Abstract
Although the ovalbumin (Ov) gene has served as a model to study tissue-specific, steroid hormone-induced gene expression in vertebrates for decades, the mechanisms responsible for regulating this gene remain elusive. Ov is repressed in non-oviduct tissue and in estrogen-deprived oviduct by a strong repressor site located from -130 to -100 and designated CAR for COUP-TF adjacent repressor. The goal of this study was to identify the CAR binding protein(s). A transcription factor database search revealed that a putative interferon-stimulated response element (ISRE), which binds interferon regulatory factors (IRFs), is located in this region. Gel mobility shift assays demonstrated that the protein(s) binding to the CAR site is recognized by an IRF antibody and that mutations in the ISRE abolish that binding. In hopes of identifying the IRF(s) responsible for the tissue-specific regulation of Ov, mRNA levels for IRFs-4, -8, and -10 were measured in seven tissues from chicks treated with or without estrogen. PCR experiments showed that both IRF-8 and -10 are expressed in all chick tissues tested whereas IRF-4 has a much more limited expression pattern. Transfection experiments with OvCAT (chloramphenicol acetyltransferase) reporter constructs demonstrated that both IRF-4 and IRF-10 are capable of repressing the Ov gene even in the presence of steroid hormones and that nucleotides in the ISRE are required for repression. These experiments indicate that the repressor activity associated with the CAR site is mediated by IRF family members and suggest that IRF members also repress Ov in non-oviduct tissues.
Keywords: oviduct, negative regulation, egg white protein, ISRE
1. Introduction
While many genes are ubiquitously expressed, a number are expressed only in a specific tissue(s) or in response to specific developmental or environmental signals. Expression of the ovalbumin (Ov) gene is among the most restricted known. It is only expressed in the tubular gland cells of the avian oviduct in response to estrogen; Ov is silent in estrogen-deprived oviduct and in all other tissues regardless of estrogen status (Dougherty and Sanders, 2005). The molecular basis for this restricted expression remains unclear. Chromosomal structure and/or nucleosomal rearrangements may play a role as estrogen treatment makes the chromatin in tubular gland cells more accessible to trans-acting factors and nucleases and allows Ov transcription (Bellard et al., 1982 and references therein). Interestingly, upon withdrawal of estrogen the chromatin in oviduct remains accessible to nucleases, yet Ov is not transcribed. Furthermore, estrogen treatment does not make Ov accessible in non-oviduct tissues. This implies that tissue-specific activators, repressors, and/or coregulators exist that allow transcription only in estrogen-stimulated oviduct and repress it in all other contexts. Data suggest that estrogen-stimulated oviduct contains proteins not present in other tissues, although their identities are unknown (Upadhyay et al., 1999; Park et al., 2006). The goal of these experiments was to gain a better understanding of the tissue-specific regulation of Ov.
Two major regulatory units control expression of Ov in primary oviduct cells (Fig. 1). The steroid-dependent regulatory element (SDRE) from -892 to -793 is required for induction by estrogen, androgen, glucocorticoids, and progesterone (Sanders and McKnight, 1988; Dean et al., 1996; Dean et al., 2001). At least four protein complexes, designated Chirp-I through -IV, bind to the SDRE (Fig. 1, panel A), although only Chirp-III has been identified. Chirp-III is the estrogen-inducible transcription factor δEF1/ZEB1 (Chamberlain and Sanders, 1999), which tethers USF to this site (Dillner and Sanders, 2002).
Fig. 1.
Regulatory elements in the Ov promoter. A. Map of the two major regulatory units in Ov, the SDRE (-892 to -793) and the NRE (-308 to -100). Genomic footprinting and/or mutagenesis indicate that protein or protein complexes bind to all of the sites that are numbered. Designated names are listed above the box depicting Ov, and real names are given in the box. The circles represent various transcription factors or factor complexes. Any circles with the same pattern represent the same factors or members of the same transcription factor family. B. The sequences in the regions of the OTE and CAR regulatory sites are shown. Sequences common to both of these are underlined. The ISRE consensus sequence and a putative unknown transcription factor binding site sequence are aligned with the common sites.
The negative regulatory element (NRE, -308 to -88) is the other major regulatory unit in Ov (Fig. 1, panel A). In the absence of steroids and/or when the SDRE is removed, the NRE represses transcription in oviduct (Sanders and McKnight, 1988; Sensenbaugh and Sanders, 1999) and non-oviduct cells (Sanders and McKnight, 1988; Monroe and Sanders, 2000). Although described as a repressive element, sequential 10 bp mutations throughout this region identified two positive sites as well as four negative sites (Sensenbaugh and Sanders, 1999). The NRE is thus bifunctional in that it cooperates with the SDRE to induce Ov expression in the presence of steroids and represses it in estrogen-deprived oviduct tissue and in non-oviduct tissues. The COUP-TF adjacent repressor (CAR) element (-130 to -100) appears to mediate much of the repressive actions exerted through the NRE. Gel mobility shift assays (GMSAs) revealed that protein binding to CAR is not affected by steroid hormone treatment (Sensenbaugh and Sanders, 1999). Interestingly, the CAR repressor site shares two conserved elements with the positive ovalbumin tissue-specific element (OTE) site (-198 to -170), suggesting that these elements may bind the same proteins or related family members (Fig. 1, panel B). One of these conserved elements is a consensus interferon-stimulated response element (ISRE), which binds interferon regulatory factors (IRFs). IRFs are involved in tissue-specific gene regulation in the immune system (Paun and Pitha, 2007; Takaoka et al., 2008). Most IRFs, in conjunction with a partner, are capable of both activating and/or repressing transcription of target genes. The goals of the following studies were to characterize the nucleotides required for protein binding to the CAR site, to determine the relationship between the OTE and CAR sites, and to ascertain whether IRF(s) bind to either of these sites.
