from the gel and analyzed by LCMS. Mass spectrometry analysis and peptide fingerprinting revealed that the eight most abundant peptides resulting from the tryptic digest covered 42% of the rat eIF4a3 sequence.
eIF4a3 Selectively Interacts with the GPx1 SECIS Element
eIF4a3 is predominantly a nuclear protein that belongs to the RNA helicase DEAD-box protein family (Li et al., 1999). Although the amino acid sequence of eIF4a3 is highly similar to the translation initiation factors eIF4a1 and eIF4a2, it fails to substitute for their function in ribosome binding, suggesting it is not part of the translation initiation complex (Li et al., 1999). Recent studies revealed that eIF4a3 is a component of the exon junction complex (EJC) (Chan et al., 2004; Shibuya et al., 2004), a marker for spliced mRNA that is deposited 20 – 24 nt upstream the exon-exon junction.
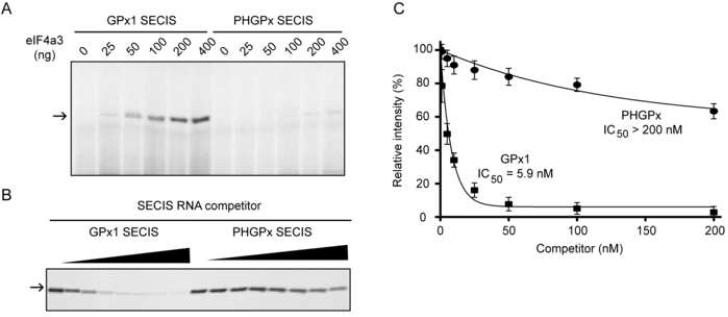
To assess whether eIF4a3 is the 48 kDa protein observed by UV-crosslinking, the rat eIF4a3 coding sequence was cloned into a bacterial expression vector. Varying amounts of purified recombinant eIF4a3 were incubated with either the 32P-labeled GPx1 or PHGPx SECIS RNAs, and the RNA-protein complexes were detected by UV-crosslinking. eIF4a3 crosslinked efficiently to the GPx1 SECIS in dose-dependent manner, while only a faint crosslinking product was detected with the PHGPx SECIS (Fig. 2A). Thus, the selective interaction of eIF4a3 with the GPx1 SECIS does not require other protein factors.
Figure 2. eIF4a3 selectively interacts with the GPx1 SECIS.
(A) Radiolabeled GPx1 or PHGPx SECIS RNAs were UV-crosslinked in the presence of varying amounts (0 to 400 ng) of recombinant rat eIF4a3. The arrow indicates the position of the crosslinked product. (B) Crosslinking of eIF4a3 to the radiolabeled GPx1 probe was competed with increasing concentrations (1 to 200 nM) of unlabeled GPx1 or PHGPx SECIS RNAs. (C) Quantification of the competition experiments. The IC50s were calculated as the concentration of RNA required to decrease the crosslinking signal by 50%. The data from 4 independent observations are represented as the mean +/− SD.
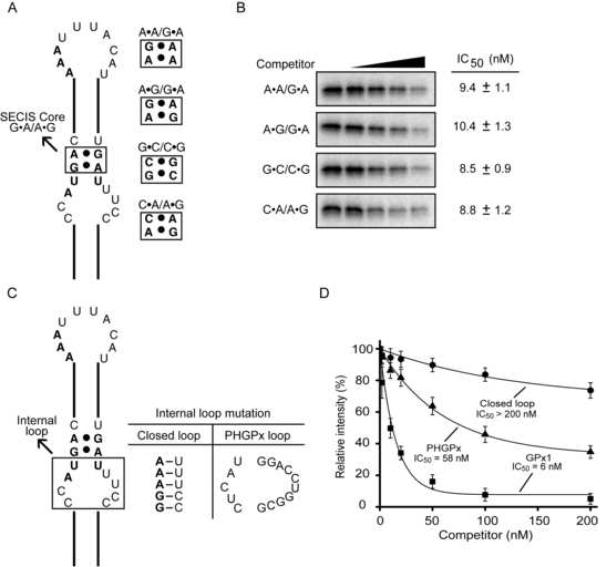
To verify that the apparent reduction in crosslinking to the PHGPx SECIS was not a result of inefficient radiolabel transfer, we performed UV-crosslinking experiments using unlabeled SECIS RNAs as competitors (Fig. 2B). The crosslinking of eIF4a3 to the 32P-labeled GPx1 SECIS was efficiently competed by unlabeled GPx1, but not PHGPx, SECIS RNA. To assess relative binding affinity, we calculated the concentration of unlabeled SECIS RNA necessary for a 50% reduction in crosslinking signal (IC50). While the GPx1 SECIS competed with an IC50 of 5.9 nM, the PHGPx SECIS was a much less effective competitor, with an IC50 > 200 nM (Fig. 2C), indicating that eIF4a3 can discriminate among different SECIS elements.
