Abstract
DNA helicases of the RECQ family are important for maintaining genome integrity, from bacteria to humans. Although progress has been made in understanding the biochemical role of some human RECQ helicases, that of RECQL5 remains elusive. We recently reported that RECQL5 interacts with RNA polymerase II (RNAPII), pointing to a role for the protein in transcription. Here, we show that RECQL5 inhibits both initiation and elongation in transcription assays reconstituted with highly purified general transcription factors and RNAPII. Such inhibition is not observed with the related, much more active RECQL1 helicase or with a version of RECQL5 that has normal helicase activity but is impaired in its ability to interact with RNAPII. Indeed, RECQL5 helicase activity is not required for inhibition. We discuss our findings in light of the fact that RECQ5−/− mice have elevated levels of DNA recombination and a higher incidence of cancer.
The RECQ family constitutes a highly conserved group of DNA helicases that play important roles in the maintenance of genome integrity, promoting cell survival in response to various types of genotoxic stress. Generally, RECQ helicases appear to be involved in maintaining functional replication forks, for example by suppressing undesired recombination events during DNA synthesis (1). Mutations in genes encoding three of the five members of the human RECQ family have been linked with severe genetic disorders, namely Bloom syndrome (2), Werner syndrome (3), and Rothmund-Thomson syndrome (4). These diseases are characterized by a predisposition to various types of cancers, suggesting a genome caretaker role for the human RECQ helicases (5).
RECQL5 is a relatively uncharacterized member of the RECQ family and has so far not been linked with a distinct genetic disease (6). However, RECQ5−/− mice exhibit an elevated level of sister chromatid exchange and are also predisposed to various types of cancer, strongly suggesting that the RECQ5 protein plays a role in the maintenance of genomic stability as well (7, 8). Recent studies indicate a role for the protein in DNA replication and cell survival after camptothecin treatment (9). RECQL5 is expressed in all human tissues examined (10), and, in contrast to BLM and WRN helicases, its protein level does not change during the cell cycle (11), suggesting that it might also have a more general, cell cycle-phase independent function.
We recently identified human RECQL5 helicase as a novel RNA polymerase II (RNAPII)2-associated protein in chromatin. RECQL5 interacts directly with the polymerase and is the only member of the human RECQ family to do so (12). A number of RNAPII binding proteins or complexes, such as TFIIF, TFIIS, elongin, ELL, and the mediator complex, can serve as positive regulators of transcription initiation and/or elongation. Thus, we considered the possibility that RECQL5 might have a previously overlooked role in RNAPII transcription.
Using RNAPII transcription assays reconstituted with highly purified factors, we now show that RECQL5 helicase is an inhibitor of transcription. We discuss the possible implications of our findings for the function of RECQL5 helicase in the maintenance of genomic stability.
EXPERIMENTAL PROCEDURES
Plasmids
Generation of the pCMV-FLAG plasmid was described previously (12). To generate pCMV-FLAG-RECQL5-(1–542), pCMV-FLAG-RECQL5 was digested with BamHI and recircularized. To generate pCMV-FLAG-RECQL5-(541–991), the DNA fragment encoding RECQL5-(541–991) was PCR-amplified using the following primers: 5′-TTTTCTCGAGAGAATCCCCAGGCTG-3′ and 5′-TTTTGGATCCTCATCTCTGGGGGCC-3′ and cloned into the pCMV-FLAG vector between the XhoI and BamHI sites. To generate pCMV-FLAG-RECQL5LID, a BamHI DNA fragment encoding RECQL5552–991 was PCR-amplified using the following primers: 5′-TTTTGGATCCACTGCCTGCGGC-3′ and 5′-TTTTGGATCCTCATCTCTGGGGGCC-3′. It was cloned into the BamHI site of pCMV-FLAG-RECQL5-(1–542). Correct orientation of the fragment was confirmed by restriction digestion. The different versions of RECQL5 were subcloned into the NdeI-SapI sites of the pTYB1 vector (New England Biolabs) for expression in Escherichia coli.
Establishment of Stable Human Cells
Stable human 293T cells were established by co-transfecting pCMV-FLAG construct with a pGK vector and selecting with 2 μg/ml puromycin as described (12).
