Abstract
The cohesin complex establishes sister chromatid cohesion during S phase. In metazoan cells, most if not all cohesin dissociates from chromatin during mitotic prophase, leading to the formation of metaphase chromosomes with two cytologically discernible chromatids. This process, known as sister chromatid resolution, is believed to be a prerequisite for synchronous separation of sister chromatids in subsequent anaphase. To dissect this process at a mechanistic level, we set up an in vitro system. Sister chromatid resolution is severely impaired upon depletion of Wapl from Xenopus egg extracts. Exogenously added human Wapl can rescue these defects and, remarkably, it can do so in a very short time window of early mitosis. A similar set of observations is made for Pds5, a factor implicated previously in the stabilization of interphase cohesion. Characteristic amino acid motifs (the FGF motifs) in Wapl coordinate its physical and functional interactions with Pds5 and cohesin subunits. We propose that Wapl and Pds5 directly modulate conformational changes of cohesin to make it competent for dissociation from chromatin during prophase. Evidence is also presented that Sgo1 plays a hitherto underappreciated role in stabilizing cohesin along chromosome arms, which is antagonized by the mitotic kinases polo-like kinsase (Plk1) and aurora B.
Keywords: Sister chromatid resolution, Wapl, Pds5, Sgo1, Xenopus egg extracts
Chromosome duplication and segregation are among the most fundamental events that must be regulated faithfully to maintain genome integrity in eukaryotic cells. Errors in these processes during mitosis or meiosis increase the probability of genome instability, potentially leading to cancer, developmental disorders, or birth defects. Extensive studies during the past decade have revealed that two structurally related protein complexes, cohesin and condensin, play central roles in controlling a series of events that makes duplicated chromosomes proficient for faithful segregation (Losada and Hirano 2005; Nasmyth and Haering 2005). Cohesin participates in holding newly duplicated chromatids together during S phase, a process known as sister chromatid cohesion (Lee and Orr-Weaver 2001; Onn et al. 2008). Condensin, on the other hand, associates with chromatin to initiate chromosome condensation during the early stage of mitosis (i.e., prophase). At the same time, the bulk of cohesin dissociates from chromosome arms, leading to the formation of metaphase chromosomes in which sister chromatids become microscopically discernible as two rod-shaped structures apposed along their lengths. Although the physiological significance of this process, often referred to as sister chromatid resolution, is not fully understood, one possibility frequently discussed is that the resolution process is a prerequisite for rapid and synchronous separation of sister chromatids in anaphase (Losada et al. 2002). Despite its fundamental importance, we are still largely ignorant of the molecular mechanisms behind this process.
The cohesin complex consists of four subunits: Smc1 and Smc3 (members of the structural maintenance of chromosomes [SMC] family of ATPases), Scc1/Mcd1/Rad21 (a member of the kleisin family of proteins), and Scc3/SA (SA1 and SA2 in vertebrates). Cohesin forms a ring-like structure that may topologically embrace two sister chromatids (Anderson et al. 2002; Haering et al. 2002, 2008; Gruber et al. 2003). While the final release of cohesin from chromosomes in anaphase accompanies proteolytic cleavage of the Scc1/Mcd1/Rad21 subunit (Uhlmann et al. 1999; Hauf et al. 2001), the release of bulk cohesin during prophase does not (Waizenegger et al. 2000). It has been shown instead that the latter process is facilitated by two mitotic kinases, polo-like kinase (Plk1) and aurora B (Losada et al. 2002; Sumara et al. 2002). While Plk1 is likely to directly phosphorylate cohesin subunits, the role of aurora B in this process is less clear. On the other hand, it has been proposed that another factor, Sgo1, protects a centromeric population of cohesin from its phosphorylation by Plk1, thereby allowing cohesion to persist at centromeres until the onset of anaphase (Kitajima et al. 2006).
Plk1 and aurora B are not the only factors that are required for sister chromatid resolution, however. Recent studies in human cells have demonstrated that Wapl, originally identified as a gene product important for heterochromatin organization and normal chromosome segregation in Drosophila melanogaster (Verni et al. 2000; Dobie et al. 2001), physically interacts with cohesin and plays a key role in promoting the resolution process (Gandhi et al. 2006; Kueng et al. 2006). It has also been shown more recently that yeast proteins related to Wapl genetically interact with cohesin regulators during interphase (Ben-Shahar et al. 2008; Bernard et al. 2008). These observations are intriguing because the process corresponding to sister chromatid resolution in early mitosis is not obvious in yeasts. Thus, Wapl is likely to be a general regulator of cohesin that functions throughout the cell cycle, although its mechanism of action remains to be elucidated.
Pds5 is another protein implicated in a complex layer of cohesin regulation. The function of Pds5 that has been best characterized so far in yeasts and humans is the maintenance of sister chromatid cohesion during G2 phase (Hartman et al. 2000; Panizza et al. 2000; Tanaka et al. 2001; Losada et al. 2005). Nevertheless, different organisms display substantially different requirements for Pds5's function. For example, Pds5 is essential for viability in budding yeast (Hartman et al. 2000; Panizza et al. 2000), but not in fission yeast (Tanaka et al. 2001). Mice lacking Pds5B, one of the two Pds5 isoforms in vertebrates, exhibit no discernible defects in sister chromatid cohesion, although they display severe developmental defects (Zhang et al. 2007). While Pds5 has been shown to physically interact with Wapl (Gandhi et al. 2006; Kueng et al. 2006), it remains to be determined whether Pds5 also has a function in sister chromatid resolution and, if so, how the two proteins may work together.
In the present study, we used a cell-free extract derived from Xenopus eggs to dissect the process of sister chromatid resolution at a mechanistic level. We show that depletion of endogenous Wapl (or Pds5) from egg extracts causes severe defects in sister chromatid resolution, which can be rescued by adding a recombinant human Wapl (or Pds5) protein back into the extracts. This experimental system, together with extensive protein–protein interaction assays, allows us to explore the molecular mechanism of how Wapl and Pds5 might interact with cohesin to facilitate its dissociation from chromatin. It is found that Wapl uses characteristic FGF motifs conserved in its N-terminal domain to coordinate an exquisite molecular cross-talk with Pds5 and cohesin. Finally, we present evidence that Sgo1 plays a negative role in cohesin release from chromosome arms, and that Plk1 and aurora B antagonize this action of Sgo1 through distinct mechanisms.
Results
Sister chromatid resolution is severely compromised in the absence of Wapl or Pds5 in Xenopus egg extracts
To gain mechanistic insights into the molecular network that regulates sister chromatid resolution, we took advantage of the cell-free system from Xenopus eggs that has proved to be powerful in studying cell cycle-dependent changes of higher-order chromosome structures, such as chromosome condensation and sister chromatid cohesion (e.g., Hirano et al. 1997; Losada et al. 1998). Basic characterization of the Xenopus ortholog of Wapl is shown in Supplemental Figure S1. In short, Xenopus Wapl was present in egg extracts as a 160-kDa polypeptide with a sedimentation coefficient of ∼7S. The majority of Wapl was not cofractionated with either Pds5 or the holocomplex of cohesin as judged by sucrose gradient centrifugation, but reciprocal immunoprecipitation revealed that a small population of Wapl did interact with Pds5 and cohesin as had been shown in human cell lysates (Gandhi et al. 2006; Kueng et al. 2006). When sperm chromatin was incubated with the cell-free extracts, Wapl was progressively accumulated on chromatin during interphase and gradually dissociated from chromatin after mitotic entry. This kinetics was similar to that of cohesin and Pds5B.