2. Materials and methods
2.1. Oviduct tubular gland cell culture and transfection
Sexually immature White Leghorn chicks were treated with estrogen and primary oviduct tubular gland cells isolated as previously described in detail (Sanders and McKnight, 1988). The cells were transfected using calcium phosphate coprecipitation as described (Sanders and McKnight, 1988). DNA concentrations were held constant by the addition of appropriate amounts of pcDNA-3.1 (Invitrogen, Carlsbad, CA USA). Cells were cultured for 24 hrs in F12/DMEM medium (1:1) with insulin (50 ng/ml) in the presence or absence of 17β-estradiol (1 × 10-7 M) and corticosterone (1 × 10-6 M) in 60 mm dishes. CAT assays were performed as previously described (Sanders and McKnight, 1985).
2.2. Nuclear protein extracts and gel mobility shift assays (GMSAs)
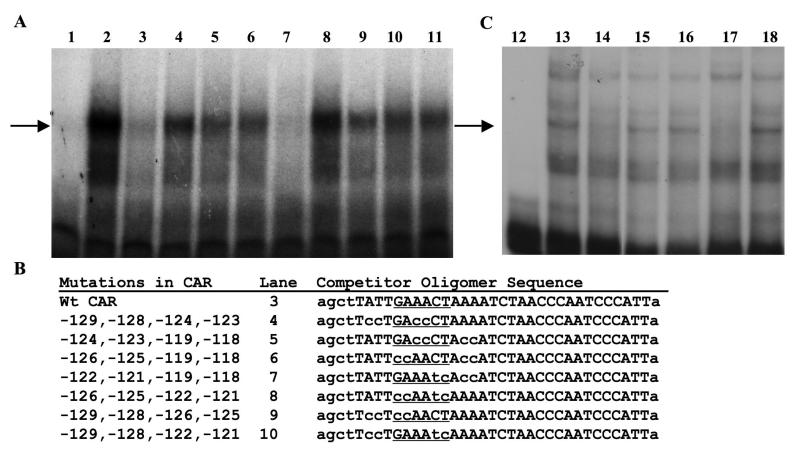
Nuclear protein extracts were prepared essentially as described (Gorski et al., 1986) using 3 g of frozen tissue. Final extracts were assayed for protein concentration, aliquoted into microfuge tubes, and stored at -80° C. All GMSAs were performed as previously described using 5 μg of nuclear protein and 50,000 cpm of Klenow P32-labeled double-stranded DNA oligomers (Nordstrom et al., 1993). For the competition experiments, unlabeled oligomers were added at the same time as the radiolabeled oligomers and before the addition of the nuclear protein. For antibody supershift experiments, nuclear protein extracts were incubated with the antibody at 4° C overnight, with rocking, before the DNA binding experiment was performed. The top strand of the oligomer sequences used for the data in Fig. 3A are shown in Fig. 3B, and the top strand of the oligomers used for Fig. 3C are below. For all oligomers, the non-wild type sequences are shown as small letters.
Fig. 3.
Oligomers with critical nucleotides mutated in the ISRE attenuate competition with the CAR oligomer. A. GMSAs were performed using the conditions described in Section 2.2 and . Lanes 1 to 11 contain the labeled CAR oligomer (-130 to -100). Lanes 2 to 11 contain 5 μg of 3 day estrogen-withdrawn oviduct nuclear protein. Lane 3 contains 25X molar unlabeled wt CAR oligomer. Lanes 4 to 10 contain 25X molar unlabeled mutated CAR oligomers that are described in Panel B. Lane 11 contains 25X molar unlabeled non-specific DNA. The arrow designates the shifted DNA-protein complex that is specific. B. The CAR competitor oligomer sequences (mutated nucleotides are shown in lower case) in the order used in the experiment. The positions of the mutated bases relative to the transcription start site are given. Comparable results were found with 50X molar excess competitors. C. GMSAs were performed using the conditions described in Section 2.2. Lanes 12 to 18 contain the labeled CAR oligomer. Lanes 13 to 18 contain 5 μg of nuclear extract protein from sexually immature chickens that had never been treated with estrogen. Lane 14 also contains 25X molar unlabeled wt CAR oligomer. Lane 15 contains 25X molar unlabeled mutated CAR oligomer. Lane 16 contains 25X molar unlabeled non-specific DNA. Lane 17 contains 25X molar unlabeled wt consensus IRF oligomer. Lane 18 contains 25X molar unlabeled mutated IRF consensus oligomer. The arrow designates the specific shifted DNA-protein complex. The GMSAs in A and C differ somewhat in their binding pattern because the hormonal treatment of the chicks differed. Also, the pattern in Fig.3A differs from that of Fig. 2 because the protein extracts used in 3A were not fresh. The upper band seen in Fig. 2 is labile.