Immunoprecipitation
0.5–1 L of 293T cells stably expressing FLAG-tagged proteins were harvested and washed three times with phosphate-buffered saline. The cell pellet was then resuspended in 2.5 volumes of hypotonic lysis buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 1 mm dithiothreitol (DTT)) in the presence of complete EDTA-free protease inhibitor mixture and phosphatase inhibitor tablet (Roche Applied Science), left on ice for 30 min, and lysed using an “A” pestle (25 strokes). After 30 min on ice, 2.5 volumes of 2× FLAG binding buffer (20 mm HEPES, pH 7.9, 3 mm MgCl2, 0.6 m NaCl, 20 mm KCl, 0.4% Triton-X-100, and 20% glycerol) was added, and the samples were incubated for 1 h at 4 °C with gentle shaking. The lysates were centrifuged for 30 min at 14,000 rpm using a microcentrifuge, and the supernatant (whole cell extract) was collected and incubated with FLAG M2-agarose beads (Sigma) at 4 °C overnight. The M2-agarose beads were washed 5× with 50 volumes of 1× FLAG binding buffer (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 0.3 m NaCl, 10 mm KCl, 0.2% Triton X-100, and 10% glycerol). Bound proteins were then eluted with 1× SDS-PAGE loading buffer and characterized by SDS-PAGE and Western blot analysis. The antibodies used were anti-FLAG (Sigma) and anti-RPB1 (4H8; Upstate Biotechnology).
Recombinant Protein Expression and Purification
The self-cleavable intein-chitin binding domain tag (New England Biolabs) was used to produce recombinant RECQL5 proteins in E. coli. BL21-CodonPlus (DE3)-RIL Competent cells (Stratagene) transformed with pTYB1-RECQL5 (or mutants) plasmids were grown in the presence of 2.5% glucose, 25 μg/ml chloramphenicol, and 100 μg/ml ampicillin until a density of A600 = 0.4 at 37 °C. Expression was induced by adding isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 0.1 mm and incubating for 18 h at 16 °C. Pelleted cells were resuspended in cell lysis buffer (50 mm Na-HEPES, pH 8.0, 500 mm NaCl, 10% glycerol, 1 mm EDTA, 0.1% Triton X-100, and protease inhibitors (Roche)) and lysed by sonication. Sonicated homogenates were cleared by centrifugation at 13,000 rpm (SS34 rotor) for 20 min at 4 °C. Purification of the soluble chitin binding domain-tagged proteins was achieved by using chitin resin (New England Biolabs) according to the manufacturer's instructions. Proteins were eluted with Buffer D (20 mm HEPES-KOH, pH 7.9, 20% glycerol, 0.1 mm EDTA, 0.1 m KCl, and 50 mm DTT). Peak fractions were combined and dialyzed against Buffer D without DTT. Recombinant RECQL1 was purified as described (13).
DNA Helicase Assays
The splayed arm substrate was generated as described (13). Helicase reactions (10 μl) were carried out in the presence of 0.2 ng of 32P-end-labeled splayed arm substrate in helicase buffer (30 mm Tris, pH 7.5, 3 mm MgCl2, 3 mm ATP, 1 mm DTT, 80 μg/ml bovine serum albumin, and 10% glycerol). Reactions were initiated by the addition of purified RECQ proteins and incubated for 30 min at 37 °C. The DNA products were deproteinized and analyzed by nondenaturing PAGE.
Reconstituted Promoter-specific Basal Transcription
The EcoRI/NdeI fragment containing an AdML promoter-driven transcription template, oligo(dC)-tailed pAd-GR220, as well as promoter-specific in vitro transcription experiments were described previously (14 and references therein). Briefly, core reaction mixtures (53 μl volume) containing 50 ng pDN-AdML EcoRI/NdeI fragment, ∼40 ng rat liver TFIIH, 20 ng recombinant TFIIF, 10 ng recombinant TFIIB, 10 ng recombinant TATA box-binding protein (TBP), 7 ng recombinant TFIIE, and 0.1 μg (0.2 pmol) of rat liver RNAPII in 20 mm Tris-HCl, pH 7.9, 20 mm HEPES-KOH, pH 7.9, 0.5 mm DTT, 0.025% Nonidet P-40, 1% polyvinyl alcohol (w/v), 60 mm KCl, 3% glycerol (v/v), 0.5 mg/ml bovine serum albumin, and 10 units RNAsin (Promega) were incubated at 28 °C for 20 min to assemble preinitiation complexes. Transcription was initiated by adding a 7-μl NTP mix to give a final concentration of 50 μm ATP, 50 μm GTP, 50 μm UTP, 6 μm CTP, 10 μCi [α-32P]CTP, and 7.5 mm MgCl2 to make a complete 60-μl reaction. RECQL5, or equimolar amounts of RECQL5ID, RECQL5D157A, or RECQL1, was titrated into the 53-μl reactions. After transcription, reactions were stopped by addition of 66 μl of STOP buffer (10 mm Tris-HCl, pH 7.2, 0.5 mm EDTA, and 0.3 m NaCl) containing 40 μg yeast RNA (Ambion) and 40 μg proteinase K (Roche). RNA was precipitated with ethanol, resuspended in 9 m urea, resolved by 6% denaturing PAGE, and analyzed by phosphorimaging. Quantification was performed using ImageQuant software.
