Abstract
Proper DNA methylation patterns are essential for mammalian development and differentiation. DNA methyltransferases (DNMTs) primarily establish and maintain global DNA methylation patterns; however, the molecular mechanisms for the generation and inheritance of methylation patterns are still poorly understood. We used sucrose density gradients of nucleosomes prepared by partial and maximum micrococcal nuclease digestion, coupled with Western blot analysis to probe for the interactions between DNMTs and native nucleosomes. This method allows for analysis of the in vivo interactions between the chromatin modification enzymes and their actual nucleosomal substrates in the native state. We show that little free DNA methyltransferase 3A and 3B (DNMT3A/3B) exist in the nucleus and that almost all of the cellular contents of DNMT3A/3B, but not DNMT1, are strongly anchored to a subset of nucleosomes. This binding of DNMT3A/3B does not require the presence of other well-known chromatin-modifying enzymes or proteins, such as proliferating cell nuclear antigen, heterochromatin protein 1, methyl-CpG binding protein 2, Enhancer of Zeste homolog 2, histone deacetylase 1, and UHRF1, but it does require an intact nucleosomal structure. We also show that nucleosomes containing methylated SINE and LINE elements and CpG islands are the main sites of DNMT3A/3B binding. These data suggest that inheritance of DNA methylation requires cues from the chromatin component in addition to hemimethylation.
Proper DNA methylation patterns are essential for mammalian development and differentiation. More than three decades ago, de novo cytosine DNA methylation and its maintenance were proposed to exist in eukaryotic cells (29, 54); however, the molecular mechanisms for the generation and inheritance of methylation patterns are still poorly understood. DNA methyltransferases (DNMTs) DNMT1, DNMT3A, and DNMT3B primarily establish and maintain global DNA methylation patterns (39, 48). DNMT1 preferentially methylates hemimethylated DNA in vitro (7) and is tethered to replication foci during S phase (38). In contrast, DNMT3A and DNMT3B (DNMT3A/3B) have no preference for hemimethylated DNA (49) and are required for de novo methylation of genomic DNA (48). It has been thought that DNMT1 acts mainly as a “maintenance methyltransferase” during DNA synthesis and that DNMT3A and DNMT3B act as “de novo” enzymes. However, more recent studies indicate that DNMT1 may also be required for de novo methylation of genomic DNA (17, 30) and that DNMT3A/3B are also required for maintenance functions (11, 40, 55). Furthermore, the different DNMTs cooperate in maintaining the methylation of some regions of the genome, particularly repetitive elements (40, 53).
Recruitment of individual DNMT enzymes to different regions of chromatin in vivo, particularly to gene regulatory regions, may require interaction with auxiliary factors (28, 36). DNMT1, which is diffusely localized throughout nuclei in non-S-phase cells (38), is targeted to replication foci by interacting with proliferating cell nuclear antigen (PCNA) (15) and also physically interacts with UHRF1 (ubiquitinlike, containing PHD and RING finger domains 1) that binds to hemimethylated DNA (3, 4, 8, 27, 62). DNMT3 enzymes are usually found localized to heterochromatin regions in most transient-expression assays (5, 12). As genomic DNA in chromatin is packaged into nucleosomes which might limit the accessibility of target sites to the enzymes, the interaction of DNMTs with nucleosomes in a chromatin context is important for the regulation of genomic methylation.
Genetic and biochemical studies have provided many insights into the distinct and cooperative functions of the DNMT enzymes; however, few of these studies have addressed how they interact with chromatin in vivo. Recombinant DNMT1 and DNMT3 enzymes can methylate the CpG sites on nucleosomes assembled in vitro (26, 50, 56, 65). Recently DNMT3L has been found to connect DNMT3A2 to nucleosomes in embryonic stem cells (52). However, DNMT3L is expressed only during gametogenesis and embryonic stages (1, 9), suggesting that other mechanisms might be necessary for directing the enzyme to specific chromatin regions in somatic cells.