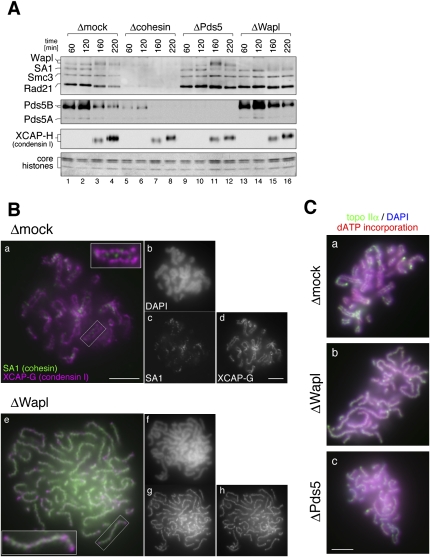
To test whether chromatin binding of Wapl, Pds5, and cohesin depends on one another, we depleted each of the three factors from interphase extracts (Fig. 1; see also Supplemental Fig. S2). Sperm chromatin was incubated with each extract for 120 min, and then cyclin B was added to convert the cell cycle state into mitosis. Chromatin fractions were isolated at various time points and analyzed by immunoblotting. We found that loading of Wapl and Pds5 onto chromatin during interphase was severely compromised in a cohesin-depleted extract (Losada et al. 2005). In a Pds5-depleted extract, cohesin and Wapl were loaded normally onto chromatin during interphase (Fig. 1A, lanes 9,10), but their dissociation from chromatin upon mitotic entry was severely impaired (Fig. 1A, lanes 11,12). Similarly, when Wapl was depleted from an extract, mitotic dissociation of cohesin and Pds5B was heavily retarded (Fig. 1A, lanes 13–16).
Figure 1.
Wapl and Pds5 are required for sister chromatid resolution in Xenopus egg extracts. (A) Sperm chromatin was incubated for 120 min with interphase egg extracts that had been depleted with control IgG (Δmock; lanes 1-4), a mixture of anti-Smc1 and anti-Smc3 (Δcohesin; lanes 5-8), a mixture of anti-Pds5A and anti-Pds5B (ΔPds5; lanes 9-12), or anti-Wapl (ΔWapl; lanes 13-16). Cyclin B was added to trigger entry into mitosis, and the mixtures were incubated for another 100 min. At the indicated time points, chromatin-bound proteins were isolated and analyzed by immunoblotting with the antibodies indicated. The lowest part of the gel was stained with Coomassie brilliant blue to determine the level of core histones as a control for chromatin recovery. The efficiency of depletion in each extract is shown in Supplemental Figure S2. (B) Metaphase chromosomes were assembled in mock-depleted (Δmock; panels a–d) or Wapl-depleted (ΔWapl; panels e–h) extracts, fixed, and double-stained with anti-SA1 (green, panels a,e; grayscale images, panels c,g) and anti-XCAP-G (magenta, panels a,e; grayscale images, panels d,h). The insets show close-ups of selected chromosomal regions (indicated by the rectangles). (Panels b,f) Bulk chromosomal DNA was counterstained with DAPI (grayscale images). Bars, 5 μm. (C) Metaphase chromosomes were assembled in mock-depleted (Δmock; panel a), Wapl-depleted (ΔWapl; panel b), or Pds5-depleted (ΔPds5; panel c) extracts that had been supplemented with biotin-dATP. The chromosomes were fixed and stained with anti-topo IIα (green). Bulk and replicated DNA were visualized with DAPI (blue) and FITC-conjugated avidin (red), respectively. Bar, 5 μm.
The same, yet more visually arresting, results were obtained by immunofluorescence analysis in which metaphase chromosomes assembled in each extract were stained with an antibody against a cohesin subunit (Fig. 1B). In chromosomes assembled in a mock-depleted extract, only a small amount of cohesin was detectable between sister chromatid axes that were visualized clearly with an antibody against a condensin I subunit (Fig. 1B, panels a–d). In chromosomes assembled in a Wapl-depleted extract, however, a large amount of cohesin remained, forming a highly characteristic, single axis-like structure on each chromosome (Fig. 1B, panel g). Sister chromatids were not discernible in these chromosomes as judged by DAPI stain (Fig. 1B, panel f), and the sister axes positive for condensin I failed to be resolved, apparently being twisted around each other to form a single axis-like structure (Fig. 1B, panel h). The distribution patterns of cohesin and condensin I in these chromosomes were similar to each other, yet they did not overlap completely (Fig. 1B, panel e). Moreover, a similar resolution defect was observed in both Wapl-depleted and Pds5-depleted extracts under a different experimental setup, in which duplication of sister DNA molecules was confirmed by incorporation of biotin-dATP and sister axes were visualized with an antibody against topoisomerase IIα (topo IIα) (Fig. 1C). These results indicate that Wapl and Pds5 facilitate release of bulk cohesin from chromatin upon mitotic entry and thereby promote sister chromatid resolution in Xenopus egg extracts.
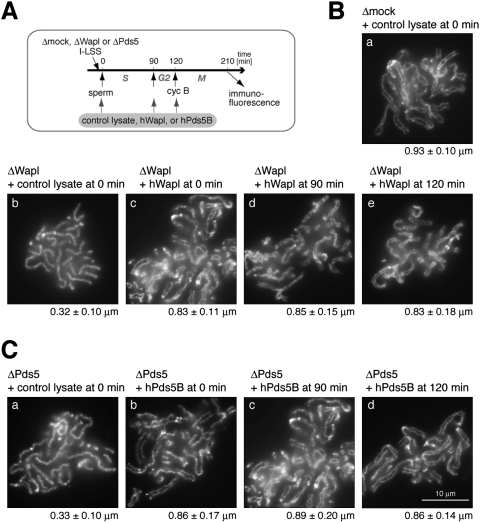
Recombinant human Wap1 (hWapl) and human Pds5B (hPds5B) can promote sister chromatid resolution in a short time window upon mitotic entry
To confirm that the resolution defects observed in Wapl-depleted extracts were due to specific depletion of Wapl rather than nonspecific depletion of other factors, we set up rescue experiments by using a recombinant hWapl protein produced in reticulocyte lysates. The protocol of the experiments is summarized in Figure 2A. We first confirmed that addition of a control reticulocyte lysate at 0 min (i.e., immediately before sperm chromatin was added) affected neither the morphology of metaphase chromosomes assembled in a mock-depleted extract (Fig. 2B, panel a) nor that of poorly resolved chromosomes observed in a Wapl-depleted extract (Fig. 2B, panel b). However, when a reticulocyte lysate containing hWapl was added into the Wapl-depleted extract at the same timing, the resolution defect was efficiently rescued (Fig. 2B, panel c). We then asked whether a persistent interaction between Wapl and cohesin during interphase is essential for promoting sister chromatid resolution in the subsequent mitosis. To this end, addition of hWapl into the Wapl-depleted extracts was delayed until either DNA replication was mostly completed or mitotic inducer cyclin B was added (at 90 min or 120 min, respectively) (see Fig. 2A). To our surprise, successful rescue of the resolution defects was observed in both cases (Fig. 2B, panels d,e).