Sarkosyl Challenge and Abortive Initiation Assays
RNAPII preinitiation complexes were assembled as described above. Sarkosyl was included to a final concentration of 0.015% either in the beginning of the reaction or just after the addition of nucleotides as indicated. 0.9 pmol RECQL5 or RECQL5ID proteins were added before or after preinitiation complex assembly. Transcription was started by the addition of NTP mix and analyzed as described above.
Abortive initiation experiments were performed as described previously (15) by using CpU dinucleotide (Sigma) as a primer for trinucleotide synthesis. 0.9 pmol RECQL5, RECQL5ID, RECQL5D157A, or RECQL1 proteins were added to the reactions as indicated.
Analysis of RNAPII Transcription Elongation
RNAPII elongation complexes were prepared by performing in vitro transcription reactions with oligo(dC)-tailed DNA template derived from pAd-GR220. The final reaction contained 26 mm Tris-HCl, pH 7.9, 10.6 mm MgCl2, 1.3 mm DTT, 0.6 mg/ml bovine serum albumin, 10 units of RNasin (Promega), 133 ng of oligo(dC)-tailed DNA template, 0.6% polyvinyl alcohol, 17.7% (v/v) buffer D (20 mm HEPES-KOH, pH 7.9, 20% glycerol, 0.1 mm EDTA, and 0.1 m KCl), and ∼0.2 μg of rat liver RNAPII. U-less transcription was started by the addition of NTP mixture to give a final concentration of 50 μm ATP, 50 μm GTP, 2 μm CTP, and 10 μCi of [α-32P]CTP and incubated at 30 °C for 40 min. Stalled elongation complexes were chased to longer transcripts by the addition of UTP and CTP to a final concentration of 2 μm and 20 μm, respectively. Transcription elongation was performed at 30 °C for increasing amounts of time as described in the legend to Fig. 4 in the presence or absence of 0.9 pmol RECQ protein or after addition of ELL/EAF (as previously described (14)). Reactions were stopped, and RNA was purified and analyzed as described above.
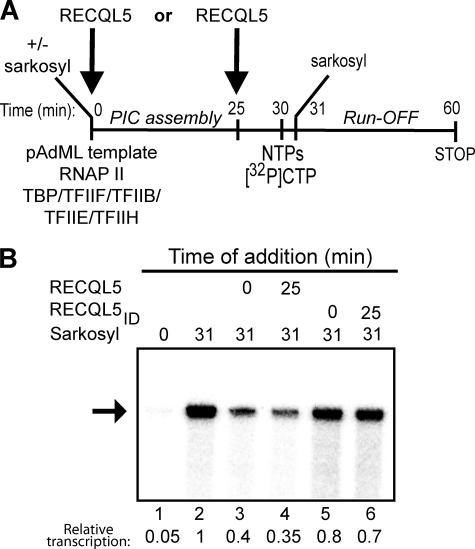
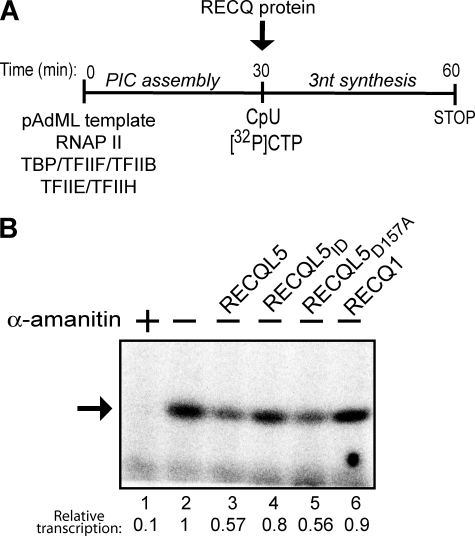
FIGURE 4.
RECQL5 inhibits an early step of RNAPII transcription. A, An outline of the experiment is shown. RECQL5 was either added during or after PIC assembly. B, Sarkosyl-treated RNAPII transcription reactions were treated with 0.9 pmol of the indicated RECQL5 protein, resolved by 6% denaturing PAGE, and visualized by phosphorimaging. The arrow indicates a 254-nt run-off transcript. Quantification of this transcript, relative to the control (lane 2, set to 1), is indicated below the lanes.