Wild type CAR oligomer: 5′-agctTATTGAAACTAAAATCTACCAATCCCATTa-3′
Mutated CAR oligomer: 5′-agctTATTGgggCTAgggTCTACCAATCCCATTa-3′
Wild Type Consensus ISRE oligomer: 5′-agcttGGAAGCGAAAATGAAATTGACa-3′
Mutated Consensus ISRE oligomer: 5′-agcttGGAAGCGAggATGAggTTGACa-3′
2.3. RNA isolation and reverse transcriptase polymerase chain reaction (RT-PCR)
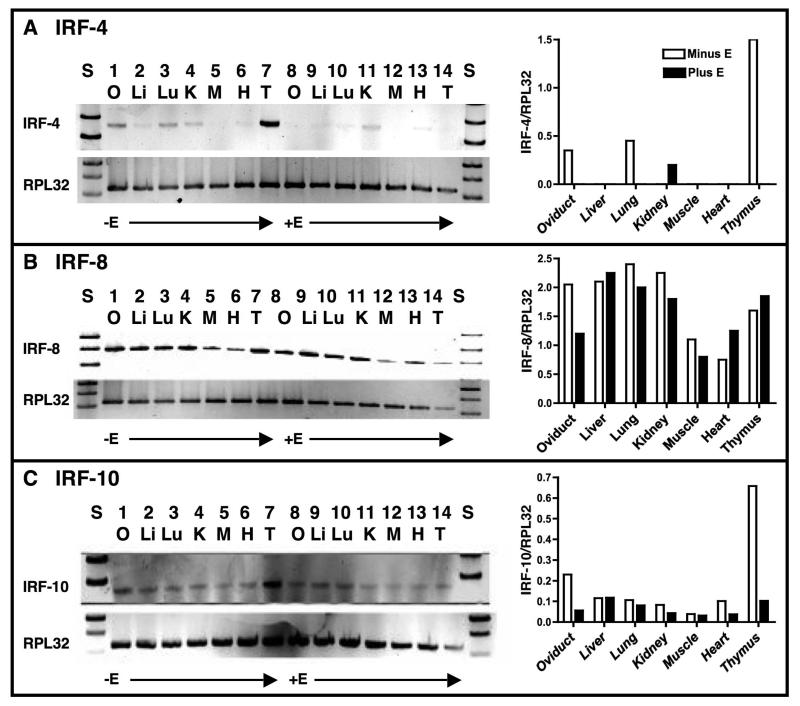
The tissues used in Fig. 5 were isolated from exsanguinated chickens to reduce the possibility that the IRFs measured represented contamination by hematopoetic cells. RNA was isolated using Trizol reagent (Invitrogen), and RT reactions were performed using 2 μg of total RNA and MMLV reverse transcriptase, both according to the manufacturer’s directions (Invitrogen). All RT-PCR reactions used 2 μl of the appropriate RT reaction as a template and recombinant Taq polymerase (Invitrogen) according to the manufacturer’s instructions. The primers and annealing temperatures are listed in Table 1. PCR cycles: 1 cycle at 95° C for 5 min., 25 cycles at 95° C for 30 sec., followed by annealing for 1 min. and an extension at 72° C for 2 min and the final cycle at 72° C for 10 min. All PCR reactions were run on a 2% metaphor gel, stained with ethidium bromide. The RT-PCR data were quantified using the Gel Logic 100 Imaging System with Kodak 1D Image Analysis software. The analysis was done on underexposed versions of the gels actually put into the paper to increase accuracy and prevent saturation. The analysis program was tested for the dynamic range with standards, and background was subtracted for all samples.
Fig. 5.
Tissue distribution of IRF-4, -8, and -10 mRNAs in estrogen-withdrawn and estrogen-stimulated chick tissues. Panel A, left side: RNA was isolated from 3 day estrogen-withdrawn (lanes 1 to 7) or chronically estrogen-stimulated (lanes 8 to 14) chicken tissues. RT-PCR reactions for IRF-4 and RPL32 were run as described in Section 2.3. Lanes 1 and 8: oviduct, Lanes 2 and 9: liver, Lanes 3 and 10: lung, Lanes 4 and 11: kidney, Lanes 5 and 12: muscle, Lanes 6 and 13: heart, and Lanes 7 and 14: thymus. Right side: IRF-4 mRNA was quantified relative to that of RPL32 (ribosomal protein L32) for each tissue. For quantification, the background was removed for each lane. When no bars are present, the ratio was too low to graph. This is representative of an experiment done three times. Panel B: As in Panel A except that IRF-8 mRNA was quantified. Panel C: As in Panel A except that IRF-10 mRNA was quantified. Note that the y-axes in histograms differ among the IRF mRNAs.
Table 1.
Primer sequences and annealing temperatures for RT-PCR
| mRNA | 5′ and 3′ Primers (shown 5′ to 3′) | Anneal T |
|---|---|---|
| IRF-4 |
ATGATTTACCAGAACTCATGACTAGTCCAGAGG CATAAGAATATAGGTATCACGCTTATCAGTATAAG |
56° |
| IRF-8 |
CTGACGTTGGAGAGACATGGAC ATTCTGGATGGCTTCGGCTG |
64° |
| IRF-10 |
GTGACCGAGGTTCTGGAGAG GAAGCACAGGTGGATTTGGT |
56° |
| RPL32 |
AGTCTGATCGCTATGTCAAGATCAAG CTTTTCCTCACTGCGCAGTC |
52° |
2.4. Site-directed mutagenesis
Primer pairs were manually designed, based upon the QuikChange® Site Directed Mutagenesis kit (Stratagene, La Jolla, CA USA), to introduce mutations into the sequences in the 5′-flanking region of the Ov gene. The PCR reactions were performed with the primers below according to the manufacturer’s (Stratagene) protocol except that the extension times were increased to 2 min per kb and the cycle number was increased from 12 to 18 cycles. Mutations were verified by DNA sequencing.
-126/-125 5′-CCATTCAATCCAAAAATGGACCTATTccAACTAAAATCTAACCCAATCCC-3′
-122/-121 5′-CCATTCAATCCAAAAATGGACCTATTAGAAtcAAAATCTAACCCAATCCC-3′
OTE 5′-GCAGAACAGGCAGtcgACACATTGTTACCCAGATTAAAAACTAATATG-3′
2.5. Plasmids
The IRF-4 cDNA was a gift from Dr. Henry Bose (University of Texas). IRF-8 was a gift from Dr. Ben-Zion Levi (Technion-Israel Institute of Technology). The IRF-10 cDNA was cloned from a cDNA library derived from estrogen-stimulated chicken oviduct. These IRF cDNAs were subcloned into the pcDNA-3.1 (Invitrogen) expression vector.
The pOvCAT-900 reporter vector was constructed using Ov sequences from -900 to +9 (GenBank Accession no. J00895), which encompass all of the elements necessary for induction of the gene by steroid hormones (Sanders and McKnight, 1988). Mutations introduced into the CAR and OTE sites are described under site-directed mutagenesis.