Figure 2.
Wapl and Pds5 promote sister chromatid resolution in a short time window upon mitotic entry. (A) Sperm chromatin was added to Xenopus egg interphase extracts that had been immunodepleted with control IgG (Δmock), anti-Wapl (ΔWapl), or anti-Pds5 (ΔPds5). Cyclin B (cyc B) was added at 120 min to drive the cell cycle state into mitosis, and chromosomes were fixed at 210 min for immunofluorescence. A control reticulocyte lysate or a lysate containing hWapl (or hPds5B) was added to the extracts at different time points. (B) Metaphase chromosomes were assembled in mock-depleted (panel a) or Wapl-depleted (panels b–e) extracts that had been supplemented with reticulocyte lysates containing no hWapl (panels a,b) or hWapl (panels c–e) at the indicated time points. The chromosomes were stained with anti-topo IIα as described in Figure 1C. Measurements of the distance between sister chromatid axes were provided below each panel. In all chromosomes shown here, duplication of sister DNA molecules was confirmed as judged by dATP incorporation (data not shown). (C) Metaphase chromosomes were assembled in Pds5-depleted extracts that had been supplemented with reticulocyte lysates containing no Pds5 (panel a) or hPds5B (panels b–d) at the indicated time points. The chromosomes were stained and analyzed as above.
We then performed the same set of experiments using the combination of recombinant hPds5B and Xenopus egg extracts depleted of endogenous Pds5. Again, the resolution defects in the Pds5-depleted extracts were efficiently rescued when hPds5B was added at 0 min (Fig. 2C, cf. panels a and b), or when the addition was delayed until 90 min or 120 min (Fig. 2C, panels c,d). Our results suggest that Wapl and Pds5 are able to promote sister chromatid resolution in a very short time window of mitosis, most likely through a transient interaction with cohesin subunits.
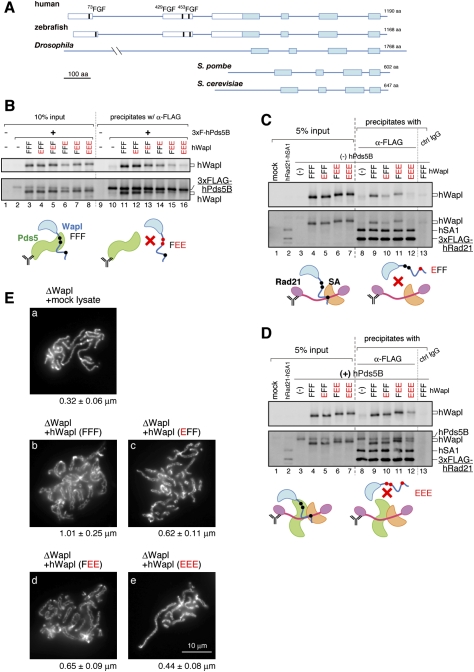
The FGF motifs conserved in the N-terminal half of vertebrate Wapl play critical roles in promoting sister chromatid resolution
The amino acid sequence of hWapl is composed of two parts. The C-terminal half is widely conserved among the Wapl family members and is predicted to be predominantly α-helical. In contrast, the N-terminal half is unique to the vertebrate members of this family and contains many putative phosphorylation sites, implicating its regulatory functions (Fig. 3A; Supplemental Fig. S3 for sequence alignments). An initial set of experiments to molecularly dissect hWapl is shown in Supplemental Figure S4. In short, subunit–subunit interaction assays demonstrated that the N-terminal half (amino acids 1–500) of hWapl was responsible primarily for interacting with Pds5 and a pair of SA1 and Rad21 (the non-SMC subunits of cohesin). Neither the N-terminal nor the C-terminal half (amino acids 501–1190) of hWapl was sufficient to support chromatin loading or to rescue the resolution defect observed in the extract depleted of endogenous Wapl.
Figure 3.
The FGF motifs in Wapl play crucial roles in sister chromatid resolution by coordinating its interaction with Pds5 and cohesin. (A) Schematic representation of the primary structure of Wapl orthologs. The filled boxes indicate regions highly conserved from yeasts to humans, whereas the open boxes indicate sequences unique to vertebrate members. Three FGF motifs (thick vertical lines) are found in all of the vertebrate orthologs investigated (see Supplemental Fig. S3). (B) A panel of mutant hWapl was translated in vitro simultaneously with 3xFlag-hPds5B, and then precipitated with anti-Flag beads. The precipitates (lanes 9–16) along with input fractions (lanes 1–8) were analyzed by immunoblotting. Representative results are depicted in the cartoons. (C) A mixture of in vitro translated 3xFlag-hRad21 and hSA1 was precipitated with anti-Flag beads. The beads were then subjected to another incubation with lysates, producing a panel of mutant hWapl. The precipitates (lanes 8–13) and input fractions (lanes 1–7) were analyzed by immunoblotting. (D) The same set of experiments as C was performed, except that hPds5 was coproduced in lysates used for the second incubation. From B–D, the membranes were probed first with anti-Wapl (top) and subsequently with a mixture of the antibodies indicated (bottom). (E) Metaphase chromosomes were assembled in a Wapl-depleted extract that had been supplemented with the hWapl mutants indicated. The chromosomes were analyzed as described in Figure 2.
We then closely inspected the amino acid sequences of vertebrate Wapl and found that a triplet sequence of FGF is highly conserved at three positions in their N-terminal halves (Fig. 3A; Supplemental Fig. S3). The motif (hereafter referred to as the FGF motif) was similar, if not identical, to the FxF/Y motif found in the double-strand repair protein Nbs1, or the FxFG and GLFG motifs found in many nuclear pore complex components (nucleoporins). It had also been shown that Nbs1 and some of the nucleoporins use these unique motifs to interact with the HEAT repeat proteins ATM and importin β, respectively (Bayliss et al. 2000; You et al. 2005). We were intrigued by these previous reports because multiple HEAT repeats are found in Pds5 (Neuwald and Hirano 2000; Panizza et al. 2000) and possibly in the SA1/SA2/Scc3 family of proteins as well (Nasmyth and Haering 2005). To test whether the FGF motifs present in Wapl play a direct role in interacting with Pds5 and/or SA1, we took a site-directed mutagenesis approach. For each FGF motif, both phenylalanine residues were replaced simultaneously with glutamic acids (i.e., being converted to EGE) according to the strategy used by You et al. (2005), and the resulting three sets of mutations were put together in different combinations. For simplicity, wild-type hWapl (73FGF–429FGF–453FGF) is referred to as FFF, whereas a protein mutated in the first FGF motif (73EGE–429FGF–453FGF) is called EFF. Likewise, a protein mutated in both the second and third FGF-motifs (73FGF–429EGE–453EGE) is referred to as FEE.
We first tested which of the FGF motifs, if any, might contribute to Wapl's interaction with Pds5. Wild-type hWapl (FFF) and the mutant protein EFF interacted efficiently with hPds5B (Fig. 3B, lanes 11,12). In contrast, the association with hPds5B was weakened substantially with FEF and FFE and compromised most severely with FEE and EEE (Fig. 3B, lanes 13–16), indicating the importance of the second and third motifs of Wapl (429FGF and 453FGF in hWapl) in its interaction with Pds5B.