RESULTS
A Central 9-Amino Acid Region of RECQL5 Important for Interaction with RNAPII
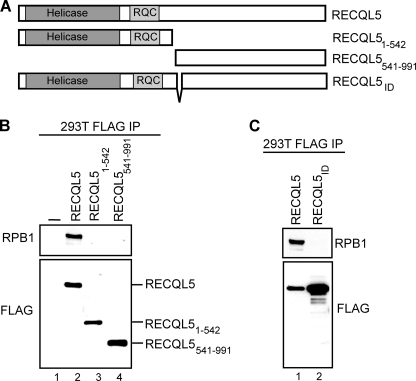
To facilitate a study of the functional consequences of the RECQL5-RNAPII interaction, we first delineated a region in the RECQL5 protein that is required for normal association with the polymerase. We previously reported that the RECQL5-RNAPII interaction requires the C-terminal half of RECQL5 (12). To more precisely map domains that contribute to this interaction, we established stable human 293T cell lines expressing similar amounts of different FLAG-tagged forms of RECQL5 (Fig. 1, A–C). The RECQL5 protein was immunoprecipitated from the individual cell lines using anti-FLAG antibody, and the association with RNAPII was analyzed by anti-RPB1 Western blotting (Fig. 1, B and C, RPB1). As reported previously (12), full-length RECQL5 protein bound RNAPII (Fig. 1B, lane 2), whereas the RECQL5 (1–542) truncation mutant did not (lane 3). The remaining C-terminal part of the protein (541–991) was unable to bind RNAPII as well (lane 4), which could suggest that residues around amino acids 542 are critical for the interaction. Indeed, we discovered that an internal deletion of nine amino acids, located between positions 542 and 552 (RECQL5ID), dramatically decreased the interaction with RNAPII (Fig. 1C, compare lanes 1 and 2).
FIGURE 1.
A central 9-amino acid sequence is essential for RNAPII-RECQL5 interaction in vivo. A, diagram describing the constructs used. B and C, Western blots of FLAG-tagged immunoprecipitations (IP) showing interactions between different forms of FLAG-tagged RECQL5 and RNAPII, using FLAG and RPB1 antibody, respectively.
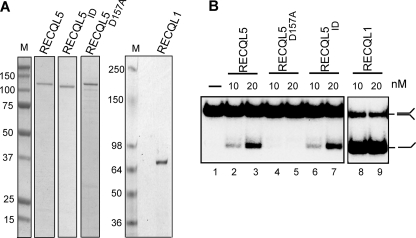
Next, we expressed wild type RECQL5, the RNAPII interaction-deficient protein (RECQL5ID), a RECQL5 helicase-deficient point mutant (RECQL5D157A), and another human RECQ family helicase, RECQL1, in E. coli, and purified the recombinant proteins to virtual homogeneity (Fig. 2A). These proteins were then tested in ATP-dependent helicase assays (Fig. 2B). As expected, the RECQL5D157A mutant did not exhibit helicase activity (Fig. 2B, lanes 4–5) but was fully capable of interacting with RNAPII (data not shown). RECQL1 exhibited robust helicase activity, much greater than that of RECQL5 (Fig. 2B, compare lanes 8–9 with 2–3). Most importantly, RECQL5 and RECQL5ID had similar activity (Fig. 2B, compare lanes 2–3 and 6–7), indicating that the internal deletion in RECQL5ID (which greatly reduced RNAPII interaction) does not significantly affect overall protein folding and enzymatic activity.
FIGURE 2.
Activity of the recombinant RECQ proteins used in this study. A, Coomassie-stained SDS-PAGE gel showing recombinant RECQ proteins. M, protein molecular weight standard marker. B, DNA helicase assays showing the relative ATP-dependent helicase activities of the recombinant proteins shown in A.
RECQL5 Inhibits RNAPII Transcription in Vitro
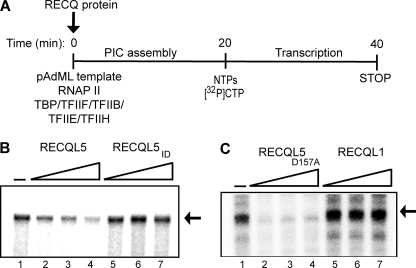
To study the potential effect of RECQL5 on transcription, we reconstituted mammalian RNAPII transcription using highly purified general transcription factors (GTFs), RNAPII, and a DNA template containing the adenovirus major late promoter (16). Then, the effect of increasing amounts of purified RECQ proteins on production of the run-off transcript was analyzed (Fig. 3A). RECQL5 protein was inhibitory for basal RNAPII transcription, and inhibition increased as larger amounts of the protein was added (Fig. 3B, compare lanes 1–4). The RECQL5ID protein had little or no effect (Fig. 3B, compare lanes 2–4 and 5–7), indicating that inhibition was not merely due to the addition of a DNA-unwinding helicase activity but required a stable RECQL5-RNAPII interaction. Indeed, the much more active RECQL1 helicase failed to affect transcription, further indicating that the effect observed with RECQL5 was specific (Fig. 3C, lanes 5–7). Finally, the RECQL5D157A helicase mutant also inhibited basal transcription (Fig. 3C, lanes 2–4), indicating that RECQL5-mediated inhibition of RNAPII basal transcription does not require its helicase activity.