3. Results
3.1. The OTE and CAR sites bind similar protein complexes
The repressive CAR element (-130 to -100) in the NRE prevents expression of Ov in the oviduct in the absence of steroid hormones (Sensenbaugh and Sanders, 1999) and at all times in other tissues (Sanders and McKnight, 1988; Monroe and Sanders, 2000). CAR shares identity with the OTE site (-198 to -170), which acts as an activator of Ov transcription, over two short sequence elements of 4 and 5 nucleotides separated by 10 bp (underlined, Fig. 1, panel B). To being to investigate whether these two elements bind the same protein(s), GMSAs were performed using nuclear protein isolated from laying hen oviduct tissue. A single major shifted band of identical migration was observed with both radiolabeled oligomers (Fig. 2. panels A and B, lane 2). Binding to the CAR oligomer is effectively competed off by 50X molar excess of self-oligomer (panel A, lane 6) or by the OTE oligomer (panel A, lane 4), but not by non-specific oligomer (panel A, lane 8). A minor upper band is competed by self-oligomer (panel A, lane 6) but not by the OTE oligomer (panel A, lane 4), suggesting that at least one protein binds to CAR that does not bind to the OTE oligomer. Conversely, both self-oligomer (panel B, lane 4) and CAR oligomer (panel B, lane 6) compete off binding to the OTE. These data raise the possibility that the CAR and OTE sites bind the same or a similar protein(s).
Fig. 2.
The OTE and CAR sites bind a similar protein(s). GMSAs were performed using the conditions described in Section 2.2. A. GMSA with the labeled CAR oligomer (-130 to -100). B. GMSA with the labeled OTE oligomer (-198 to -170). Lane 1: probe alone. Lanes 2-8 contain 5 μg of laying hen oviduct nuclear protein extract. Lanes 3 and 4 contain 10X and 50X molar unlabeled OTE oligomer, respectively. Lanes 5 and 6 contain 10X and 50X molar unlabeled CAR oligomer, respectively. Lanes 7 and 8 contain 10X and 50X molar unlabeled nonspecific competitor, respectively.
3.2. Protein binding to the CAR oligomer requires an intact ISRE
In order to determine which nucleotides are necessary for protein binding to the CAR site, 2 bp mutations were introduced into the CAR oligomer beginning at -129 and ending at -118. These mutated oligomers were tested in GMSAs for binding (data not shown). Interestingly, oligomers containing mutations in a potential interferon-stimulated response element (ISRE) from -126 to -121 were unable to compete effectively for binding. IRFs bind a core consensus sequence of GAAANN (Paun and Pitha, 2007), although other nucleotides may be important, depending on which IRF complex is binding. Subsequently, combinations of these 2 bp mutations were combined and tested in GMSAs using the labeled CAR probe (Fig. 3A). The protein complex binding to the CAR oligomer, designated by the arrow, is specific because cold self-competitor competes for the complex at 25X molar excess (Fig. 3A, Lane 3). None of the oligomers except the one containing the combined mutations at -122/-121 and -119/-118 are able to compete effectively with the wt (wild type) CAR oligomer at this low molar excess (Fig. 3A, Lane 7). The fact that this oligomer can compete is not too surprising as the ISRE can apparently tolerate any nucleotides in positions occupied by -122/-121 (GAAANN). The oligomer used for the competition in lane 8 only retains two of the six core ISRE nucleotides (at positions -124 and -123) and is ineffective at competing for the binding protein(s). Thus, these data provide strong support for the contention that the protein(s) binding to the CAR element is binding in the putative ISRE.
The data thus far suggest that the protein binding to the CAR element is doing so via nucleotides found in the ISRE. To investigate whether this putative ISRE is in fact binding a protein that recognizes a consensus ISRE, additional GMSAs were done using nuclear protein extracts from sexually immature chicken oviduct (Fig. 3C), which is why the banding pattern appears somewhat different from the other GMSA. The CAR probe was labeled and, as expected, an unlabeled wt CAR oligomer was able to compete off binding (Fig. 3C, lane 14) whereas a mutated CAR oligomer was not (lane 15) nor was an unrelated oligomer (lane 16). However, a wt consensus ISRE was able to compete (lane 17) and its mutated counterpart was not (lane 18). This supports the idea the CAR element harbors a bone fide ISRE.
3.3. The protein complex that binds to the CAR site cross reacts with an IRF antibody
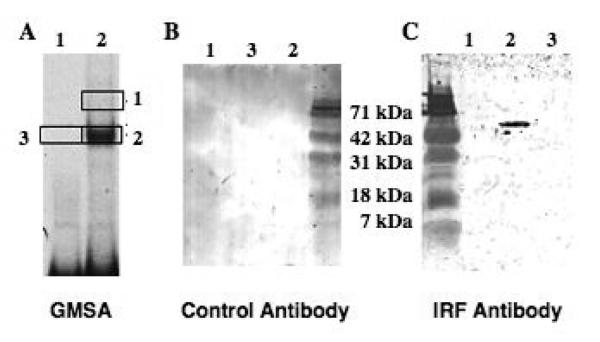
To ascertain whether the protein binding to the CAR oligomer is in fact an IRF, GMSAs were done, shifted bands were excised (Fig. 4, panel A, areas 1, 2, and 3), and the eluted proteins were analyzed by western blotting with an IRF or a control antibody. No bands were detected with the control antibody nor with the control area 3 (panel B). However, western blotting with the IRF antibody revealed that the protein in shifted Band 2 cross-reacted and migrated at approximately 50 kDa, the expected size of an IRF family member (Takaoka et al., 2008). This suggests that the binding protein is an IRF, although we cannot rule out the possibility that an IRF protein comigrated in the gel with the shifted complex. Excision and western blot analysis of shifted Band 1 did not yield a protein that cross reacts with the IRF antibody, although the mass was so much lower that the assay likely would not detect IRF protein. Although this antibody was supposedly selective for IRF-4, it also recognizes IRF-8 and -10 transfected into cells where they are not normally expressed (data not shown). Because few antibodies are available that are selective for chicken IRFs, we were unable to definitively determine which IRF might be binding. Nonetheless, these data strongly suggest that an IRF family member binds the CAR oligomer in vitro and raise the possibility that an IRF may also bind to the OTE.
Fig. 4.