We then asked whether any one of the FGF motifs of Wapl also participate in its interaction with the non-SMC subunits (SA1–Rad21) of cohesin. The assay was performed in the absence (Fig. 3C) or presence (Fig. 3D) of hPds5B. Wild-type hWapl (FFF) interacted efficiently with hSA1–hRad21 in the absence (Fig. 3C, lane 9) or presence (Fig. 3D, lane 9) of hPds5B. In the absence of hPds5B, hWapl's ability to interact with hSA1–hRad21 was compromised with EFF, less severely with FEE, and yet was completely abolished with EEE (Fig. 3C, lanes 10–12). Inclusion of hPds5B in the mixtures partially restored the weakened interactions observed with EFF and FEE, but failed to fully restore the interaction between EEE and hSA1–hRad21 (Fig. 3D, lanes 10–12). Thus, although the mode of interactions among hWapl, hPds5B, and hSA1–hRad21 is most likely synergistic, our results suggest that the first FGF motif of Wapl (73FGF in hWapl) contributes primarily to its interaction with SA1–Rad21.
Finally and most importantly, the wild-type and mutant forms of hWapl were added back into Wapl-depleted extracts, and their ability to rescue the resolution defect was tested. We found that wild-type hWapl (FFF) was able to efficiently rescue the defect (Fig. 3E, panel b), but the EEE mutant protein failed to do so (Fig. 3E, panel e). The addition of EFF and FEE into the Wapl-depleted extracts produced intermediate phenotypes as expected (Fig. 3E, panels c,d). Taken all together, our results demonstrate that the three FGF motifs present in the N-terminal domain of Wapl contribute to its productive interaction with Pds5 and cohesin, thereby playing a crucial role in promoting sister chromatid resolution.
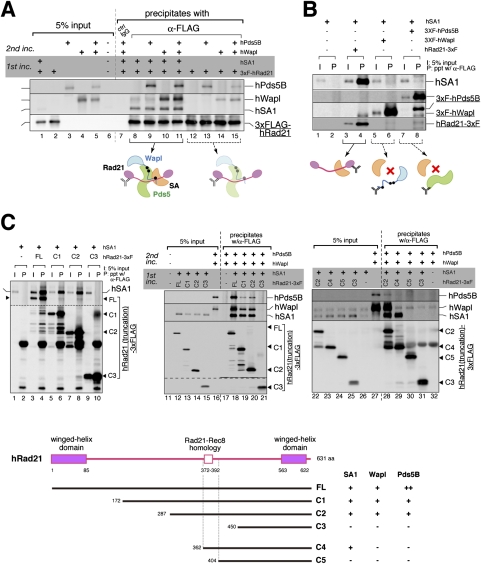
The major target of SA1, Wapl, and Pds5 encompasses a conserved block in the central region of Rad21
We next asked which part of the SA1–Rad21 dimer might make a contact with Wapl and Pds5. We first found that hWapl and hPds5B could each bind to hRad21 in a mutually independent manner (Fig. 4A, lanes 12–15), and that these interactions were greatly stimulated by hSA1 (Fig. 4A, lanes 8–11). Nevertheless, in the absence of hRad21, virtually no interaction was detectable between hWapl and hSA1 (Fig. 4B, lane 6) or between hPds5B and hSA1 (Fig. 4B, lane 8). These results suggest that, although Rad21 has an intrinsic property to interact with Wapl, Pds5, and SA1 individually, an SA1-bound form of Rad21 offers a better context for recruiting Wapl and Pds5.
Figure 4.
The major target of SA1, Wapl, and Pds5 is the central region of Rad21. (A) 3xFlag-hRad21 was translated in vitro individually or simultaneously with hSA1, and then precipitated with anti-Flag beads. The beads were subjected to another incubation with hWapl and hPds5B, which had been translated in various combinations as indicated. After the second incubation, the precipitates (lanes 7–15) and input fractions (lanes 1–6) were analyzed by immunoblotting. (B) hSA1 was translated in vitro alone (lanes 1,2) or simultaneously with 3xFlag-tagged hRad21 (lanes 3,4), 3xFlag-tagged hWapl (lanes 5,6), or 3xFlag-tagged hPds5B (lanes 7,8), and was subjected to immunoprecipitation with anti-Flag. Aliquots of input fractions (lanes 1,3,5,7) and precipitates (lanes 2,4,6,8) were analyzed by immunoblotting. (C) Full-length hRad21-3xFlag (FL) or its truncated versions (C1, C2, C3, C4, and C5) were translated with hSA1 and then precipitated with anti-Flag beads. (Left panel) Five percent of the input fraction (I) and the bead-bound fractions (P) was analyzed by immunoblotting. (Middle and right panels) Alternatively, the beads were subjected to another incubation with hPds5B and hWapl before being analyzed by immunoblotting. A summary of the results is shown in the bottom panel. For sequence alignment of Rad21 and Rec8 in vertebrates, see Supplemental Figure S5. We notice that, compared with hSA1 or hWapl, hPds5B was more sensitive to any truncations of hRad21. Thus, hPds5B may make additional contacts with hRad21 outside of its central region.
To localize the domain(s) required for this composite array of subunit–subunit interactions, a truncation series of hRad21 was constructed and used for similar analyses (Fig. 4C). In an initial set of experiments, we found that a central region of hRad21 (amino acids 287–449) was required for its interaction with hSA1 (Fig. 4C, left panel), and that hWapl and hPds5B bound to the same region in the presence of hSA1 (Fig. 4C, middle panel). In a second set of experiments, the hSA1-binding region was further narrowed down to a small region of hRad21 (amino acids 362–403), whereas binding of hWapl–hPds5 was found to require a larger region (amino acids 287–403) containing the hSA1-binding region (Fig. 4C, right panel). Close inspection of the amino acid sequences revealed that the region required for SA1 binding contained a block of residues (372–392) highly conserved among members of the kleisin α (Rad21–Rec8) family (Fig. 4C, bottom panel; Supplemental Fig. S5). Thus, these analyses uncover a hierarchy of protein assembly that centers around the previously unrecognized sequence conserved in the central region of Rad21.