FIGURE 3.
RECQL5 inhibits reconstituted RNAPII transcription. A, outline of the experiment. PIC, pre-initiation complex. B, 0.113, 0.225, and 0.9 pmol of RECQL5 (lanes 2–4) or RECQL5ID (lanes 5–7) were added during preinitiation complex formation. Transcripts were resolved by 6% denaturing PAGE and visualized by phosphorimaging. The arrow indicates a 254-nt run-off transcript. C, effect of RECQL5D157A and RECQL1 as described in B. The experiments in B and C were performed at different times and thus cannot be used for precise quantitative comparisons of the relative effects of RECQL5 and RECQL5D157A. Please note that the experiments in Figs. 5 and 6 show that RECQL5D157A does not inhibit transcription more potently than wild type RECQL5.
RECQL5 Inhibits an Early Initiation Step of RNAPII Transcription
We next investigated which step or steps of the RNAPII transcription cycle can be targeted by RECQL5. RNAPII transcription starts with the assembly of a preinitiation complex at the promoter, followed by formation of the first phosphodiester bond (initiation), promoter escape, and transcript elongation (17). To test whether RECQL5 is able to inhibit early steps of the transcription cycle, we restricted transcription to a single round by using a sarkosyl challenge approach (18)(Fig. 4). 0.015% sarkosyl specifically inhibits the preinitiation/initiation stage of RNAPII transcription without significantly affecting the processivity of an already formed elongation complex (18). If, for example, RECQL5 inhibits transcription by interfering with assembly of the preinitiation complex, it would be expected to affect transcription only when added together with the GTFs. RECQL5 (or RECQL5ID) was therefore added to the reconstituted RNAPII transcription reactions either before, or just after, preinitiation complex assembly (Fig. 4A). As expected, sarkosyl prevented transcription when included at the beginning of the reaction (Fig. 4B, lane 1). In contrast, because sarkosyl at this concentration has little or no effect on transcript elongation, addition immediately after the nucleotides merely restricted transcription to a single round (Fig. 4B, lane 2). Addition of RECQL5 to such single-round transcription reactions significantly inhibited transcription whether it was added at 0 or 25 min (Fig. 4B, lanes 3 and 4), indicating that the protein does not need to be present during preinitiation complex assembly to exert its inhibitory effect (Fig. 4B, lanes 3 and 4). In contrast, RECQL5ID protein did not inhibit transcription to the same extent, further indicating that the observed inhibitory effect was specific (Fig. 4B, lanes 5 and 6).
Because the results above suggested a potential inhibitory effect for RECQL5 on an early step of RNAPII transcription, we next investigated whether it is capable of specifically inhibiting initiation. RNAPII can initiate transcription by using a dinucleotide primer and a radiolabeled third nucleotide around the normal transcription start site (15). This abortive transcription initiation assay has been used extensively to investigate the effect of different factors on the formation of the first phosphodiester bond (19). RNAPII transcription in this assay was initiated by the addition of the dinucleotide CpU and radiolabeled CTP. The respective RECQ proteins were added with the nucleotides after preinitiation complex assembly to test their effect specifically on formation of the trinucleotide transcript (Fig. 5A). As expected, RNAPII-initiated transcription in this assay was inhibited by α-amanitin (Fig. 5B, compare lanes 1 and 2). More importantly, although the effect was not dramatic, addition of RECQL5 and RECQL5D157A, but not RECQL5ID or RECQL1, consistently significantly inhibited the reaction (Fig. 5B, compare lanes 3–6 with lane 2).
FIGURE 5.
RECQL5 inhibits first phosphodiester bond formation by RNAPII. A, outline of the abortive initiation experiment. B, abortive transcription producing a trinucleotide transcript (indicated by an arrow) resolved by 25% denaturing PAGE. 0.9 pmol of the indicated RECQ protein was added after preinitiation complex formation, together with the nucleotides. The arrow indicates a 3-nt transcript. Quantification of the trinucleotide transcript, relative to the control (lane 2, set to 1), is indicated below the lanes.