The CAR complex contains a protein that cross reacts with an IRF-4 antibody. A. GMSA with the wt CAR oligogomer (-130 to -100) with (lane 2) and without (lane 1) nuclear protein from estrogen-withdrawn chicks. B. Gel regions as indicated by the boxes from a gel comparable to that in A were run on an SDS/PAGE gel and subjected to western blotting with an irrelevant (δEF1) antibody. C. Areas 1, 2 and 3 were excised from the gel in A and analyzed by western blotting using an IRF-4 antibody (Santa Cruz). This antibody also recognizes chicken IRFs-8, and -10 (data not shown). Area 3 was used as a control.
3.4. Tissue distribution of IRFs-4, -8 and -10
The mammalian IRF family consists of 9 members, IRFs-1 through -9 (Takaoka et al., 2008). While IRFs-1, -2, -5, and -7 are fairly ubiquitous, expression of IRFs-4 and -8 is often considered to be unique to hematopoietic cells (Takaoka et al., 2008). However, in our attempts to identify the IRF(s) binding to Ov, we cloned a novel IRF, now called IRF-10, out of an estrogen-stimulated oviduct library (Park and Sanders, unpublished observations). This was not published because another group reported the cloning of IRF-10 out of a chicken splenic cDNA library (Nehyba et al., 2002) while our manuscript was in preparation. Nonetheless, it led us to speculate that other IRFs might be expressed in oviduct. Because we cloned IRF-10 out of chick oviduct and because IRFs-4, -8 and -10 are in the same IRF family subgroup (the hematopoietic IRFs, Takaoka et al., 2008) and are highly homologous in the DNA binding domains, we hypothesized that one or more of these IRFs may be regulating Ov. This seemed logical as IRF-4 and IRF-8 are present in at least some non-immune tissues (Li et al., 1999; Dadoune et al., 2005; Eguchi et al., 2008).
In hopes of identifying which IRF(s) might regulate Ov, we examined the tissue distribution of IRF-4, -8 and -10 in chicken by RT-PCR (Fig. 5). Interestingly, IRF-4 mRNA was detected in estrogen-withdrawn oviduct, lung, kidney, thymus, and faintly in liver (Fig. 5A). Much less IRF-4 mRNA was observed in estrogen-stimulated animals, with the highest amount being in kidney. While IRF-4 mRNA is high in thymus from estrogen-withdrawn chicks, it is exceptionally low in estrogen-treated thymus. Although this might be ascribed to the atrophic effect that estrogen has on thymus (Wang et al., 2008 and references therein), IRF-8 mRNA was not similarly affected. IRF-4 mRNA is also very low in estrogen-stimulated oviduct, suggesting that estrogen selectively represses transcription of Irf4.
IRF-8 mRNA is relatively abundant in all tissues analyzed regardless of estrogen treatment with the exception of the oviduct (Fig. 5B). In estrogen-stimulated oviduct, the amount of IRF-8 mRNA is about 50% less than that observed in estrogen-withdrawn oviduct, suggesting estrogen directly or indirectly represses Irf8 in this tissue (Fig. 5B, histogram). IRF-10 mRNA, like IRF-8 mRNA, was found in all tissues analyzed, although, at lower levels (Figure 5C). As with IRFs-4 and -8, IRF-10 mRNA is lower in estrogen-stimulated oviduct. These data indicate that at least these three IRFs are expressed in oviduct tissue and that their expression is repressed by estrogen. Furthermore, they suggest that IRFs-4, -8, and/or -10 may play a role in repressing the Ov gene in absence of estrogen in oviduct tissue.
3.5. IRF-4 and IRF-10 repress Ov
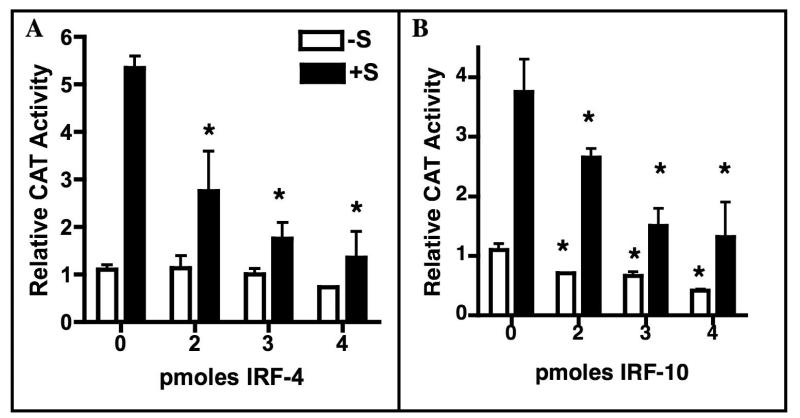
To ascertain whether IRFs-4, -8 and -10 are capable of repressing Ov transcription, increasing amounts of these IRFs were cotransfected with pOvCAT-900 (Ov sequences -900 to +9), which contains all of the Ov sequences required for induction by steroid hormones (Sanders and McKnight, 1988). The amount of expression vector DNA was held constant at 4 pmol by using the empty expression vector. The cells were then cultured for 24 hrs without or with 17β-estradiol and corticosterone (+S). The results show that both IRF-4 and IRF-10 repress the steroid hormone-dependent expression of pOvCAT-900 in a dose-dependent manner (Fig. 6), with maximal repression being about 70%. IRF-10 also represses basal expression of pOvCAT-900 by about the same amount at the highest dose (panel B). Although IRF-8 was tested, we later found that several amino acids were missing from the cDNA, so those data are invalid. Nonetheless, these functional studies reveal for the first time that IRFs repress expression of Ov.
Fig. 6.