In the present study, we investigated how different DNMT enzymes interact with chromatin at the nucleosomal level in somatic cell lines. Micrococcal nuclease (MNase) treatment of nuclei in a low-ionic-strength buffer digests nucleosomal linker DNA regions, thereby minimizing the disruption of protein complexes on the nucleosomes. We prepared nucleosomes from partial or maximum MNase-digested nuclei and resolved them on sucrose density gradients to analyze their interactions with chromatin proteins. The results indicate that while DNMT1 interacts primarily with linker DNA, DNMT3A/3B enzymes interact strongly with nucleosomes containing methylated repetitive elements and also containing methylated CpG islands (CGIs) and may not require additional proteins for this strong binding. These data are particularly intriguing in that they provide insights into the mechanisms of the interaction of DNMTs with chromatin and maintenance of DNA methylation in somatic cells.
MATERIALS AND METHODS
Cell culture.
HCT116, a human colon cancer cell line, and 293T cells were maintained in McCoy's 5A medium and Dulbecco modified Eagle medium, respectively, containing 10% inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Puromycin was included in the culture medium at 3 μg/ml to maintain transfected 293T cells.
Expression vector construction.
A modified version of the pIRESpuro3 vector (Clontech), pIRESpuro/Myc, was constructed by ligating the Myc tag DNA sequence into the NheI-EcoRI site of the pIRESpuro3 vector. Human DNMT3A1, -3B1, and -3B2 cDNA were kindly provided by A. Riggs (the City of Hope). DNMT3A1, -3B1, and -3B2 cDNA were cloned into the EcoRI-NotI site of the pIRESpuro/Myc vector and used for expressing N-terminal Myc-tagged DNMTs in mammalian cells. Human ΔDNMT3B2 cDNA was amplified from pcDNA3/Myc-DNMT3B1 using PCR and ligated into the EcoRI-BstXI site of pIRESpuro/Myc-DNMT3B2. To generate ΔDNMT3B4 cDNA, the corresponding N-terminal sequence of ΔDNMT3B4 was amplified from HCT116 cDNA, which expresses ΔDNMT3B4, using reverse transcription-PCR and ligated into the EcoRI-MscI site of IRESpuro/Myc. All expression vector constructs were transfected into 293T cells using Lipofectamine 2000 (Invitrogen), and the cells stably expressing DNMTs were selected in the presence of 3 μg/ml puromycin for 3 weeks.
Nucleus preparation.
The nuclei were prepared according to the procedure described previously (23). Briefly, the cells were trypsinized and washed once with phosphate-buffered saline. The cells were then resuspended in ice-cold RSB buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2) containing protease inhibitors and kept on ice for 10 min before Dounce homogenization in the presence of 0.5% NP-40 to break up cell membranes. The nuclei were washed twice with RSB buffer plus the protease inhibitors (Roche) without the detergent.
Salt extraction of nuclei.
The nuclei from 5 × 106 cells were resuspended in 500 μl of ice-cold RSB buffer containing 0.25 M sucrose and protease inhibitors and various concentrations of NaCl and kept at 4°C for 5 min. The nuclei were then harvested by microcentrifugation, dissolved in sodium dodecyl sulfate (SDS) loading buffer, and subjected to Western blotting.
MNase digestion and sucrose density gradient centrifugation.
Purified nuclei (1 × 108) resuspended in 1 ml of RSB buffer containing 0.25 M sucrose, 3 mM CaCl2, and 100 μM phenylmethylsulfonyl fluoride were digested with 5 units of MNase (Worthington) for partial digestion or with 500 units of MNase for maximum digestion for 15 min at 37°C, and then the reaction was stopped with EDTA and EGTA (up to 10 mM). After microcentrifugation at 5,000 rpm for 5 min, the nuclear pellet was resuspended in 0.3 ml of the buffer (10 mM Tris-HCl [pH 7.4], 10 mM NaCl) containing 5 mM EDTA and EGTA, gently rocked for 1 h at 4°C, and followed by microcentrifugation to obtain soluble nucleosomes, which were then fractionated through a sucrose density gradient solution (5 to 25% sucrose, 10 mM Tris-HCl [pH 7.4], 0.25 mM EDTA) containing the indicated concentrations of NaCl at 30,000 rpm for 16 h at 4°C. For the control experiment, purified nuclei (1 × 108) were incubated in 650 μl of RSB buffer containing 300 mM NaCl for 5 min at 4°C. Nuclear extract (supernatant) was then taken out after centrifuging the incubated nuclei at 13,000 rpm for 10 min followed by loading 550 μl onto the sucrose gradients. Nucleosomes for a native chromatin immunoprecipitation (ChIP) assay were prepared in the digestion buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 8 mM MgCl2, 3 mM CaCl2) for 15 min at 37°C. The reaction was stopped with EDTA at a concentration up to 10 mM and left at room temperature for 20 min, before the soluble fraction of nucleosomes was collected. The ethidium bromide (EtBr) treatment of the mononucleosome samples was done by adding 20 mg/ml EtBr to the samples (final concentration of 300 μg/ml) and then incubating the samples at room temperature for 10 min before loading them onto the gradient. Sixteen fractions were taken from the centrifuge tube starting from the top of the tube.