eIF4a3 Selectively Inhibits UGA Recoding Activity
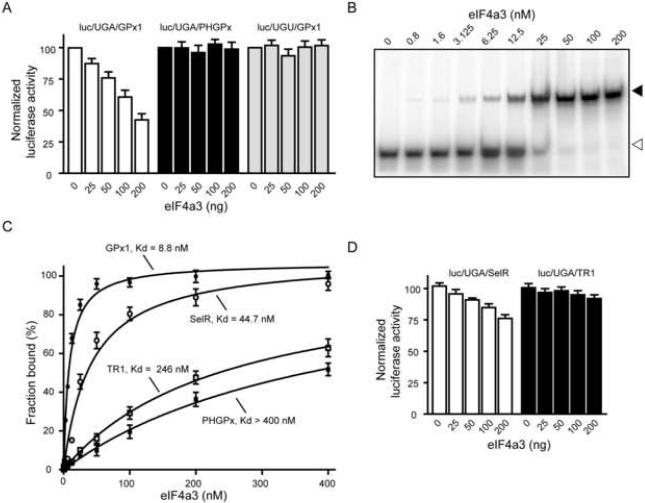
The rat GPx1 gene contains two exons, and the SECIS is positioned within the second exon (Ho et al., 1988). Given that the SECIS is ~400 nt downstream of the predicted location of the EJC in the spliced transcript, it is not expected to serve as a binding site for eIF4a3. We wondered whether this association represented a specialized function of eIF4a3, possibly in regulating the translatability of the GPx1 mRNA. To test this hypothesis, we performed in vitro UGA recoding assays using rabbit reticulocyte lysate and a modified luciferase reporter construct containing a UGA codon at position 258 and a SECIS element in its 3′-UTR. This construct has been previously validated to be specific for UGA recoding, and the luciferase expression depends on the presence of a functional SECIS (Mehta et al., 2004). Addition of eIF4a3 to the in vitro translation assay containing the reporter harboring the GPx1 SECIS (luc/UGA/GPx1) resulted in a reduction of luciferase activity in a dose-dependent manner (Fig. 3A). When 200 ng of eIF4a3 was added, we observed a ~45% decrease in UGA recoding activity. This effect was not due to degradation of the reporter RNA during the reaction (Supplementary Fig. 1). The inhibition was specific for GPx1-dependent UGA recoding since the luciferase activity from the construct containing the PHGPx SECIS (luc/UGA/PHGPx) was not affected by eIF4a3 (Fig. 3A). As the addition of large amounts (4–8 μg) of eIF4a3 to rabbit reticulocyte lysate has been shown to nonspecifically reduce translation in vitro (Li et al., 1999), we repeated the assays using a reporter containing a UGU (Cys) codon in place of the UGA. Addition of eIF4a3 up to 200 ng did not reduce luciferase activity from luc/UGU/GPx1 (Fig. 3A), suggesting that the interaction between eIF4a3 and the GPx1 SECIS did not interfere with the general translation mechanism.
Figure 3. Selective inhibition of UGA recoding activity by eIF4a3.
(A) Varying amounts of eIF4a3 (25 to 200 ng) were added to in vitro translation assays containing 100 ng of luc/UGA/GPx1, luc/UGA/PHGPx, or luc/UGU/GPx1 reporter mRNA. The luciferase results were expressed relative to reactions that were performed in the absence of eIF4a3. The data are represented as mean +/− SEM. (B) REMSA assay using the 32P-labeled GPx1 SECIS element, which was incubated with increasing amounts of eIF4a3 protein as indicated. The samples were then analyzed by native gel electrophoresis and autoradiography. Representative REMSA assays for the SelR, TR1, and PHGPx SECIS elements are shown in Supplementary Fig. 2. (C) Graph illustrating the apparent Kds of eIF4a3 for the GPx1, SelR, TR1, and PHGPx SECIS elements, which were calculated as the concentration of protein required to achieve 50% binding of the RNA. (D) Recoding assays were performed as described in (A) using reporter constructs containing the SelR and TR1 SECIS elements.
We then investigated whether eIF4a3 is a GPx1-specific binding protein, or whether it binds to other SECIS elements. As discussed above, the synthesis of stress-related selenoproteins, including GPx1 and Selenoprotein R (SelR), is regulated at the translational level in mice that lack the Um34 isoform of Sec-tRNASec. In contrast, the translation of PHGPx and Thioredoxin Reductase 1 (TR1) mRNAs is unaffected by this defect (Carlson et al., 2005). We analyzed the affinity of eIF4a3 for various SECIS elements using RNA Electrophoretic Mobility Shift Assays (REMSA). The 32P-labeled SECIS RNAs were incubated with increasing amounts of eIF4a3 and the resulting RNA-protein complexes were analyzed by native gel electrophoresis. Representative REMSA experiments are shown in Fig. 3B (GPx1 SECIS) and Supplementary Fig. 2 (SelR, PHGPx and TR1 SECIS elements). The apparent Kd was calculated as the protein concentration at which 50% of the RNA was bound (Fig. 3C). eIF4a3 bound to the GPx1 and SelR SECIS elements with apparent Kds of 8.8. and 44.7 nM, respectively. In contrast, the TR1 and PHGPx SECIS elements were bound with much lower affinity (> 200 nM).
We also performed functional assays as shown in Fig. 3D. The addition of eIF4a3 to the luc/UGA/SelR reporter mRNA resulted in the reduction of UGA recoding in a dose-dependent manner. The effect was not as dramatic as for the luc/UGA/GPx1 reporter, which is consistent with the fact that eIF4a3 has a lower binding affinity for the SelR SECIS. In agreement with the in vitro binding experiment (Fig. 3C), eIF4a3 did not inhibit UGA recoding directed by the TR1 SECIS (Fig. 3D).
Helicase Activity of eIF4a3 is not Required to Inhibit UGA Recoding
Our finding that binding of eIF4a3 to a SECIS element correlates with a reduction in UGA recoding activity raises questions regarding its mechanism. As a member of the DEAD-box protein family, eIF4a3 exhibits an RNA helicase activity in vitro (Li et al., 1999). Interestingly, this helicase activity is not required for binding of eIF4a3 to the spliced mRNAs (Shibuya et al., 2006), which is its canonical function. The helicase activity of eIF4a3 could contribute to its ability to reduce UGA recoding activity, presumably by unwinding the SECIS element so that it could no longer bind SBP2. We generated a mutation in the canonical Walker motif II (DEAD→DQAD) of eIF4a3. This mutation has been shown to negatively impact the ability of eIF4a3 to hydrolyze ATP and consequently abolishes its helicase activity in vitro (Shibuya et al., 2006). We found that DQAD mutant protein bound to the GPx1 SECIS with an apparent Kd of 20.7 nM, which is only 2.4-fold higher than the wild-type protein (Supplementary Fig. 3A). The mutant eIF4a3 protein retained the ability to inhibit luciferase activity from the luc/UGA/GPx1 construct in a dose-dependent manner similar to the wild-type protein (Supplementary Fig. 3B). We also tested ATP, ADP and AMP-PNP in REMSA assays and found that they had no effect on the eIF4a3-GPx1 SECIS interaction (Supplementary Fig. 4).