Overexpression of IRF-4 or IRF-10 represses Ov. Primary oviduct cells were cultured for 24 hrs without and with estrogen and corticosterone (+S). The pOvCAT-900 reporter vector was cotransfected with increasing amounts of an IRF-4 (panel A) or IRF-10 (panel B) expression plasmid. Total expression plasmid DNA was held constant at 4 pmol using the empty vector. Data are presented as CAT activity relative to pOvCAT-900 in the absence of steroid hormones (-S). Statistical significance was determined using one way ANOVA with Tukey’s Multiple Comparison post test. Stars indicate samples that are significantly different from pOvCAT with the corresponding hormonal treatment in the absence of added IRF (0 pmoles IRF). For IRF-4 +S, p<0.021; for IRF-10 -S, p<0.05; for IRF-10 +S, p<0.01
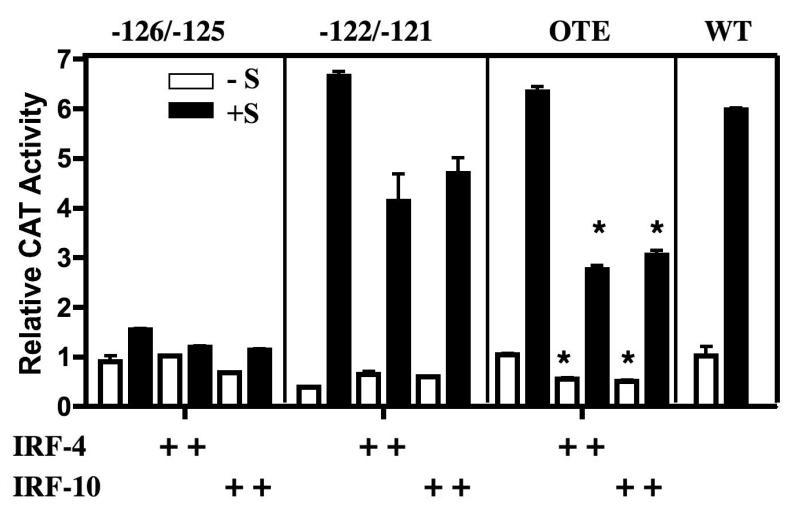
Having identified nucleotides within the CAR element ISRE that are critical for protein binding (Fig. 3), the functional significance of these nucleotides was analyzed in transfection experiments (Fig. 7). Site-directed mutagenesis was utilized to introduce mutations into the CAR site at -126 and -125 or at -122 and -121, and the constructs were transfected into primary oviduct cells, which were subsequently cultured with or without estrogen and corticosterone. Because the CAR element as a whole appears to be a repressive site, we anticipated that IRF binding to the ISRE would repress Ov and that mutation of those critical nucleotides in the ISRE (Paun and Pitha, 2007) would relieve repression. Surprisingly, mutation of nucleotides -126 and -125 abolished steroid-dependent induction of the Ov promoter (Fig. 7, first panel), so we were unable to assess whether those two nucleotides are required for repression of Ov by the IRFs, as predicted. Although transcription of the construct with mutations in Ov at -122 and -121 appears to be repressed by the IRFs, this decrease was not statistically significant (Fig 7, second panel). This is not surprising as those two nucleotides do not affect the ability of the CAR oligomer to compete for binding in GMSAs (Fig 3, lane 7) and as any nucleotide can be in those two positions in a consensus ISRE. Because the OTE has some sequence identity to CAR (Fig. 1), because the OTE oligomer competes for proteins bound by the CAR oligomer in GMSA (Fig. 2), and because the OTE region plays a role in the activation of Ov, this element may be confounding the overexpression experiments.
Fig. 7.
Mutation of critical nucleotides in the ISRE located in the CAR site reveal roles for both positive and negative regulation of Ov. Primary oviduct cells were cultured and transfected as described in the legend to Fig. 6. The pOvCAT reporter vectors with mutations in the indicated sites were cotransfected with IRF-4 or IRF-10 expression plasmid. Mutations were made at -126 and -125, at -122 and -121, or at -193 to -191 in the OTE ISRE as indicated in Section 2.4. Statistical significance was determines using one way ANOVA with Tukey’s Multiple Comparison post test. Stars indicate samples that are significantly different from the corresponding vector and hormonal treatment in the absence of added IRF. For OTE -S, p<0.05; for OTE +S, p<0.01
To investigate the role of the OTE, three nucleotides in the ISRE in the OTE element were mutated, and the resultant construct was transfected into primary oviduct cells. Expression of this construct did not differ from wt, and the IRFs were able to repress both steroid-stimulated and basal expression by about 50% (Figure 7, third panel). A construct was made in which both the CAR and OTE ISREs were mutated, but this proved uninformative because its expression was at background levels (data not shown). These data indicate that nucleotides -126 and -125 are required for Ov expression and suggest that only one functional ISRE is necessary for partial repression of Ov by IRFs. More extensive repression is seen when both ISREs are present (Fig. 6).
4. Discussion
Although some success has been achieved in predicting tissue-specific enhancers (Pennachhio et al., 2007), the molecular basis for tissue-specific gene expression has been determined for a relatively small number of genes. Many factors have made understanding tissue-specific gene regulation difficult, including the observations that more than half of all human genes rely on regulatory elements that are at considerable distance from the promoters, that multiple regulatory elements are often used to control a single transcription unit, that tissue-specific regulation is often difficult to recapitulate in cell culture, and that tissue-specific and target gene-specific coactivator complexes modulate transcriptional activity (Levine and Tjian, 2003; Nobrega et al., 2003). In addition, activities of tissue-specific transcription factors and coactivator complexes are frequently regulated by environmental signals, hormones and growth factors, and developmental signals. Even core promoter complexes are variable and contribute to tissue-specific gene expression (Levine and Tjian, 2003).
The molecular basis for the tissue-specific expression of the Ov gene has eluded investigators. Its expression is strictly limited to the tubular gland cells in the magnum portion of the estrogen-stimulated oviduct. The region from -900 to +9 directs expression in primary tubular gland cell cultures but not in other tissue culture cells, although a region between -3100 and -2800 might contribute to tissue specificity in vivo (Park et al., 2006). Unfortunately, transgenic chicken technology is not available to assess the elements required for oviduct-specific expression in vivo. Removal of all sequences upstream of -80 abolishes tissue-specific expression of Ov in primary tubular gland cell cultures, although basal expression is high (Sanders and McKnight, 1988). Dissecting the region from -900 to -80 is difficult as all of the SDRE (-892 to -793) is absolutely required for estrogen-dependent expression of the gene and for any expression above basal levels when the NRE (-308 to -88) is present. Thus, in the presence of estrogen the SDRE overcomes the repressive effects of the NRE, but only in oviduct. Removal of sequences upstream of -308 reduces expression to background levels (Sanders and McKnight, 1988). Site-directed mutagenesis every 10 bp through the NRE identified a strong repressive site from -130 to -100, which was called the CAR element for COUP-TF adjacent repressor (Sensenbaugh and Sanders, 1999). CAR was subsequently shown to contribute to the tissue-specific expression of Ov (Monroe and Sanders, 2000).