Western blot analysis.
Proteins from the same volume of each fraction (150 to 200 μl) were concentrated by trichloroacetic acid precipitation, dissolved in SDS-β-mercaptoethanol loading buffer, and resolved on a 4 to 15% gradient SDS-polyacrylamide gel (Bio-Rad, Hercules, CA). Antibodies against H3 (ab1791), histone H1 (ab7789), DNMT3A (ab2850), histone deacetylase 1 (HDAC1) (ab7028), and heterochromatin protein 1α (HP1α) (ab9057) were purchased from Abcam Inc. (Cambridge, United Kingdom). Antibodies against EZH2 (ac22) were purchased from Cell Signaling Technology, Inc. (Danvers, MA), and antibodies against MeCP2 (07-013) were purchased from Upstate, Inc. (Charlottesville, VA). Antibodies against DNMT1 (sc-20701), DNMT3B (sc-10235), and PCNA (sc-56) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), while antibodies against the Myc epitope tag (05-724) were from Upstate (now Millipore, Billerica, MA) and anti-UHRF1 antibodies (catalog no. 612264) were from BD Biosciences. Image of individual proteins was visualized using the ECL detection system (Amersham Biosciences, Piscataway, NJ).
Quantification of DNA methylation levels.
The methylation-sensitive single-nucleotide primer extension assay was performed as described previously (71). Genomic DNA was prepared from 293T cells transfected with myc-DNMT3A/B expression vectors. The primers used for this assay are available on request.
ChIP assay.
ChIP assays were performed as described previously (42). The following antibodies were used: 10 μg of anti-Myc antibody (05-724; Upstate), anti-DNMT3A (ab2850; Abcam), and anti-CD8 (sc-32812; Santa Cruz Biotech) as a nonspecific control antibody. The primers and probes used are available on request.
For the native ChIP assay, we used DNMT3A-specific antibodies (ab2850), DNMT3B-specific antibodies (ab2851), and CD8-specific antibodies (sc-32812). The nucleosomes were incubated with antibodies overnight at 4°C in MNase digestion buffer containing 10 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, and 0.1% NP-40 and then incubated with protein A-agarose beads for 2 h. The beads were washed two times with 150 mM NaCl- and 300 mM NaCl-containing buffer containing 20 mM Tris-HCl (pH 7.4), 1 mM EDTA, and 0.1% NP-40. Proteins pulled down by different antibodies were analyzed by Western blot analysis.
To ascertain the possible role of nuclear RNA in the binding of DNMT3s to nucleosomes, native IP was performed using DNMT3A and CD8 antibodies on pooled polynucleosomal fractions from a partial digest sucrose gradient. Nucleic acids (DNA and/or RNA, if any) pulled down by the DNMT3A and CD8 antibodies were 32P end labeled, treated with DNase or RNase or not treated, and then subjected to the denaturing polyacrylamide-urea gel electrophoresis.
AP-PCR assay.
Sixty nanograms of DNA (input and unbound DNA) and two microliters of antibody-bound DNA were used for each arbitrarily primed PCR (AP-PCR) to make sure that the amounts of the input and unbound DNA were much more than the amount of bound DNA. Four random primers (GC-rich) were added to each PCR mixture. The detailed procedure of methylation-sensitive AP-PCR was previously described (40, 41).
RESULTS
DNMT enzymes associate with chromatin with different affinities.