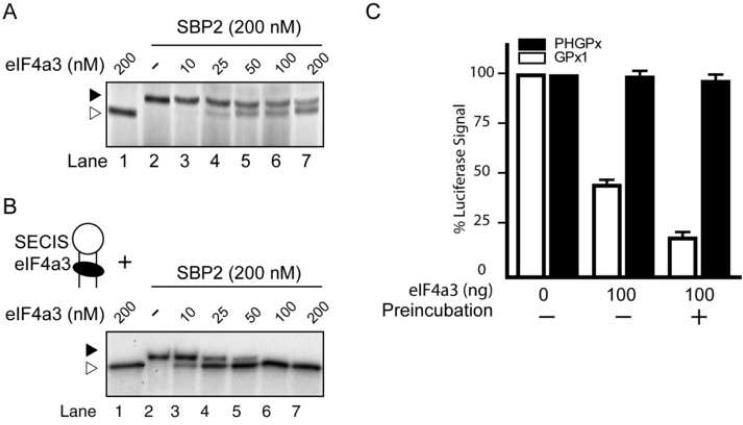
eIF4a3 Prevents Binding of SBP2 to the GPx1 SECIS
As the SBP2-SECIS interaction is essential for UGA recoding, we investigated whether this interaction is hindered in the presence of eIF4a3. UV-crosslinking experiments were performed using the 32P-labeled GPx1 SECIS, which was incubated with eIF4a3, SBP2 or both proteins. A fixed amount of SBP2 (200 nM) was used, which represents the condition in which almost all of the GPx1 SECIS RNA molecules were bound by SBP2 (Supplementary Fig. 5A). When eIF4a3, SBP2, and the GPx1 SECIS were incubated together, we observed that as the amount of eIF4a3 was increased, the SBP2-SECIS interaction decreased in favor of the eIF4a3-SECIS complex (Fig. 4A). When equimolar amounts of eIF4a3 and SBP2 were present, the protein-SECIS complexes were distributed near equally between eIF4a3 and SBP2 (lane 7, Fig. 4A). This observation is in agreement with the results of the UGA recoding assays, which showed that the translation of luc/UGA/GPx1 is reduced by ~45% when equivalent amounts of SBP2 and eIF4a3 were added to the reaction (Fig. 3A). We also attempted to confirm these results using REMSA. Although we were able to detect the GPx1-SBP2 and GPx1-eIF4a3 complexes individually, it was difficult to distinguish between these interactions when both proteins were present since the two complexes migrated close to each other (Supplementary Fig. 5B). Importantly, we did not observe formation of a new and slower migrating complex when the two proteins were present, suggesting that there was no ternary complex formation (Supplementary Fig. 5B).
Figure 4. GPx1 SECIS-binding activities of eIF4a3 and SBP2 in vitro.
(A) The radiolabeled GPx1 SECIS was incubated with SBP2 (0 or 200 nM) and increasing concentrations of eIF4a3 (0 to 200 nM), as indicated, and analyzed by UV-crosslinking. Two different crosslinked products are indicated: eIF4a3-GPx1 SECIS, open triangle; SBP2-GPx1 SECIS, filled triangle. (B) Pre-formed complexes between radiolabeled GPx1 SECIS (1 nM) and varying amounts of eIF4a3 (0 to 200 nM) were challenged with SBP2 (200 nM). Samples were analyzed by UV-crosslinking. (C) Recoding assays with the luc/UGA/GPx1 and luc/UGA/PHGPx reporter RNAs were performed in the absence or presence of 100 ng purified eIF4a3. The RNAs and eIF4a3 were not preincubated or preincubated for 20 minutes prior to in vitro translation, as indicated.
Since eIF4a3 is localized predominantly in the nucleus, one would expect that the eIF4a3-SECIS interaction in vivo may occur prior to export of the GPx1 mRNA to the cytoplasm. Consequently, in order to support Sec insertion, SBP2 would need to displace eIF4a3 from the SECIS complex. To mimic this situation in vitro, UV-crosslinking assays were performed using a two-step incubation. The 32P-labeled GPx1 SECIS RNA was incubated with various concentrations of eIF4a3 (0 to 200 nM). The preformed complexes were then challenged with near-saturating amounts of SBP2 (200 nM). When the amounts of eIF4a3 and SBP2 were equivalent (lane 7, Fig. 4B), no SBP2 crosslinking products were detected, which was not the case in the previous UV-crosslinking experiment that was performed without preincubation (lane 7, Fig. 4A).
We also tested the effect of eIF4a3 preincubation on UGA recoding using the luc/UGA258/GPx1 construct. We observed that under the same conditions, luciferase activity was ~70% lower when eIF4a3 was preincubated with the reporter mRNA compared to when eIF4a3, SBP2, and the reporter mRNA were incubated together (Fig. 4C). These observations suggest that eIF4a3-bound selenoprotein mRNAs are unable to interact with SBP2 and therefore cannot serve as a template for UGA recoding.
The Internal Loop of the GPx1 SECIS is Required for eIF4a3 Binding
Since binding of eIF4a3 and SBP2 to the GPx1 SECIS RNA appears to be mutually exclusive, we wondered whether the two proteins share the same RNA binding requirements. Four point mutations (A•A/G•A, A•G/G•A, G•C/C•G, and C•A/A•G) were made in the GPx1 SECIS core (Fig. 5A), and the mutant RNAs were used as competitors in UV-crosslinking experiments (Fig. 5B). While these mutations abolished SBP2 binding (Fletcher et al., 2001), all four mutant RNAs competed for eIF4a3 binding almost as effectively as the wild-type GPx1 SECIS element. The IC50s of the mutant RNAs ranged from 8.8 to 10.4 nM (Fig. 5B), compared to 6 nM for the wild-type GPx1 SECIS (Fig. 5D). Thus, unlike SBP2, eIF4a3 does not require the SECIS core for interacting with the GPx1 SECIS.
Figure 5. Characterization of the eIF4a3-GPx1 SECIS interaction.
(A) Schematic representation of the GPx1 SECIS, with the SECIS core mutations indicated. (B) The mutant RNAs in (A) were tested as competitors (0 to 200 nM) in UV-crosslinking assays containing eIF4a3 and the radiolabeled wild-type GPx1 SECIS probe. The IC50s of the mutant RNAs, which were calculated as described in the legend to Fig. 2, are shown. (C) Schematic representation of the mutations in the internal loop of the GPx1 SECIS. (D) Graphic representation of the results of competition experiments in which cold wild-type and mutant SECIS RNAs were tested for their ability to compete for binding of eIF4a3 to the 32P-labeled GPx1 SECIS element. The IC50s were calculated as described in (B).