Identification of the CAR binding protein was attempted using mass spectrometry, which pulled out hits for IRF-4. This was very puzzling because at that time expression of IRF-4 was thought to be restricted to the immune system. Because the chicken genome had not been sequenced, we attempted to clone IRF family members out of oviduct using low-stringency hybridization. We cloned, but did not publish, what would later be called IRF-10 (Nehyba et al., 2002). Inspection of the sequence in CAR revealed a putative binding site for members of the IRF transcription factor family. GMSA analysis revealed that specific binding to CAR involved nucleotides in the putative IRF binding site (-126 to -121, GAAACT) (Fig. 3). Crystallographic analysis demonstrated that IRF-4 recognizes the GAA nucleotides in positions 1 - 3 and the G on the opposite strand of the C in position 5 (Escalante et al., 2002). Only mutations not involving those core 3 nucleotides were able to compete for binding with the wt CAR oligomer (Fig. 3, lane 7). This strongly suggests that an IRF family member binds to the CAR element. This was supported by GMSA followed by western blotting with an IRF-4 antibody (Fig. 4), although we find this antibody also recognizes chicken IRFs-8 and -10 (data not shown). IRFs-4 and -10 repress Ov in a dose-dependent manner (Fig. 6), and mutation of the core nucleotides interferes with proper expression of Ov (Fig. 7). Surprisingly, mutation of -126 and -125 abolished induction of the gene by steroids, so the effects of IRFs on this mutated construct could not be assessed. We expected that they would completely relieve repression. However, the loss of expression suggests that an IRF functions as an activator when steroid hormones are present. This is consistent with the known abilities of IRFs to both activate and repress transcription (Paun and Pitha, 2007; Takaoka et al., 2008). Mutation of nucleotides -122 and -121, which are in the variable positions of the ISRE (GAAANN) (Paun and Pitha, 2007), did not attenuate the repression by either IRF. This was expected as mutation of these sites did not reduce the ability of an oligomer to compete with the wt CAR oligomer (Fig. 3, lane 7).
Because the OTE shares sequence identity with CAR and because they can cross compete for protein binding (Fig. 2) we reasoned that it could be confounding results in our functional analysis of the CAR element. However, mutation of the 3 ISRE core nucleotides in the OTE relieved the repression only by about 25%, presumably because the ISRE in CAR was still intact. Unfortunately, mutation of both ISREs reduced expression to that seen with the -126/-125 mutations (data not shown), so it was impossible to confirm in the context of the Ov promoter that the repressive effects of the IRFs is occurring through the CAR and/or the OTE sites. Nonetheless, the GMSA experiments and the GMSA followed by western blotting strongly suggest that at least one IRF family member represses Ov in the absence of steroid hormones by binding to the CAR element. There, in the absence of estrogen, one or more IRFs repress Ov in oviduct, and likely in other tissues. When estrogen is present, expression of IRFs is repressed (Fig. 5), allowing activation of Ov through as yet unidentified tissue-specific mechanisms involving the CAR element.
Although expression of IRFs-4, IRF-8, and IRF-10 was thought to be restricted to immune tissue, we find that both IRF-8 and -10 are widely distributed in chicken (Fig. 5B and 5C). IRF-4 appears to be expressed primarily in estrogen-withdrawn oviduct and lung but to lesser extent in liver and kidney (Fig. 5A). This is the first report of such widespread transcription of these IRF genes. IRFs-4 and -10 mRNAs are repressed by estrogen in the thymus, which might be due to estrogen-induced thymic atrophy (Wang et al., 2008 and references therein), but this explanation is not consistent with observation that expression of IRF-8 mRNA in thymus is estrogen-independent. All three IRFs are reduced in estrogen-treated oviduct, lending credence to the idea that one or more of them repress Ov in the absence of estrogen. Which IRF represses Ov in non-oviduct tissue remains unclear but it is likely to be IRF-8 or -10 based on their expression patterns. Unfortunately, due to the lack of specific antibodies, we were unable to determine how much of each of these IRFs is expressed in the various tissues. Only IRF-3 and -9 can be absolutely ruled out as potential regulators of Ov as those genes are not present in chicken.
The role of IRFs in gene regulation has largely been studied in the context of the immune system and in host defense against invading pathogens. However, evidence is accruing to indicate that these family members play pivotal roles in other events linked to cellular differentiation, tissue-specific gene expression, and oncogenesis (Paun and Pitha, 2007). The link to oncogenesis is the most obvious, particularly with hematological malignancies. All of the IRFs except IRF-6 are associated with promoting or inhibiting oncogenesis in one or more cellular contexts (Takaoka et al., 2008). IRF-6 is critical for skin, limb, and craniofacial development (Ingraham et al., 2006). Most recently, all 9 IRFs were shown to be expressed in mouse adipocytes and to vary during the differentiation of preadipocytes to adipocytes in cell culture (Eguchi et al., 2008). More importantly, many of these IRFs repress adipogenesis. Thus, there is some precedence for essential roles for IRFs in non-immune contexts. The understanding of the importance of this protein family in gene regulatory events beyond the scope of the immune system is expanded by our observations that it plays a central role in the tissue-specific and estrogen-induced expression of Ov.
Acknowledgements
This work was supported by NIH T32 HL07741 (DCD) and by NIH R01-DK061913 (MMS).