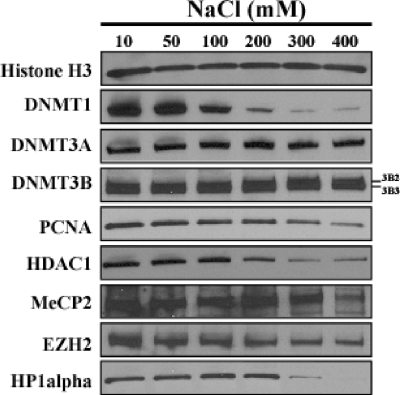
The DNMTs have been shown to be localized to distinct regions of chromatin (5, 12, 38). To determine whether they have different affinities for chromatin, we first incubated purified nuclei from HCT116 human colon cancer cells in the presence of protease inhibitors in buffers with increasing NaCl concentrations from 10 mM up to 400 mM. Using Western blots, we measured the amounts of various proteins remaining in the nuclei after washing them with buffers containing increasing salt concentrations (Fig. 1). Similar amounts of histone H3 remained in the extracted nuclei under all salt conditions as expected. Residual DNMT1 was reduced to about half at 100 mM salt, and the majority of it disappeared at 200 mM salt. Strikingly, both DNMT3A/3B protein levels remained almost constant within the nuclei up to 400 mM NaCl. The relative amounts of released proteins at 300 mM salt were estimated to be 100% for DNMT1, 6% for DNMT3B, and 9% for DNMT3A (data not shown). HCT116 cells are known to express at least two different isoforms of DNMT3B; DNMT3B2 is the top band, and DNMT3B3 is the bottom band (69). DNMT3B3 is known to be catalytically inactive (2, 13).
FIG. 1.
DNMT3A/3B, but not DNMT1, are strongly associated with chromatin in nuclei. Nuclei purified from 5 × 106 HCT116 cells were incubated in nondenaturing extraction buffers containing 10 to 400 mM NaCl in the presence of protease inhibitors for 5 min. The nuclei were purified by centrifugation, and the proteins remaining within the nuclei were resolved by SDS-polyacrylamide gel electrophoresis and probed by Western blotting with various antibodies. The positions of two DNMT3B isoforms (DNMT3B2 and DNMT3B3) are indicated to the right of the blots.
As DNMTs are known to interact with various chromatin-associating proteins, we wanted to test whether these proteins are involved in the strong interaction between DNMT3 and chromatin. To do this, we probed for several proteins that are known to interact with DNMTs, such as PCNA (15), HDAC1 (20, 21, 57), methyl-CpG binding protein 2 (MeCP2) (35), Enhancer of Zeste homolog 2 (EZH2) (66), and HP1α (22, 37, 63). The levels of PCNA and HDAC1 retained inside the nuclei decreased with increasing salt concentrations, leaving some residual protein at the higher salt concentrations. The amounts of MeCP2, EZH2, and HP1α showed constant levels of the protein up to 200 mM salt but decreased at higher salt concentrations.
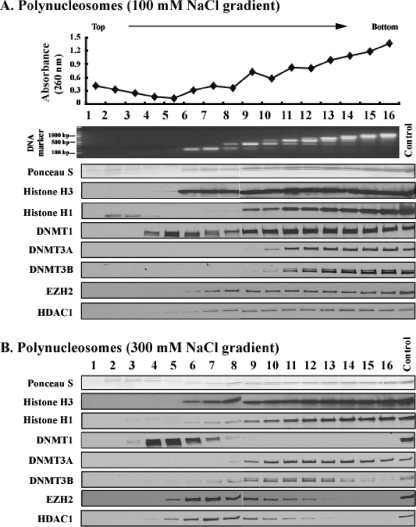
These differential interactions of DNMTs with chromatin prompted us to analyze how the enzymes are associated with nucleosomes. The nuclei were purified and partially digested with a low concentration of MNase, which cuts the linker DNA regions to generate nucleosomal fragments of various sizes under low-salt conditions (10 mM NaCl), so that no artificial rearrangements of nuclear proteins would occur. The digested nuclei were then incubated in low ionic buffer to collect diffused nucleosomes, which were loaded onto sucrose gradients and separated by ultracentrifugation. Since both DNMT1 and DNMT3 remained associated with chromatin at 100 mM salt (Fig. 1), we first used sucrose gradients containing 100 mM NaCl (Fig. 2A). The relative amounts and sizes of DNA in consecutive gradient fractions increased from fractions 6 to 16, and mononucleosomal DNA fragments were enriched largely in fractions 6 and 7. Dinucleosomes formed a main peak in fraction 9, and trinucleosomes formed a main peak in fractions 11 and 12, and so forth. The distribution of histones H3 and H1 matched that of the nucleosomal DNA fragments, and the bulk histone proteins stained with Ponceau S solution.