Another region of the SECIS implicated in SBP2 binding is the internal loop (Fletcher et al., 2001). We assessed the importance of this region for eIFa43 binding by replacing the internal loop in the GPx1 SECIS with five Watson-Crick base pairs (Fig. 5C). Compared to the wild-type SECIS, the mutant lacking the internal loop was a very poor competitor (Fig. 5D). To test whether the binding of eIF4A3 depends on the single-stranded nature or the nucleotide sequence of the internal loop, we replaced this region in the GPx1 SECIS with the internal loop from the PHGPx SECIS (Fig. 5C). Surprisingly, eIF4a3 was able to bind to the mutant containing the PHGPx internal loop (Fig. 5D). Thus the presence of an internal loop is necessary but not sufficient to mediate binding of eIF4a3 to a SECIS element.
eIF4a3 Expression Is Regulated by Selenium
Given the ability of eIF4a3 to prevent SBP2 binding to the SECIS, the ratio of these two proteins may be critical for determining UGA recoding efficiency. GPx1 expression is exquisitely sensitive to selenium concentration, and its SECIS element is selectively bound by eIF4a3. Therefore, we tested whether the expression of eIF4a3 changes in response to selenium. We first established conditions of moderate selenium insufficiency in which GPx1 expression is mainly regulated at the level of translation. Since media supplemented by 10% FBS contains ~29 nM selenium, we grew McArdle 7777 in the absence (−Se) or presence (+Se) of additional sodium selenite (30 nM) to imitate conditions of selenium insufficiency and selenium sufficiency, respectively. After three days, we isolated RNA for quantitative real time PCR assay (qRT-PCR) and proteins (nuclear and cytoplasmic) for immunoblotting analysis.
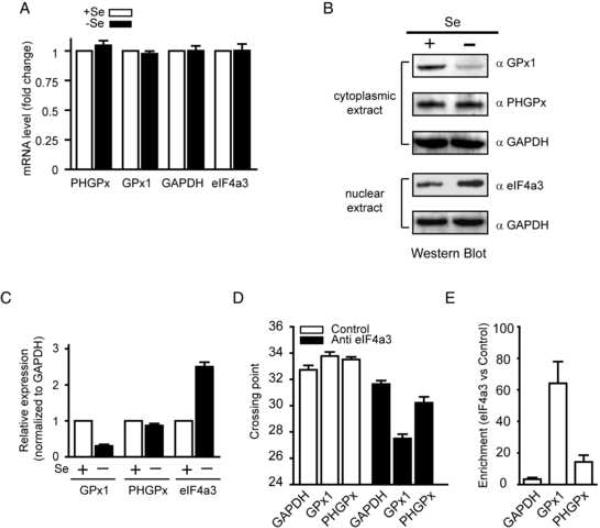
As shown in Fig. 6A, the GPx1, PHGPx, and GAPDH mRNA levels were similar in −Se and +Se cells. In contrast, GPx1 protein was 3-fold lower in the cytoplasmic extract from −Se cells than from +Se cells (Fig. 6B and 6C). Selenium status had little or no effect on PHGPx and GAPDH protein levels (Fig. 6B). To analyze eIF4a3 protein expression, we used 75 μg nuclear extract which is in the linear range for detecting changes in eIF4a3 protein levels, based on dose response experiments (Supplementary Fig. 6). We found that the level of eIF4a3 protein in the nuclear extract was increased 2.5-fold in −Se cells compared to +Se cells (Fig. 6B and 6C). Since selenium had no effect on eIF4a3 mRNA levels (Fig. 6A), these results suggest that the expression of eIFA4a3 is regulated at the level of protein synthesis or degradation.
Figure 6. Effect of selenium on mRNA and protein expression.
(A) Total RNA was extracted from McArdle 7777 cells grown with (+Se) or without (−Se) 30 nM sodium selenite supplementation after 3 days. GPx1, PHGPx, GAPDH, and eIF4a3 mRNA levels were determined by qRT-PCR. The level of mRNA was expressed relative to the amount of mRNA from +Se cells (dashed line). The data are represented as the mean +/− SEM. (B) Cytoplasmic or nuclear extracts were analyzed by Western blotting using different antibodies as indicated. (C) The graph represents quantification of Western blot data from three independent experiments. The results are normalized to GAPDH expression. (D) qRT-PCR crossing points values of GAPDH, GPx1 and PHGPx mRNAs co-immunoprecipitated with anti-eIF4a3 or isotype control antibody. (E) Relative enrichment of GAPDH, GPx1 and PHGPx mRNAs in the eIF4a3 immunoprecipitation compared to the isotype control.
Given the inverse correlation between eIF4a3 and GPx1 protein levels, we wondered whether eIF4a3 is associated with GPx1 mRNA in selenium insufficient cells, thereby inhibiting UGA recoding. Cytoplasmic extracts from −Se cells were co-immunoprecipitated with an anti-eIF4a3 antibody or an isotype control antibody, and the immunoprecipitated RNAs were analyzed by qRT-PCR. The mean PCR crossing points were calculated for the anti-eIF4a3 and control immunoprecipitations (Fig. 6D). The results were also expressed as fold-enrichment of a transcript in the eIF4a3 immunoprecipitation compared to the control (Fig. 6E). The anti-eIF4a3 antibody but not the control antibody was able to co-immunoprecipitate the GPx1, PHGPx and GAPDH mRNAs, suggesting that eIF4a3 was associated with these transcripts in cell extracts. This result is not surprising as eIF4a3 binds to exon-exon junctions in spliced mRNAs. However, the GPx1 transcript was preferentially immunoprecipitated by the anti-eIF4a3 antibody compared the GAPDH and PHGPx mRNAs. This enrichment of GPx1 in the eIF4a3 immunoprecipitation is not due to inherent differences in transcript abundance as GPx1 mRNA levels are several fold lower than PHGPx1 and GAPDH mRNA levels in McArdle 7777 cells (Supplementary Fig. 7) and most mouse tissues (Hoffmann et al., 2007). In order to establish that the eIF4a3-GPx1 SECIS interaction is physiologically relevant, we manipulated eIF4a3 levels in cells using siRNA and overexpression approach.