RECQL5 Also Inhibits Transcript Elongation by RNAPII
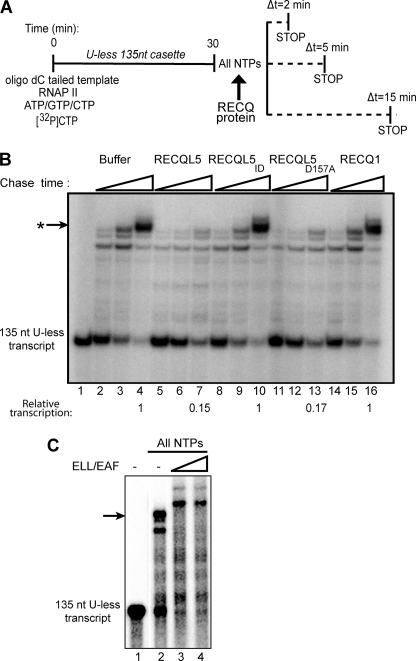
Although the results above show that RECQL5 can inhibit the initiation stage of RNAPII transcription, this does not rule out the possibility that it also affects transcript elongation. We therefore prepared RNAPII elongation complexes using an oligo(dC)-tailed DNA template containing a 5′ ‘U-less cassette’, which can be transcribed by RNAPII (without the GTFs) to produce a 135-nt transcript in the absence of UTP (14). Subsequently, upon addition of all nucleotides (including UTP), the stalled elongation complexes continue elongation to produce transcripts of increasing length with time, making it possible to test the effect of RECQL5 specifically on the process of transcript elongation (Fig. 6A). Addition of RECQL5 protein significantly inhibited elongation, so that instead of long transcripts, a more general background smear was observed (Fig. 6B, compare lanes 2–4 and 5–7). As expected from the data above, RECQL5ID protein was unable to inhibit transcript elongation, indicating that also in this assay (which did not contain GTFs) the observed effect is due to specific interaction of RECQL5 with RNAPII (compare lanes 5–7 and 8–10). Likewise, as observed previously, addition of RECQL1 had no effect (lanes 14–16), whereas the RECQL5D157A mutant was still able to inhibit transcript elongation (lanes 11–13). These inhibitory effects were in contrast to that observed when another RNAPII-binding factor, ELL/EAF, was added to the assay. As observed previously (14), this factor stimulated transcript elongation (Fig. 6C).
FIGURE 6.
RECQL5 inhibits RNAPII elongation. A, an outline of the RNAPII elongation experiment is shown. B, 135-nt RNAPII elongation complexes, stalled by UTP omission, were incubated with all NTPs and the indicated RECQ protein (0.9 pmol) for 2, 5, and 15 min, respectively. The resulting transcripts were resolved by 6% denaturing PAGE and analyzed by phosphorimaging. The arrow indicates the strongest, long transcript obtained. Quantification of this transcript, relative to the control (lane 4, set to 1), is indicated below the lanes. C, as in B, except that ELL/EAF was added, or not added, as indicated, and incubation with all NTPs was for 30 min.
DISCUSSION
Here, we report that RECQL5 inhibits reconstituted, promoter-driven RNAPII transcription in a dose-dependent manner. Moreover, by dissecting the different stages of RNAPII transcription, we show that RECQL5 inhibits transcript elongation and formation of the first phosphodiester bond independently of each other. Inhibition requires a stable interaction of RECQL5 with RNAPII, but not the ATPase/helicase activity of RECQL5.
RECQL5 Helicase as a Novel Inhibitor of RNAPII Transcription
The discovery of RECQL5 as a novel inhibitor of RNAPII transcription adds a potential tumor suppressor and regulator of genome integrity to the list of proteins that generally affect RNAPII transcription. It is important to point out that not all large proteins that interact with RNAPII are inhibitory. Indeed, the majority of RNAPII binding proteins that have been described either have no effect or stimulate transcription in assays such as those used in our studies. For example, RNAPII binding proteins such as elongin, ELL/EAF, TFIIF, and, under certain assay conditions, DSIF, have previously been found to stimulate transcript elongation in the systems used here (20–23), and the very large, multisubunit Mediator complex stimulates basal and activator-dependent transcription in related systems (24, 25). Other large RNAPII-binding factors, such as Elongator, Integrator, and the polymerase-associated factor (PAF) complex have little or no effect on transcription reconstituted on naked DNA templates (26, 27).