Abbreviations
- IRF
interferon regulatory factor
- Ov
ovalbumin
- bp
base pair(s)
- RT
reverse transcriptase
- NRE
negative regulatory element
- GMSA
gel mobility shift assay(s)
- CAR
COUP-TF adjacent repressor
- OTE
ovalbumin tissue-specific element
- CAT
chloramphenicol acetyl transferase
- EDTA
ethylenediaminetetraacetic acid
- CHIRP
chicken induced regulatory protein
- ISRE
interferon-stimulated response element
- SDRE
steroid dependent regulatory element
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction(s)
- RPL32
ribosomal protein L32
- IRF
interferon regulatory factor(s)
- C
corticosterone
- kb
kilobase(s)
- E
17β-estradiol
- +S, -S
plus steroids, minus steroids
- wt
wild type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bellard M, Dretzen G, Belard F, Oudet P, Chambon P. Disruption of the typical chromatin structure in a 2500 base-pair region at the 5′-end of the actively transcribed ovalbumin gene. EMBO J. 1982;1:223–230. doi: 10.1002/j.1460-2075.1982.tb01151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain EM, Sanders MM. Identification of the novel player ™EF1 in estrogen transcriptional cascades. Mol. Cell. Biol. 1999;19:3600–3606. doi: 10.1128/mcb.19.5.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadoune JP, Pawlak A, Alfonsi MF, Siffroi JP. Identification of transcripts by macroarrays, RT-PCR and in situ hybridization in human ejaculate spermatozoa. Mol. Human Reprod. 2005;11:133–140. doi: 10.1093/molehr/gah137. [DOI] [PubMed] [Google Scholar]
- Dean DM, Jones PM, Sanders MM. Alterations in chromatin structure are implicated in the activation of the steroid hormone response unit in the ovalbumin gene. DNA Cell Biol. 2001;20:27–39. doi: 10.1089/10445490150504675. [DOI] [PubMed] [Google Scholar]
- Dean DM, Jones PS, Sanders MM. Regulation of the chick ovalbumin gene by estrogen and corticosterone requires a novel DNA element that binds a labile protein, Chirp-1. Mol. Cell. Biol. 1996;16:2015–2024. doi: 10.1128/mcb.16.5.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillner NB, Sanders MM. Upstream Stimulatory Factor (USF) is recruited into a steroid hormone-triggered regulatory circuit by the estrogen-inducible transcription factor ™EF1. J. Biol. Chem. 2002;277:33890–33894. doi: 10.1074/jbc.M204399200. [DOI] [PubMed] [Google Scholar]
- Dougherty DC, Sanders MM. Estrogen action: revitalization of the chick oviduct model. Trends Endocrinol. Metab. 2005;16(2005):414–419. doi: 10.1016/j.tem.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Eguchi J, Yan Q-W, Schones DE, Kamal M, Hsu C-H, Zhang MQ, Crawford GE, Rosen ED. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab. 2008;7:86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante CR, Brass AL, Pongubala JMR, Shatova E, Shen L, Singh H, Aggarwal AK. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol. Cell. 2002;10:1097–1105. doi: 10.1016/s1097-2765(02)00703-7. [DOI] [PubMed] [Google Scholar]
- Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC. Abnormal skin, limb, and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nature Genet. 2006;38:1335–1340. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Li W, Nagineni CH, Efiok B, Chepelinsky AB, Egwuagu CE. Interferon regulatory transcription factors are constitutively expressed and spatially regulated in the mouse lens. Dev. Biol. 1999;210:44–55. doi: 10.1006/dbio.1999.9267. [DOI] [PubMed] [Google Scholar]
- Monroe DG, Sanders MM. The COUP-adjacent repressor (CAR) element participates in the tissue-specific expression of the ovalbumin gene. Biochim. Biophys. Acta. 2000;1517:27–32. doi: 10.1016/s0167-4781(00)00241-4. [DOI] [PubMed] [Google Scholar]
- Nehyba J, Hrdlickova R, Burnside J, Bose JH. A novel interferon regulatory factor (IRF), IRF-10, has a unique role in immune defense and is induced by the v-rel oncoprotein. Mol. Cell. Biol. 2002;22:3942–3957. doi: 10.1128/MCB.22.11.3942-3957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Nordstrom LA, Dean DM, Sanders MM. A complex array of double-stranded and single-stranded DNA-binding proteins mediates induction of the ovalbumin gene by steroid hormones. J. Biol. Chem. 1993;268:13193–13202. [PubMed] [Google Scholar]
- Park H-M, Sanders MM, Suzuki T, Muramatsu T. An oviduct-specific and enhancer-like element resides at about -3000 in the chicken ovalbumin gene. Biochimie. 2006;88:1909–1914. doi: 10.1016/j.biochi.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennachhio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MM, McKnight GS. Chicken egg white genes: multihormonal regulation in a primary cell culture system. Endocrinol. 1985;116:398–405. doi: 10.1210/endo-116-1-398. [DOI] [PubMed] [Google Scholar]
- Sanders MM, McKnight GS. Positive and negative regulatory elements control the steroid-responsive ovalbumin promoter. Biochem. 1988;27:6550–6557. doi: 10.1021/bi00417a053. [DOI] [PubMed] [Google Scholar]
- Sensenbaugh KR, Sanders MM. Multiple promoter elements including a novel repressor site modulate expression of the chick ovalbumin gene. DNA Cell Biol. 1999;18:147–156. doi: 10.1089/104454999315538. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Tamura T, Taniguchi T. Interferon regulatory factor family of transcription factors and regulation of oncogenesis. Cancer Sci. 2008;99:467–478. doi: 10.1111/j.1349-7006.2007.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay R, Gupta S, Kanungo MS. Trans-acting factors that interact with the proximal promoter sequences of ovalbumin gene are tissue-specific and age-related. Mol. Cell. Biochem. 1999;201:65–72. doi: 10.1023/a:1007000215524. [DOI] [PubMed] [Google Scholar]
- Wang C, Dehghani B, Magrisso IJ, Rick EA, Bonhomme E, Cody DB, Elenich LA, Subramanian S, Murphy SJ, Kelly MJ, Rosenbaum JS, Vandenbark AA, Offner H. GPR30 contributes to estrogen-induced thymic atrophy. Mol. Endocrinol. 2008;22:636–648. doi: 10.1210/me.2007-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]