Manipulation of eIF4a3 Levels Regulates GPx1 Expression in Cells
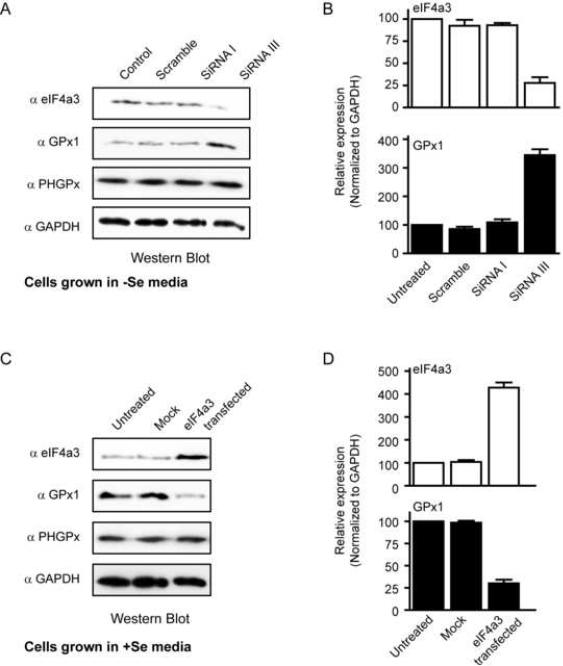
The above results suggested that eIF4a3 acts as a regulatory protein for selenoprotein expression when selenium is limiting. To directly test this hypothesis, we used siRNA to knockdown the expression of eIF4a3 in the −Se cells. McArdle 7777 cells grown in the absence of sodium selenite were treated with various siRNAs for 72 hours. We identified one siRNA (siRNA III) which inhibited eIF4a3 protein expression by ~ 4-fold (Fig. 7A and 7B). The levels of eIF4a3 protein were not reduced by treatment with the control, or scramble siRNAs, or by siRNA I, which was directed against eIF4a3 but ineffective. As shown in Figures 7A and 7B, there was a 3.5-fold increase in GPx1 protein levels in cells treated with siRNA III, which was not observed in the other siRNA-treated cells. This effect was specific as PHGPx and GAPDH protein levels did not change (Fig. 7A). The knockdown of eIF4a3 did not reduce GPx1 mRNA levels as determined by qRT-PCR (Supplementary Fig. 8A). Thus, eIF4a3 is required to inhibit GPx1 expression in selenium insufficiency.
Figure 7. Manipulation of eIF4a3 regulates GPx1 expression in cells.
(A) McArdle 7777 cells were treated with different siRNAs for 72 hours as described in Materials and Methods. Cell lysates were analyzed by Western blotting using the indicated antibodies. (B) Quantification of the Western blot data were from 3 independent experiments. The results are normalized to GAPDH and represented as mean +/− SD. (C) McArdle 7777 cells were transiently transfected with empty vector DNA (mock) or a plasmid encoding the human eIF4a3 cDNA. The Western blots (C) and quantification (D) were performed as described in (A) and (B)
We also performed overexpression studies in +Se cells, which were transiently transfected with vector or plasmid DNA encoding rat eIF4a3. Transfections were done under conditions where > 80% of the cells were transfected. The amount of plasmid DNA was titrated in pilot experiments in order to obtain only a modest increase of eIF4a3 protein. As shown in Figures 7C and 7D, we obtained a ~4-fold overexpression of eIF4a3 in +Se cells, which is comparable to the level observed in −Se cells. The overexpression of eIF4a3 resulted in a ~3-fold decrease in GPx1 protein levels (Fig. 7C and 7D) but had no effect on PHGPx or GAPDH (Fig. 7C). This reduction in GPx1 expression was not mediated through a change in GPx1 mRNA levels (Supplementary Fig. 8B). These results suggest that an increase in eIF4a3 protein is sufficient to regulate GPx1 expression even when selenium is not limiting.
DISCUSSION
The differential expression of selenoproteins in response to selenium deficiency has long been observed (Lei et al., 1995) yet the mechanism underlying this regulation has not been elucidated. In this study, we demonstrate that eIF4a3 is a selenium-regulated SECIS binding protein that links selenium status and differential selenoprotein expression. The selective binding of eIF4a3 to a subset of SECIS elements limits the accessibility of those transcripts for SBP2 binding, thereby preventing Sec insertion. Based on siRNA experiments, eIF4a3 is necessary to reduce the expression of GPx1 in selenium deficient cells. Furthermore, the overexpression of eIF4a3 is sufficient to induce downregulation of GPx1 when selenium is adequate. Thus, while SBP2 acts as the master regulator in selenoprotein synthesis, eIF4a3 serves to selectively modulate the availability of selenoprotein mRNAs for SBP2 binding. Our results provide new insight into how the expression of a subset of selenoprotein mRNAs is preferentially inhibited to allow the synthesis of essential housekeeping selenoproteins such as PHGPx and TR1.
eIF4a3 is a member of the DEAD-box protein family which shares ~70% amino acid identity with the initiation factor eIF4a1 (Li et al., 1999). While eIF4a3, like eIF4a1, exhibits an RNA-dependent ATPase activity and an ATP-dependent RNA helicase activity, it does not substitute for eIF4a1 in an in vitro-reconstituted 40S ribosome binding assay (Li et al., 1999). In contrast to eIF4a1, which resides in the cytoplasm, eIF4a3 is found predominantly in the nucleus, although it has also been implicated as a nucleocytoplasmic shuttling protein (Li et al., 1999; Shibuya et al., 2004). In cells, eIF4a3 plays an important role in the NMD pathway and associates preferentially with nuclear complexes containing the EJC proteins Magoh, Y14, and MLN51 (Ballut et al., 2005; Chan et al., 2004; Palacios et al., 2004). In this complex, eIF4a3 is stably bound with the mRNA and serves as a molecular placeholder for the subsequent assembly of the EJC (Andersen et al., 2006; Ballut et al., 2005; Le Hir and Andersen, 2008; Shibuya et al., 2006). Mutations in the canonical Walker A and B motifs of human eIF4a3 eliminated RNA-dependent ATP hydrolysis in vitro but had no effect on EJC formation and NMD activation (Shibuya et al., 2006). While its function in protein synthesis is unclear, eIF4A3 has been shown to associate with both neuronal and dendritic mRNAs and modulate their translation (Giorgi et al., 2007). Each subfamily of DEAD-box proteins contains several conserved signature motifs (Tanner and Linder, 2001). These motifs, which are highly divergent between subfamilies, are thought to provide functional specificity through interaction with distinct RNA substrates or other factors (Cordin et al., 2006). Thus it is been suggested that eIF4a3 may participate in selective mRNA translational inhibition caused by distinctive physiological perturbations (Li et al., 1999).