RECQL5 thus joins a short list of factors that includes TTF2 (28), DSIF (23), and NELF (29), which can negatively regulate transcription by directly affecting RNAPII. The exact mechanism by which DSIF and NELF inhibit transcription is still not completely understood, whereas TTF2 uses its ATP-dependent activity to disrupt RNAPII transcription elongation complexes (30). Interestingly, in contrast to TTF2, the ATPase activity of RECQL5 does not seem to be required for the inhibition of transcription reported here, although it may have a slight effect under certain conditions. RECQL5 helicase activity may be required for aspects of transcriptional inhibition not uncovered in our assays or for coupling transcription to other cellular processes such as recombination or replication or possibly only for its function in these latter processes (see below). Because the ATPase activity of RECQL5 is not essential for inhibition, RECQL5 may exert its effect primarily by binding to RNAPII and changing its conformation to a state that is less favorable for transcription. During initiation, such interaction might affect the stability of interactions with one or more GTFs or somehow change the configuration or activity of RNAPII itself to a state that is less favorable for phosphodiester bond formation. The fact that RECQL5 also inhibits RNAPII transcript elongation in the absence of additional factors seems to favor the second possibility. Future studies will focus on understanding the structural aspects of RECQL5-RNAPII association, which may help elucidate the precise mechanism by which RECQL5 inhibits transcription.
Transcription Inhibition by RECQL5 and Genome Stability
RECQL5 was previously proposed to suppress tumor formation by preventing undesired recombination events in vivo (7, 8), and recent evidence points to a role in replication-associated events (9). Although RECQL5 (8), and presumably other helicases like it, is capable of dissociating RAD51 filaments in vitro, the exact mechanism and cellular pathway that RECQL5 utilizes to suppress recombination remains unclear. Our recent discovery of RECQL5 as a novel RNAPII-associated factor in human chromatin suggested a potential role for RECQL5 in transcription (12). In the same experiments, RECQL5 was also found to associate with RAD51 and PCNA, confirming interactions that were previously reported by others (8, 31). The inhibitory role of RECQL5 in transcription reported here may be linked to its role in the suppression of undesired recombination events, or the protein may have separate functions in transcription and DNA recombination.
Interestingly, work primarily in budding yeast has suggested that transcription is associated with replication- dependent DNA hyper-recombination, primarily in the context of head-on-collision between the transcription and DNA replication machineries (32, 33). In apparent agreement with this idea, a reduction in RNA polymerase II initiation rate suppresses hyper-recombination (34). Transcription-associated, replication-dependent, DNA recombination also occurs in mammalian cells (35). The precise molecular mechanisms underlying such transcription-associated recombination is not yet clear, but given the results reported here, it is an intriguing possibility that RECQL5 is involved in inhibiting transcription (initiation and/or elongation) to somehow allow safe replication of transcribed regions of the genome. Alternatively, or additionally, RECQL5 might be involved in the regulation of a transcriptional program that is important for cell cycle progression and/or genome stability. In this model, RECQL5 represses genes encoding factors that increase genome instability if inappropriately expressed. As mentioned above, RECQL5 might also conceivably play distinct roles in the cell: one in the context of transcription and a separate one in maintaining genome stability by affecting DNA recombination. The finding that the ATPase/helicase activity of RECQL5 is not required for its inhibitory role in transcription could be construed as support for this hypothesis. In any case, the RECQL5ID mutant, lacking amino acids whose presence is important for normal RECQL5-RNAPII interaction but not the helicase activity of the protein, will be useful for testing whether the ability of RECQL5 to associate with RNAPII is required for its ability to suppress crossover events during DNA recombination (7, 8) and for its involvement in replication during genotoxic stress (9).
Taken together, the observations reported here establish a direct inhibitory role for RECQL5 in RNAPII transcription. Further studies will be needed to elucidate the mechanistic details of inhibition as well as its physiological consequence.
Acknowledgment
We thank members of the Svejstrup lab for helpful discussions.
This work was supported, in whole or in part, by NIGMS, National Institutes of Health Grant R37 41628 (to R. C. C). This work was also supported by Integrated Project DNA Repair Grant LSHG-CT-2005-512113 from the European Community, an in-house grant from Cancer Research UK (to J. Q. S.), the Bloom's Syndrome Foundation, and by the American Cancer Society (to Y. L.).
- RNAPII
- RNA polymerase II
- GTF
- general transcription factor
- DTT
- dithiothreitol.