Sgo1 localizes to the intersister regions in poorly resolved chromosomes
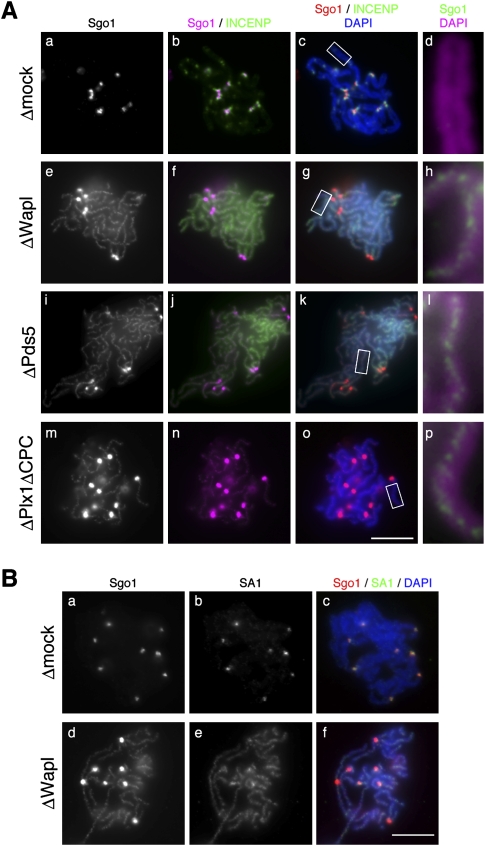
Previous studies showed that Plk1 and aurora B kinase, a component of the chromosome passenger complex (CPC), are required for cohesin release from chromosome arms, thereby contributing to sister chromatid resolution (for review, see Peters et al. 2008). According to a prevailing scenario, Plk1 phosphorylates the SA subunit to facilitate release of cohesin from arms, whereas Sgo1 protects a centromeric population of cohesin against Plk1-mediated phosphorylation by recruiting protein phosphatase 2A (PP2A) to centromeres. To gain more insight into the functional relationship between arm cohesion and centromeric cohesion, we examined the localization of Sgo1 and INCENP (another component of the CPC) in metaphase chromosomes assembled under various conditions. Consistent with previous reports (Boyarchuk et al. 2007; Rivera and Losada 2009), both Sgo1 and INCENP were found to be restricted to centromeric regions of metaphase chromosomes assembled in a mock-depleted extract (Fig. 5A, panels a–d). Unexpectedly, however, in Wapl-depleted or Pds5-depleted extracts, we observed discrete axial distribution of Sgo1 along the entire length of poorly resolved chromatids in addition to its enhanced centromeric accumulation (Fig. 5A, panels e–l). The axial distribution of Sgo1 was reminiscent of that of cohesin observed in Wapl-depleted chromosomes (Fig. 5B, panels d–f), most likely corresponding to the “intersister” regions where sister chromatids are glued together. On the other hand, INCENP failed to be enriched at centromeres in the absence of Wapl or Pds5 and, rather, displayed diffuse signals along the length of chromosomes (Fig. 5A, panels f,g,j,k), suggesting that subchromosomal localization of Sgo1 and the CPC is intimately related with each other (see the Discussion). Similarly, axial distribution of Sgo1 was observed in resolution-deficient chromosomes assembled in an extract that had been depleted of both Plx1 (the Xenopus ortholog of Plk1) and the CPC (Fig. 5A, panels m–p). Thus, these results implicate that the target of action of Sgo1 is not restricted to centromeres, and raise the possibility that Sgo1 plays a hitherto underappreciated role in stabilizing cohesion along chromosome arms.
Figure 5.
Sgo1 localizes to the intersister regions in poorly resolved chromosomes. (A) Metaphase chromosomes were assembled in the extracts that had been depleted with control IgG (Δmock; panels a–d), anti-Wapl (ΔWapl; panels e–h), anti-Pds5 (ΔPds5; panels i–l), or a mixture of anti-Plx1, anti-aurora B, and anti-INCENP (ΔPlx1ΔCPC; panels m–p). The chromosomes were fixed and double-stained with anti-Sgo1 (grayscale images, panels a,e,i,m; magenta, panels b,f,j,n; red, panels c,g,k,o; green, panels d,h,l,p) and anti-INCENP (green, panels b,c,f,g,j,k,n,o). DNA was counterstained with DAPI (blue, panels c,g,k,o; magenta, panels d,h,l,p). Close-ups of selected regions of chromosomal arms (indicated by the rectangles in panels c,g,k,o, respectively) are shown in panels d, h, l, and p. (B) Chromosomes that had been assembled in mock-depleted and Wapl-depleted extracts were double-stained with anti-Sgo1 (grayscale, panels a,d; red, panels c,f) and anti-SA1 (grayscale, panels b,e; green, panels c,f). (Panels c,f) DNA was counterstained with DAPI (blue). Bars, 10 μm. The efficiency of depletion in each extract is shown in Supplemental Figure S2.
Sgo1 plays a role in stabilizing arm cohesion and functionally interacts with Plk1 and Wapl in distinct manners
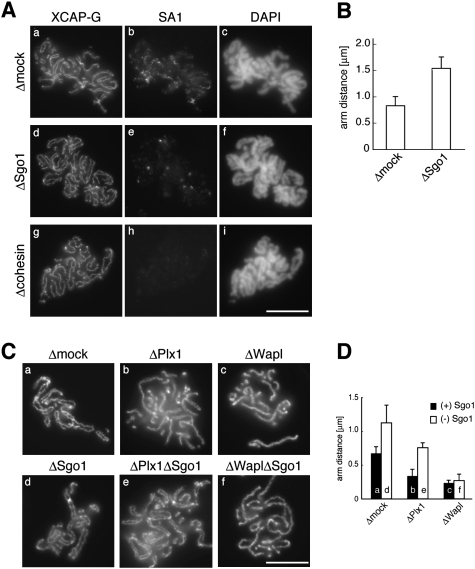
To test directly whether Sgo1 plays a role in stabilizing arm cohesion, metaphase chromosomes were assembled in extracts depleted of Sgo1 or cohesin, and stained with antibodies against XCAP-G (a condensin I subunit) and SA1. We found that arm cohesion was loosened substantially in Sgo1-depleted chromosomes compared with control chromosomes (Fig. 6A, panels a,d). Cohesin depletion caused much more severe defects in cohesion, frequently making it difficult to discern the pair of sister chromatids within individual chromosomes (Fig. 6A, panel g). These observations were confirmed by quantitative measurements of the distance between sister chromatids assembled in mock-depleted and Sgo1-depleted chromosomes (Fig. 6B). Consistently, the Sgo1-depleted chromosomes had a much smaller amount of SA1 left between chromatid arms than the control chromosomes (Fig. 6A, panels b,e). As expected, virtually no SA1 was detectable in cohesin-depleted chromosomes (Fig. 6A, panel h). These results substantiate the idea that Sgo1 may act as a negative regulator of cohesin release and the resolution of sister chromatid arms.
Figure 6.
Sgo1 plays a role in stabilizing arm cohesion. (A) Metaphase chromosomes were assembled in a control extract (Δmock; panels a–c), Sgo1-depleted extract (ΔSgo1; panels d–f), or cohesin-depleted extract (Δcohesin; panels g–i), and double-stained with anti-XCAP-G (panels a,d,g) and anti-SA1 (panels b,e,h). (Panels c,f,i) The bulk DNA was counterstained with DAPI. (B) The distance between sister chromatid axes was measured for >50 chromosomes from each condition and plotted. (C) Replicated chromosomes were assembled in a control extract (panel a) or extracts depleted of various factors (individually or in combinations) as indicated (panels b–f), and stained with anti-topo IIα. (D) The distance between sister chromatid axes was measured and plotted as in B. Bars, 10 μm. The efficiency of depletion in each extract is shown in Supplemental Figure S2.