In contrast to the sequence-independent binding of eIF4a3 to the mRNA in the EJC, the protein distinguishes among SECIS elements. Based on mutagenesis studies, the integrity of the SECIS internal loop, but not the SECIS core, is essential for eIF4a3 binding (Fig. 5), whereas both of these regions are required for SBP2-SECIS interaction (Fletcher et al., 2001). Thus, we believe the SECIS binding requirements for eIF4a3 and SBP2 are likely to be overlapping but not identical. The simplest explanation for the observed mutually exclusive binding of eIF4a3 and SBP2 to the SECIS is steric inhibition, although other models need to be considered. Structural studies of eIF4a3 show that its conserved helicase cores can be largely superimposed on other DEAD-box proteins, including eIF4a1, which adopt a “dumbbell” structure consisting of two domains tethered by a linker (Bird et al., 1998; Caruthers et al., 2000; Korolev et al., 1997). The eIF4a3-SECIS interaction might resemble the proposed model for selected RNA aptamers that inhibit eIF4a1 ATPase activity, in which the two domains of the protein interact with two separate loop regions in the RNA (Oguro et al., 2003). We speculate that the interaction of eIF4a3 with the SECIS element requires two binding sites as well. Such a model could explain the preferential binding of eIF4a3 to the GPx1 and SelR SECIS elements, which contain an internal loop as well as a large apical loop. In contrast, eIF4a3 binds with low affinity to the PHGPx and TR1 SECIS elements, which have small apical loops. Our finding that the PHGPx internal loop supports eIF4a3 binding in the context of the GPx1 SECIS element is consistent with a two-site interaction model.
The effects of selenium deficiency are not the same for each selenoprotein, creating a hierarchy in selenoprotein synthesis. This complex pattern of regulation is not likely to be explained by a single mechanism. The availability of selenium regulates not only the levels of the Sec-tRNASec, but also its Um34 isoform (Diamond et al., 1993). While the 2′-O-ribose methylation at position 34 has been shown to dramatically alter the tertiary structure of Sec-tRNASec (Kim et al., 2000), how such modification affects the expression of only certain selenoproteins is not fully understood. Many proteins whose synthesis heavily relies on the Um34 isoform appear to be involved in responses to stress, and their expression is also highly sensitive to selenium in the diet (Carlson et al., 2005; Sengupta et al., 2008). In the absence of the Um34 isoform, these transcripts are not competent for UGA recoding; however, they are still able to bind SBP2, which is a limiting factor in cells (Squires et al., 2007). In addition to inhibiting UGA recoding, binding of eIF4a3 to the stress-related selenoprotein mRNAs will prevent the formation of nonproductive SBP2-SECIS complexes. We speculate that the upregulation of eIF4a3 protein together with the decrease in the Um34 isoform of Sec-tRNASec are utilized to efficiently inhibit Sec incorporation into certain selenoproteins in selenium deprivation. This also ensures that SBP2 will be available for the synthesis of the necessary housekeeping selenoproteins.
In summary, our studies have identified an unexpected function of eIF4a3 in selectively regulating selenoprotein mRNA translation. During the initial stages of selenium deficiency, changes in the translational profile of the selenoproteome must occur rapidly to preserve the expression of selenoproteins with essential functions. Thus, binding of eIF4a3 to a subset of selenoprotein mRNAs could act as a brake to create a temporary translational arrest for those non-essential stress-related selenoproteins. Upon restoration of the selenium status, the translational repression of these selenoproteins could be alleviated promptly, presumably by releasing eIF4a3 from the SECIS element and making it available for SBP2 binding. Whether selenium modulates the expression of eIF4a3 at the level of mRNA translation or protein stability and the mechanism by which this regulation occurs are important areas for future investigation.
EXPERIMENTAL PROCEDURES
The SECIS and luciferase reporter constructs, cloning and expression of recombinant protein, RNA synthesis and immunoblotting are described in The Supplementary Experimental Procedures.
Preparation of cell extracts
McArdle 7777 cells were grown in DMEM/F12 media containing 10% FBS in the absence or presence of 30 nM sodium selenite. Nuclear and cytosolic extracts were isolated as previously described (Bubenik et al., 2009). Protein concentrations were determined using the Biorad Protein Assay.
UV-crosslinking
Crosslinking with McArdle 7777 nuclear extracts was done by incubating 20 fmol 32P-labeled SECIS RNA with 20 μg of extract in 20 μL binding buffer (20 mM HEPES pH 7.9, 100 mM KCl, 2 mM DTT, 10 μM tRNA and 5% glycerol) for 30 minutes at 37°C. The reactions were UV-crosslinked using a GS Gene Linker (Biorad) at 125 mJoules for 10 minutes followed by RNase A (Fermentas) treatment for 60 minutes at 37°C. Samples were separated by 12% SDS-PAGE and analyzed using a Storm PhosphorImager (GE Healthcare). Crosslinking with purified recombinant protein was done as described above using the amount of proteins indicated. OriginPro software (Origin Lab) was used to calculate the IC50 values and performed statistical analyses for the competition assays.