REFERENCES
- 1.Bachrati C. Z., Hickson I. D. (2008) Chromosoma 117,219–233 [DOI] [PubMed] [Google Scholar]
- 2.Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., Ciocci S., Proytcheva M., German J. (1995) Cell 83,655–666 [DOI] [PubMed] [Google Scholar]
- 3.Yu C. E., Oshima J., Fu Y. H., Wijsman E. M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., Martin G. M., Mulligan J., Schellenberg G. D. (1996) Science 272,258–262 [DOI] [PubMed] [Google Scholar]
- 4.Kitao S., Lindor N. M., Shiratori M., Furuichi Y., Shimamoto A. (1999) Genomics 61,268–276 [DOI] [PubMed] [Google Scholar]
- 5.Hickson I. D. (2003) Nat. Rev. Cancer 3,169–178 [DOI] [PubMed] [Google Scholar]
- 6.Bohr V. A. (2008) Trends Biochem. Sci. 33,609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y., Lu X., Barnes E., Yan M., Lou H., Luo G. (2005) Mol. Cell. Biol. 25,3431–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y., Raynard S., Sehorn M. G., Lu X., Bussen W., Zheng L., Stark J. M., Barnes E. L., Chi P., Janscak P., Jasin M., Vogel H., Sung P., Luo G. (2007) Genes Dev. 21,3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y., Lu X., Zhou G., Barnes E. L., Luo G. (2009) Mol. Biol. Cell 20,114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimamoto A., Nishikawa K., Kitao S., Furuichi Y. (2000) Nucleic Acids Res. 28,1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawabe T., Tsuyama N., Kitao S., Nishikawa K., Shimamoto A., Shiratori M., Matsumoto T., Anno K., Sato T., Mitsui Y., Seki M., Enomoto T., Goto M., Ellis N. A., Ide T., Furuichi Y., Sugimoto M. (2000) Oncogene 19,4764–4772 [DOI] [PubMed] [Google Scholar]
- 12.Aygün O., Svejstrup J., Liu Y. (2008) Proc. Natl. Acad. Sci. U.S.A. 105,8580–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X., Liu Y. (2009) EMBO J. 28,568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong S. E., Banks C. A., Shilatifard A., Conaway J. W., Conaway R. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102,10094–10098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dvir A., Garrett K. P., Chalut C., Egly J. M., Conaway J. W., Conaway R. C. (1996) J. Biol. Chem. 271,7245–7248 [DOI] [PubMed] [Google Scholar]
- 16.Conaway J. W., Hanley J. P., Garrett K. P., Conaway R. C. (1991) J. Biol. Chem. 266,7804–7811 [PubMed] [Google Scholar]
- 17.Svejstrup J. Q. (2004) Biochim. Biophys. Acta. 1677,64–73 [DOI] [PubMed] [Google Scholar]
- 18.Hawley D. K., Roeder R. G. (1985) J. Biol. Chem. 260,8163–8172 [PubMed] [Google Scholar]
- 19.Dvir A. (2002) Biochim. Biophys. Acta. 1577,208–223 [DOI] [PubMed] [Google Scholar]
- 20.Shilatifard A., Lane W. S., Jackson K. W., Conaway R. C., Conaway J. W. (1996) Science 271,1873–1876 [DOI] [PubMed] [Google Scholar]
- 21.Aso T., Lane W. S., Conaway J. W., Conaway R. C. (1995) Science 269,1439–1443 [DOI] [PubMed] [Google Scholar]
- 22.Tan S., Aso T., Conaway R. C., Conaway J. W. (1994) J. Biol. Chem. 269,25684–25691 [PubMed] [Google Scholar]
- 23.Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G. A., Winston F., Buratowski S., Handa H. (1998) Genes Dev. 12,343–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Björklund S., Gustafsson C. M. (2005) Trends Biochem. Sci. 30,240–244 [DOI] [PubMed] [Google Scholar]
- 25.Malik S., Roeder R. G. (2005) Trends Biochem. Sci. 30,256–263 [DOI] [PubMed] [Google Scholar]
- 26.Kim J. H., Lane W. S., Reinberg D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99,1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baillat D., Hakimi M. A., Näär A. M., Shilatifard A., Cooch N., Shiekhattar R. (2005) Cell 123,265–276 [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y., Liu M., Spencer C. A., Price D. H. (2004) Mol. Cell 14,375–385 [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H. (1999) Cell 97,41–51 [DOI] [PubMed] [Google Scholar]
- 30.Xie Z., Price D. H. (1996) J. Biol. Chem. 271,11043–11046 [DOI] [PubMed] [Google Scholar]
- 31.Kanagaraj R., Saydam N., Garcia P. L., Zheng L., Janscak P. (2006) Nucleic Acids Res. 34,5217–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aguilera A. (2002) EMBO J. 21,195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prado F., Aguilera A. (2005) EMBO J. 24,1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jimeno S., García-Rubio M., Luna R., Aguilera A. (2008) Mol. Genet. Genomics 280,327–336 [DOI] [PubMed] [Google Scholar]
- 35.Gottipati P., Cassel T. N., Savolainen L., Helleday T. (2008) Mol. Cell. Biol. 28,154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]