Finally, we wished to test how Sgo1 might functionally interact with positive regulators of sister chromatid resolution such as Plk1 and Wapl. Depletion of Plx1 from the extracts caused resolution defects that were slightly milder than those observed in Wapl-depleted extracts (Fig. 6C, panels b,c; see also Losada et al. 2002). Again, depletion of Sgo1 resulted in loosely paired sister chromatids (Fig. 6C, panel d). We found that simultaneous depletion of the two factors produced an intermediate phenotype in which the degree of resolution was comparable with that found in a mock-depleted extract (Fig. 6C, cf. panels a and e). In contrast, however, additional depletion of Sgo1 failed to relieve the defects caused by Wapl depletion (Fig. 6C, panel f). Measurements of the distance between sister chromatids confirmed the morphological examination (Fig. 6D). Our data suggest that Wapl and Plk1 promote sister chromatid resolution through distinct mechanisms, and that one function of Plk1 may be to antagonize the action of Sgo1.
Discussion
Sister chromatid resolution, which occurs from prophase through metaphase, is assumed to be a preparatory step that allows rapid and synchronous separation of sister chromatids in subsequent anaphase. In the present study, we used a cell-free system from Xenopus eggs to demonstrate that Wapl and Pds5 cooperate to promote sister chromatid resolution. By acting on the central region of the kleisin subunit of cohesin, Wapl and Pds5 may directly modulate the conformation of the cohesin complex and convert it into a form competent for dissociation from chromatin. This process is exquisitely coordinated by three FGF motifs present in the N-terminal domain of Wapl. We also present evidence that Sgo1 plays a hitherto underappreciated role in stabilizing cohesin along chromosome arms, which is antagonized by distinct actions of Plk1 and aurora B.
The FGF motifs in Wapl coordinate its physical and functional interactions with Pds5 and cohesin
The present results show that there exists a specific and composite mode of protein–protein interactions among the two regulatory subunits of cohesin (SA1 and Rad21) and its two regulatory factors (Wapl and Pds5). Among the three FGF motifs conserved in the N-terminal domain of human Wapl, the first one contributes primarily to its interaction with SA1–Rad21, whereas the second and third ones contribute to its interaction with Pds5. Our results demonstrate clearly that the FGF motif-mediated interactions are functionally important because combinations of mutations in these motifs progressively reduce its ability to promote sister chromatid resolution in Xenopus egg extracts.
How might the FGF motifs contribute to this process at a mechanistic level? Interestingly, previous structural and functional studies have revealed some examples of specific interactions between HEAT repeats and short amino acid motifs related to FGF (Bayliss et al. 2000; You et al. 2005). For instance, an FxFG repeat from Nsp1 (a component of the nuclear pore complex) binds to a convex face between two successive HEAT repeats in importin β. Binding of Ran-GTP to the other surface of importin β induces its conformational change, which in turn weakens the FxFG–HEAT interaction, thereby releasing importin β from Nsp1 (Bayliss et al. 2000). Similarly, an FxF/Y motif from Nbs1 (a subunit of the Mre11–Rad50–Nbs1 complex) interacts with HEAT repeats in the checkpoint kinase ATM and stimulates its activity (You et al. 2005). By analogy, it is tempting to speculate that the FGF motifs of Wapl may bind to specific parts of HEAT repeats in Pds5 (and possibly SA1) and trigger a series of conformational changes of the cohesin complex, eventually leading to its dissociation from chromatin. Such interactions among Wapl, Pds5, and cohesin would occur most efficiently on chromatin (Fig. 1A) rather than in soluble extracts (Supplemental Fig. S1). Yet, a transient action of Wapl and Pds5 upon mitotic entry appears sufficient to convert cohesin into a dissociation-proficient form (Fig. 2), further emphasizing the highly dynamic and elaborate nature of this molecular process.
A model for the action of Wapl and Pds5 on cohesin release
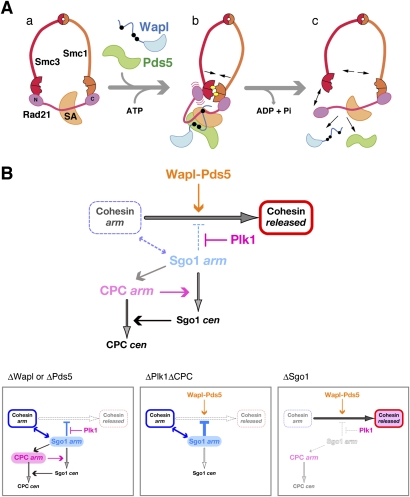
How might the postulated action of Wapl and Pds5 render cohesin competent for dissociation from chromatin, and how might this process be coupled with the ATP-binding and hydrolysis cycle of cohesin? The embrace model for the action of cohesin predicts that its dissociation from chromatin would require transient opening of the cohesin ring. Our results suggest that SA1 binds to a conserved block within the central region of Rad21, which in turn creates the primary target of actions of Wapl and Pds5. One possible scenario may be that Wapl and Pds5, in collaboration with SA1, bend the Rad21 molecule around its central region so that its N-terminal and C-terminal winged helix domains are brought together into close proximity (Fig. 7A, panels a, b). This configuration naturally increases the probability of head–head engagement of SMC dimers when their heads are bound to ATP. Alternatively or additionally, Wapl or Pds5 may have an activity that loads ATP on empty SMC heads or exchanges prebound ADP with ATP. Such ATP-sandwiched head–head engagement could help create a situation in which one of the linkages between Rad21 and the SMC dimer is destabilized transiently (Fig. 7A, panel b), leading to subsequent opening of the SMC–kleisin ring upon hydrolysis of the bound ATP (Fig. 7A, panel c). While this scenario is admittedly speculative, it is supported by a recent structural study of a bacterial SMC–kleisin protein complex (Woo et al. 2009). Whatever the actual mechanisms might be, it is most likely that there exists an intricate cross-talk between the ATPase cycle of the SMC subunits and the conformational change of the non-SMC regulatory subunits, both of which may be the targets of action of cohesin regulators such as Wapl and Pds5.
Figure 7.
Opposing actions of Wapl–Pds5 and Sgo1 on releasing cohesin from chromosome arms. (A) A speculative model for the action of Wapl and Pds5 on cohesin release. (Panels a,b) Wapl and Pds5 interact with the central region of Rad21 to induce its conformational changes upon entry into mitosis, which may in turn facilitate ATP-mediated engagement of the head domains of Smc1 and Smc3. (Panel c) The engaged configuration could transiently destabilize either one of the two SMC–kleisin interfaces, and subsequent ATP hydrolysis allows opening of the cohesin ring, thereby allowing its dissociation from chromatin. ATP molecules are shown by the yellow ovals. (B, top panel) Schematic presentation of a regulatory network for cohesin release from chromosome arms during early mitosis. The two populations of Sgo1 (and the CPC) along chromosome arms and at centromeres are indicated by “arm” and “cen,” respectively. (Bottom panels) This model nicely predicts the behavior of cohesin and its regulators observed under different conditions. See the text for details.
Evolutionary aspects of Wapl and Pds5 in cohesin regulation
Although the primary structure of Pds5 is conserved from yeast to humans, the degree of sequence conservation among members of the Wapl family of proteins is very low (see Peters et al. 2008). In budding and fission yeast, the Wapl orthologs are nonessential proteins that play rather cryptic roles in regulating cohesin dynamics during interphase (Ben-Shahar et al. 2008; Bernard et al. 2008). Notably, they lack N-terminal extensions (Fig. 3A), and the process corresponding to sister chromatid resolution is not prominent or absent in these organisms. We speculate that, during evolution, acquisition of N-terminal extensions might have conferred on Wapl an ability to interact with cohesin and Pds5 in a more efficient and productive manner, thereby modifying or fine-tuning its original function. In this way, Wapl might have gained an ability to rapidly release cohesin during early mitosis, thereby contributing to the establishment of the so-called prophase pathway in metazoan species.