Biotinylated RNA pulldown
Reactions containing either 25 nmol of biotinylated GPx1 SECIS or biotinylated PHGPx SECIS RNA were incubated with 200 μg of McArdle 7777 nuclear extract in 200 μL binding buffer for 30 minutes at 37°C to allow RNA-protein complexes to form. Following the incubation, 100 μL of prewashed streptavidin coated magnetic beads were added to each reaction for 30 minutes at 25°C. The RNA-protein complexes were extracted from the mixture using a magnetic stand, washed with 100 μL binding buffer 5 times and resuspended in 15 μL binding buffer. The resuspended complexes were mixed with SDS loading dye, boiled for 5 minutes and then centrifuged. The supernatants were analyzed by 10% SDS-PAGE. The gel was stained with Coomassie Blue and the band of interest was excised, washed, reduced, alkylated and digested with trypsin. Peptides were extracted from the gel and analyzed by LCMS. The CID spectra were used for protein identification using primary sequence databases (MASCOT).
UGA recoding assay
In vitro UGA recoding assays were performed in rabbit reticulocyte lysate (Promega) by adding 100 ng of reporter mRNA and 100 ng of purified recombinant SBP2 in the absence or presence of eIF4a3. The reactions were incubated at 37°C for 30 minutes and aliquots were analyzed for luciferase activity by adding luciferase substrate (Promega). Luciferase activities were measured using Victor3 Multilabel Plate Reader (Perkin Elmer). Statistical analyses were done with Graphpad Prism 5.
REMSA assay
32P-labeled SECIS RNAs (10 fmol) were heated at 95°C, slow cooled to room temperature and then incubated with the indicated amount of protein in 20 μL binding buffer (10 mM HEPES, 100 mM KCl, 2.5% glycerol, 0.1 mM EDTA and 1 mM DTT) supplemented with 6.5 μg of tRNA. Incubations were done at 37°C for 30 minutes and the formed complexes were resolved on 6% nondenaturing polyacrylamide gels. The gels were dried and analyzed using a PhosphorImager. Dissociation constants (Kd) were determined by quantified the protein-bound fractions using ImageQuant software (GE Healthcare) and plotted against protein concentration (nM). Kd values were extracted from plots fitted to a hyperbolic function in Origin 7 software (OriginLab).
Immunoprecipitation
MagnaBind Protein A beads (Pierce) were incubated with either an α-eIF4a3 (3F1) mouse monoclonal antibody (SantaCruz) or an isotype control antibody α-CUGBP (3B1, SantaCruz) for 1 hour at 25°C and then washed twice with Na-phosphate buffer (100 mM, pH 8.1). The conjugated beads were added to cytoplasmic extracts from McArdle cells grown in DMEM/F12 media with 10% FBS and then placed in the rotating platform at 4°C. After 45 minutes, the mixtures were placed on a magnetic stand and the supernatants were removed. The beads were washed with NET 2 buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.03% NP40) three times and then extracted for the total RNA, which was analyzed by qRT-PCR.
Quantitative real time PCR
Total RNA was extracted from McArdle 7777 cells using Trizol reagent (Invitrogen) following the manufacturer's instruction. SuperScript III Reverse Transcription kit (Invitrogen) was used to generate cDNA and qRT-PCR analysis was done using SYBR Green PCR Core Reagents in an ABI Prism 7000 Real-Time PCR system (Applied Biosystem). Relative gene expression was calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Primers used for qRT-PCR are listed in Table S1.
siRNA experiments
The sequences for siRNA I and III targeting the rat eIF4a3 mRNA were 5′- UGACCAAAGUGGAGUUCGAUU-3′ and 5′-CGGGAGAUCAGGUCGAUAUUU-3′, respectively (Dharmacon). Accell non-targeting siRNA #1 (Dharmacon) was used as a control. McArdle 7777 cells were seeded in 6 well plates (5×105 cells/well) containing DMEM/F12 media supplemented with 10% FBS. Cells were grown for 16 hours and transfected with 100 nM siRNA using Dharmafect 4 (Dharmacon) according to manufacturer's protocol. After 72 hours, cells were harvested and extracted for total protein using cell culture lysis reagent (Promega) or total RNA using Trizol (Invitrogen).
Transient transfections
McArdle 7777 cells were seeded in 6 well plates (3.5×105 cells/well) containing DMEM/F12 media supplemented with 10% FBS and 30 nM Na2SeO3. Cells were grown for 16 hours and transfected with either plasmid containing rat eIF4a3 gene or with empty vector using lipofectamine (Invitrogen) in DMEM/F12 media according to manufacturer's protocol. Equal volume of DMEM-F12 media supplemented with 20% FBS and 60 nM Na2SeO3 was added to the cells 5 hours after transfection. Cells were grown for 48 hours and harvested for total protein or total RNA as described above.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants RO1 DK078591 (D.M.D.) and F32 DK083154 (M.E.B.). We thank Dr. Mike Kinter and Dr. Belinda Willard from the Lerner Research Institute Proteomics Laboratory for the mass spectrometry analysis. The authors declare they have no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allamand V, Richard P, Lescure A, Ledeuil C, Desjardin D, Petit N, Gartioux C, Ferreiro A, Krol A, Pellegrini N, et al. A single homozygous point mutation in a 3'untranslated region motif of selenoprotein N mRNA causes SEPN1-related myopathy. EMBO Rep. 2006;7:450–454. doi: 10.1038/sj.embor.7400648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Bermano G, Nicol F, Dyer JA, Sunde RA, Beckett GJ, Arthur JR, Hesketh JE. Selenoprotein gene expression during selenium-repletion of selenium-deficient rats. Biol Trace Elem Res. 1996;51:211–223. doi: 10.1007/BF02784076. [DOI] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- Bird LE, Subramanya HS, Wigley DB. Helicases: a unifying structural theme? Curr Opin Struct Biol. 1998;8:14–18. doi: 10.1016/s0959-440x(98)80004-3. [DOI] [PubMed] [Google Scholar]
- Bubenik JL, Driscoll DM. Altered RNA binding activity underlies abnormal thyroid hormone metabolism linked to a mutation in selenocysteine insertion sequence-binding protein 2. J Biol Chem. 2007;282:34653–34662. doi: 10.1074/jbc.M707059200. [DOI] [PubMed] [Google Scholar]
- Bubenik JL, Ladd AN, Gerber CA, Budiman ME, Driscoll DM. Known turnover and translation regulatory RNA-binding proteins interact with the 3' UTR of SECIS-binding protein 2. RNA Biol. 2009;6:73–83. doi: 10.4161/rna.6.1.7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BA, Xu XM, Gladyshev VN, Hatfield DL. Selective rescue of selenoprotein expression in mice lacking a highly specialized methyl group in selenocysteine tRNA. J Biol Chem. 2005;280:5542–5548. doi: 10.1074/jbc.M411725200. [DOI] [PubMed] [Google Scholar]
- Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CC, Dostie J, Diem MD, Feng W, Mann M, Rappsilber J, Dreyfuss G. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10:200–209. doi: 10.1261/rna.5230104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavatte L, Brown BA, Driscoll DM. Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat Struct Mol Biol. 2005;12:408–416. doi: 10.1038/nsmb922. [DOI] [PubMed] [Google Scholar]
- Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Diamond AM, Choi IS, Crain PF, Hashizume T, Pomerantz SC, Cruz R, Steer CJ, Hill KE, Burk RF, McCloskey JA, et al. Dietary selenium affects methylation of the wobble nucleoside in the anticodon of selenocysteine tRNA([Ser]Sec) J Biol Chem. 1993;268:14215–14223. [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR. Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- Dumitrescu AM, Liao XH, Abdullah MS, Lado-Abeal J, Majed FA, Moeller LC, Boran G, Schomburg L, Weiss RE, Refetoff S. Mutations in SECISBP2 result in abnormal thyroid hormone metabolism. Nat Genet. 2005;37:1247–1252. doi: 10.1038/ng1654. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JE, Copeland PR, Driscoll DM, Krol A. The selenocysteine incorporation machinery: interactions between the SECIS RNA and the SECIS-binding protein SBP2. RNA. 2001;7:1442–1453. [PMC free article] [PubMed] [Google Scholar]
- Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Hill KE, Lyons PR, Burk RF. Differential regulation of rat liver selenoprotein mRNAs in selenium deficiency. Biochem Biophys Res Commun. 1992;185:260–263. doi: 10.1016/s0006-291x(05)80984-2. [DOI] [PubMed] [Google Scholar]
- Ho YS, Howard AJ, Crapo JD. Nucleotide sequence of a rat glutathione peroxidase cDNA. Nucleic Acids Res. 1988;16:5207. doi: 10.1093/nar/16.11.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LK, Matsufuji T, Matsufuji S, Carlson BA, Kim SS, Hatfield DL, Lee BJ. Methylation of the ribosyl moiety at position 34 of selenocysteine tRNA[Ser]Sec is governed by both primary and tertiary structure. RNA. 2000;6:1306–1315. doi: 10.1017/s1355838200000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Andersen GR. Structural insights into the exon junction complex. Curr Opin Struct Biol. 2008;18:112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Lei XG, Evenson JK, Thompson KM, Sunde RA. Glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase are differentially regulated in rats by dietary selenium. J Nutr. 1995;125:1438–1446. doi: 10.1093/jn/125.6.1438. [DOI] [PubMed] [Google Scholar]
- Li Q, Imataka H, Morino S, Rogers GW, Jr, Richter-Cook NJ, Merrick WC, Sonenberg N. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol Cell Biol. 1999;19:7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann PR, Hoge SC, Li PA, Hoffmann FW, Hashimoto AC, Berry MJ. The selenoproteome exhibits widely varying, tissue-specific dependence on selenoprotein P for selenium supply. Nucleic Acids Res. 2007;35:3963–3973. doi: 10.1093/nar/gkm355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Low SC, Grundner-Culemann E, Harney JW, Berry MJ. SECIS-SBP2 interactions dictate selenocysteine incorporation efficiency and selenoprotein hierarchy. EMBO J. 2000;19:6882–6890. doi: 10.1093/emboj/19.24.6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Rebsch CM, Kinzy SA, Fletcher JE, Copeland PR. Efficiency of mammalian selenocysteine incorporation. J Biol Chem. 2004;279:37852–37859. doi: 10.1074/jbc.M404639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C, Wingler K, Brigelius-Flohe R. 3'UTRs of glutathione peroxidases differentially affect selenium-dependent mRNA stability and selenocysteine incorporation efficiency. Biol Chem. 2003;384:11–18. doi: 10.1515/BC.2003.002. [DOI] [PubMed] [Google Scholar]
- Oguro A, Ohtsu T, Svitkin YV, Sonenberg N, Nakamura Y. RNA aptamers to initiation factor 4A helicase hinder cap-dependent translation by blocking ATP hydrolysis. RNA. 2003;9:394–407. doi: 10.1261/rna.2161303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Carlson BA, Weaver JA, Novoselov SV, Fomenko DE, Gladyshev VN, Hatfield DL. A functional link between housekeeping selenoproteins and phase II enzymes. Biochem J. 2008;413:151–161. doi: 10.1042/BJ20080277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Stroupe ME, Moore MJ. Mutational analysis of human eIF4AIII identifies regions necessary for exon junction complex formation and nonsense-mediated mRNA decay. RNA. 2006;12:360–374. doi: 10.1261/rna.2190706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires JE, Stoytchev I, Forry EP, Berry MJ. SBP2 binding affinity is a major determinant in differential selenoprotein mRNA translation and sensitivity to nonsense-mediated decay. Mol Cell Biol. 2007;27:7848–7855. doi: 10.1128/MCB.00793-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunde RA. Regulation of glutathione peroxidase-1 expression. In: Hatfield DL, Berry MJ, V.N. G, editors. Selenium: Its molecular biology and role in human health. Kluwer Academic Publishers; Norwell, MA: 2006. pp. 149–160. [Google Scholar]
- Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Tujebajeva RM, Copeland PR, Xu XM, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss-Sachdev S, Sunde RA. Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver. Biochem J. 2001;357:851–858. doi: 10.1042/0264-6021:3570851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SL, Sunde RA. Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels. Rna. 1998;4:816–827. doi: 10.1017/s1355838298971990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler K, Bocher M, Flohe L, Kollmus H, Brigelius-Flohe R. mRNA stability and selenocysteine insertion sequence efficiency rank gastrointestinal glutathione peroxidase high in the hierarchy of selenoproteins. Eur J Biochem. 1999;259:149–157. doi: 10.1046/j.1432-1327.1999.00012.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.