Our previous study (Losada et al. 2005) reported that depletion of Pds5 caused only a subtle set of defects in cohesin regulation in Xenopus egg extracts. In the present study, however, careful re-evaluation of depletion conditions (Supplemental Fig. S2) followed by successful rescue assays (Fig. 2) provides a compelling set of evidence that Pds5 facilitates cohesin release and promotes sister chromatid resolution during mitosis in Xenopus egg extracts. At first glance, this conclusion is inconsistent with the so far frequently discussed role for Pds5 in the maintenance of sister chromatid cohesion during interphase in yeast and humans (for review, see Losada and Hirano 2005; Peters et al. 2008). We suggest that the apparent discrepancy may simply reflect different faces of Pds5, whose actions potentially regulate cohesin dynamics either positively or negatively depending on the context (before or after cohesin's loading onto chromatin) and/or on different stages of the cell cycle. This argument may also be compatible with the recent findings that, in budding yeast, Wapl and Pds5 counteract the establishment of cohesion (Rowland et al. 2009; Sutani et al. 2009). It is clear, however, that future work is required to understand to what extent the interphase function of Wapl–Pds5 reported in yeast might mechanistically be related to its mitotic function observed in vertebrates.
Regulatory network of sister chromatid resolution
Although the role of Sgo1 in stabilizing centromeric cohesion during mitosis and meiosis has been characterized extensively, its potential contribution to stabilizing arm cohesion has largely been unrecognized. One of the unexpected findings in the present study is that, whenever cohesin release from chromatid arms is severely impaired, a substantial amount of Sgo1 is concentrated on a discrete axis-like structure along the entire length of chromosomes, presumably corresponding to the intersister regions where cohesin is also enriched under these conditions (Fig. 5). This observation prompted us to examine the morphology of Sgo1-depleted chromosomes in detail, and it was found that Sgo1 depletion caused enhanced release of cohesin from chromatid arms and loosened arm cohesion (Fig. 6A). Taken together, our results provide a convincing set of evidence that Sgo1 in fact plays a role in stabilizing arm cohesion, at least in the Xenopus egg cell-free extracts. Curiously enough, defects in centromeric cohesion, if any, are very subtle in Sgo1-depleted egg extracts (Rivera and Losada 2009; this study). It is well known that metaphase chromosomes in early embryonic cells tend to be longer and thinner than those in somatic chromosomes (Belmont et al. 1987), and chromosomes assembled in Xenopus egg extracts recapitulate such embryonic properties (Ono et al. 2003). Accordingly, we speculate that the relative contribution of arm and centromeric cohesion may be different between the two types of chromosomes, and that somatic ones rely more on centromeric cohesion than on arm cohesion. If this idea were right, measuring the minor contribution of arm cohesion would be rather difficult in somatic cells, providing a reasonable explanation for why the potential contribution of Sgo1 to arm cohesion has not been fully appreciated so far. Nonetheless, a recent study has provided evidence in favor of such a cryptic role of Sgo1 in stabilizing arm cohesion in HeLa cells (Nakajima et al. 2007).
Our model for the regulatory network of sister chromatid resolution is shown in Figure 7B (top panel). It should be emphasized that loss of Sgo1 from chromatid arms is not sufficient to promote cohesin release and arm resolution, and that Wapl–Pds5 is absolutely essential for these processes regardless of the presence or absence of Sgo1. In contrast, Sgo1 depletion partially negates the requirement for Plk1 in the resolution process, implicating that one function of Plk1 is to antagonize the action of Sgo1 along arms. While how Sgo1 might stabilize arm cohesion remains to be elucidated, PP2A was not found concentrated in the intersister regions within resolution-deficient chromosomes (Supplemental Fig. S6). Thus, the action of Sgo1 along arms, unlike its proposed action at centromeres, could be independent of PP2A. On the other hand, subchromosomal localization of Sgo1 and the CPC is heavily dependent on each other (Supplemental Fig. S7; see also Boyarchuk et al. 2007; Rivera and Losada 2009). The CPC is therefore most likely to contribute to the resolution process as part of the regulatory loop that localizes Sgo1 properly to centromeres. Our model nicely illustrates how these positive and negative factors constitute the regulatory network of sister chromatid resolution, and helps explain the consequences that are observed when each of them is removed individually or in combination from the system (Fig. 7B, bottom panels). It will be an exciting challenge in the future to understand how this elaborate network actually works and how aberrations of its components potentially lead to chromosome instability.
Materials and methods
Antibodies
We previously reported antibodies raised against three different antigens for human Wapl (Gandhi et al. 2006). It was found that all of them specifically recognized Xenopus Wapl present in egg extracts (Supplemental Fig. S1), and the antibody against a C-terminal recombinant fragment of human Wapl was used throughout the present study unless otherwise stated. Rabbit polyclonal antisera were raised against a synthetic peptide corresponding to the C-terminal amino acid sequence of Xenopus Sgo1 (RQSLSRYNEVFVGCRR) and a recombinant full-length Cdc45 expressed in insect cells. Immunization and affinity purification were carried out as described previously (Hirano et al. 1997). Other antibodies used in this study were as follows: anti-Smc1, anti-Smc3, and anti-Rad21 (Losada et al. 1998); anti-SA1 (Losada et al. 2000); anti-Pds5A and anti-Pds5B (Losada et al. 2005); anti-XCAP-G and XCAP-H (Hirano et al. 1997); anti-Plx1, anti-aurora B, and anti-INCENP (Losada et al. 2002); and anti-topo II α (Hirano and Mitchison 1993).
Preparation of Xenopus egg extracts
Interphase low-speed supernatant (LSS) of Xenopus eggs and demembranated sperm chromatin were prepared according to Murray (1991) with some modifications. Briefly, dejellied eggs were activated by treatment with the Ca2+ ionophore A23187 (Sigma) at 50 nM in MMR (5 mM Na-HEPES at pH 7.8, 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 0.1 mM Na-EDTA) for 3 min. The eggs were washed with MMR, incubated with KMH (20 mM K-HEPES at pH 7.7, 100 mM KCl, 2.5 mM MgCl2) containing 100 μg/mL cyclohexamide and 50 μg/mL cytochalasin D at 22°C until 20 min after activation, and finally chilled for 5 min on ice. The cytoplasmic fraction of these eggs was prepared by centrifugation as described previously (Losada et al. 1998). The interphase LSS was supplemented with recombinant sea urchin cyclin B (100 nM) to convert the cell cycle into mitosis. All egg extracts were supplemented with ATP and creatine phosphate at final concentrations of 1 mM and 10 mM, respectively.
Immunodepletion and immunoprecipitation
Affinity-purified antibodies were mixed with rProtein A-Sepharose beads (GE Healthcare) and incubated for 1 h at room temperature. The antibody-coupled beads were washed and equilibrated with KMH. For immunodepletion, 100 μL of egg extract were incubated with 25 μL of antibody beads for 30 min on ice with occasional mixing. After two successive rounds of incubation, the supernatants were recovered by brief spin and used as depleted extracts. The amounts of each antibody incubated with 100 μL of beads were as follows: for Wapl depletion, 75 μg of anti-Wapl; for Pds5 depletion, a mixture of 25 μg of anti-Pds5A and 25 μg of anti-Pds5B; for cohesin depletion, a mixture of 15 μg of anti-Smc1 and 15 μg of anti-Smc3; for Plx1 depletion, 50 μg of anti-Plx1; for CPC depletion, a mixture of 65 μg of anti-aurora B and 65 μg of anti-INCENP; for Sgo1 depletion, 60 μg of anti-Sgo1. When multiple components were depleted simultaneously from egg extracts, the same volume of beads was coupled with a mixture of corresponding antibodies. For immunoprecipitation, 10 μL of egg extract were incubated with 5 μL of beads (coupled with 2.5 μg of IgG) for 30 min at 4°C with occasional mixing. The beads were washed with 500 μL of KMH containing 0.1% Triton X-100 five times and subjected to SDS-PAGE. For mock depletion and precipitation, the same amount of control rabbit IgG (Sigma) was used.
Biochemical and morphological analyses of chromatin and chromosomes assembled in egg extracts
Demembranated sperm chromatin was mixed with interphase egg extracts at a final concentration of 4000 nuclei per microliter for biochemical assays or 1000 nuclei per microliter for morphological assays. After incubation for 100 or 120 min at 22°C, the mixture was supplemented with recombinant sea urchin cyclin B (100 nM) to convert the cell cycle state into mitosis, and incubated for another 100 or 120 min. For biochemical analysis, aliquots were taken at time intervals and diluted with 10 vol of ice-cold KMH containing 0.2% Triton X-100. The diluted fractions were overlaid on a 30% sucrose cushion made in KMH, and spun at 1500g for 10 min at 4°C. After removing soluble fractions, chromatin fractions recovered in the pellets were washed by resuspending with KMH and spun again at 4500g, before they were processed for SDS-PAGE. For morphological analysis, metaphase chromosomes were assembled as described above. In the experiment shown in Figure 1C, the reactions were supplemented with 4 μM biotin-14-dATP (Invitrogen) to check the efficiency of DNA replication. The assembly mixtures were fixed with 10 vol of 2% formaldehyde in KMH containing 0.1% Triton X-100 for 15 min at room temperature, and centrifuged at 7000g for 10 min onto coverslips through a 30% glycerol cushion made in KMH. Immunofluorescence was performed as described previously (Losada et al. 1998, 2000, 2002; Ono et al. 2003). The distance between sister chromatid axes was analyzed by using ImageJ software (http://rsb.info.nih.gov/ij).
In vitro transcription and translation
DNA fragments encoding different regions of hWapl, hPds5B, hRad21, and hSA1 were amplified by PCR and inserted into a pTnT vector (Promega). Point mutations were introduced with the QuikChange XL site-directed mutagenesis kit (Stratagene). 3xFlag-tagged constructs were made by insertion of annealed oligonucleotides encoding the corresponding sequence. In vitro transcription and translation reactions were carried out with the TNT Quick Coupled Transcription/Translation System (Promega) according to the manufacturer's instruction. The details of plasmid construction and the primer sequences used are available in the Supplemental Material. A typical reaction mixture (25 μL) containing 0.5 μg of plasmid DNA for each construct was supplemented with methionine (25 μM) and incubated for 90 min at 30°C. A control lysate was prepared using the same protocol with an empty pTnT vector. For immunoprecipitation analyses, 10 μL of the reaction mixture were incubated with 5 μL of rProtein A-Sepharose that had been coupled with 5 μg of control mouse IgG or anti-Flag M2 mouse monoclonal antibody (Sigma). For complementation assays, depleted egg extract was supplemented with 1/10 vol of reticulocyte lysates.
Acknowledgments
We thank A. Arnaoutov and M. Dasso (NIH) for an aliquot of antiserum against Xenopus Sgo1, H. Takisawa (Osaka University, Japan) for a Cdc45 expression vector, and Kazusa DNA Research Institute (Japan) for human Pds5B cDNA. We are grateful to Tom Hartl and members of the Hirano laboratory for critically reading the manuscript. This work was supported by the RIKEN President's Discretionary Fund and Grant-in-Aid for Specially Promoted Research (20002010) (to T.H.). K.S. is a RIKEN Special Post-doctoral Researcher and was also supported by an Incentive Research Grant. This paper is dedicated to the memory of Michiko Hirano, who made great contributions to the original identification of cohesin and condensin and to the subsequent development of the SMC field.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1844309.
Supplemental material is available at http://www.genesdev.org.
References
- Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R, Littlewood T, Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- Belmont AS, Sedat JW, Agard DA. A three-dimensional approach to mitotic chromosome structure: Evidence for a complex hierarchical organization. J Cell Biol. 1987;105:77–92. doi: 10.1083/jcb.105.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar TR, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, Ekwall K, Uhlmann F, Javerzat JP. Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J. 2008;27:111–121. doi: 10.1038/sj.emboj.7601955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk Y, Salic A, Dasso M, Arnaoutov A. Bub1 is essential for assembly of the functional inner centromere. J Cell Biol. 2007;176:919–928. doi: 10.1083/jcb.200609044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie KW, Kennedy CD, Velasco VM, McGrath TL, Weko J, Patterson RW, Karpen GH. Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics. 2001;157:1623–1637. doi: 10.1093/genetics/157.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Haering CH, Farcas AM, Arumugam P, Metson J, Nasmyth K. The cohesin ring concatenates sister DNA molecules. Nature. 2008;454:297–301. doi: 10.1038/nature07098. [DOI] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Waizenegger IC, Peters JM. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 2001;293:1320–1323. doi: 10.1126/science.1061376. [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Lee JY, Orr-Weaver TL. The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol. 2001;17:753–777. doi: 10.1146/annurev.cellbio.17.1.753. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano T. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes & Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes & Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes & Dev. 2002;16:3004–3016. doi: 10.1101/gad.249202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Murray AW. Cell cycle extracts. Methods Cell Biol. 1991;36:581–605. [PubMed] [Google Scholar]
- Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T. The complete removal of cohesin from chromosome arms depends on separase. J Cell Sci. 2007;120:4188–4196. doi: 10.1242/jcs.011528. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: A simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes & Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- Rivera T, Losada A. Shugoshin regulates cohesion by driving relocalization of PP2A in Xenopus extracts. Chromosoma. 2009;118:223–233. doi: 10.1007/s00412-008-0190-4. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N, Nigg EA, Peters JM. The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell. 2002;9:515–525. doi: 10.1016/s1097-2765(02)00473-2. [DOI] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)–Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Lottspeich F, Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Verni F, Gandhi R, Goldberg ML, Gatti M. Genetic and molecular analysis of wings apart-like (wapl), a gene controlling heterochromatin organization in Drosophila melanogaster. Genetics. 2000;154:1693–1710. doi: 10.1093/genetics/154.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development. 2007;134:3191–3201. doi: 10.1242/dev.005884. [DOI] [PubMed] [Google Scholar]