FIG. 2.
DNMT3A/3B are bound to polynucleosomes at high ionic strength. Nucleosomes released from nuclei partially digested with MNase at low ionic strength (20 mM) were resolved by ultracentrifugation on a sucrose density gradient (5% to 25%) containing 100 mM NaCl (A) and 300 mM NaCl (B). Gradients were fractionated into 16 aliquots numbered 1 to 16 starting from the top of the centrifuge tube. The absorbance of each fraction was read at 260 nm. DNA purified from each fraction was resolved by agarose gel electrophoresis and stained with EtBr. To probe the distribution of proteins in each fraction, Western blotting was performed with various antibodies. Ponceau S staining shows core histones transferred onto the membrane from the SDS-polyacrylamide gel. The control lanes on the gels were loaded with unfractionated nuclear extract loaded on the gels to monitor the quality of immunostaining of the membranes.
DNMT1 sedimented as a main peak at fraction 5 where there were no nucleosomes and was also distributed with bulk nucleosomes. DNMT1 in fraction 5 may correspond to the enzyme bound to the linker DNA digested by MNase, or it may also represent a fraction of DNMT1 not associated with chromatin. In contrast, DNMT3A and DNMT3B were tightly associated with nucleosomal fractions, and their distributions were quite similar to that of histone H1. EZH2 and HDAC1 also cosedimented with nucleosomes in the 100 mM salt gradient.
We next increased the salt concentration to 300 mM to determine the strength of association of these proteins with nucleosomes (Fig. 2B). The sedimentation profile of nucleosomes was not changed relative to the 100 mM salt-containing gradient (Fig. 2A). However, DNMT1 was now detected in fractions 4 to 7 only, separate from the fractions containing nucleosomes, showing that DNMT1 was not able to bind to polynucleosomes at high salt concentrations. In contrast, DNMT3A/3B enzymes were still associated with the nucleosomal fractions. Due to the relative abundance of polynucleosomes compared to mononucleosomes in a partial digest, the majority of the DNMT3A/3B enzymes were detected in polynucleosomal fractions 8 to 16 and not in mononucleosomal fractions 6 and 7 (Fig. 2B). Overexposure of the immunoblot showed small amounts of DNMT3A/3B sedimenting in fraction 6 and 7 also (data not shown). However, the patterns of DNMT3A and DNMT3B distribution differed from each other in 300 mM NaCl and did not completely mirror each other or the distribution of bulk nucleosomes. EZH2 and HDAC1, which cosedimented with nucleosomes at 100 mM salt, were separated from the polynucleosomes and showed main peaks in fractions 6 to 8 at 300 mM NaCl. These two proteins or complexes of these proteins, unlike DNMT3A/3B, were therefore not able to bind to polynucleosomes at high salt concentrations.
To confirm that EZH2 and HDAC1 which cosedimented with nucleosomes in fractions 6 to 8 at 300 mM NaCl were not physically bound to chromatin, we prepared nuclear extracts by eluting native proteins and protein complexes from nuclei incubated in high-salt buffer (300 mM) and subjected them to sucrose gradient fractionation at 300 mM NaCl (data available on request). Even in a chromatin-free environment, EZH2 and HDAC1 still sedimented in the same fractions, showing main peaks in fraction 6 to 8 similar to their sedimentation profiles in the presence of nucleosomes. This control experiment thus proves that EZH2 and HDAC1 do not bind to nucleosomes at high salt concentrations even though they sediment in the same fractions, possibly as part of large multiprotein complexes. Likewise, the sedimentation profiles of DNMT1 and PCNA were similar to those in the presence of polynucleosomes at 300 mM NaCl. DNMT3A could not be detected in any of the fractions (data not shown). These data indicate that the DNMT3A sedimentation profile is governed by the presence or absence of chromatin and that it is physically bound to nucleosomes at 300 mM salt (Fig. 2B).
DNMT1 interacts with the linker DNA, whereas DNMT3A/3B cosediment with intact nucleosomes.
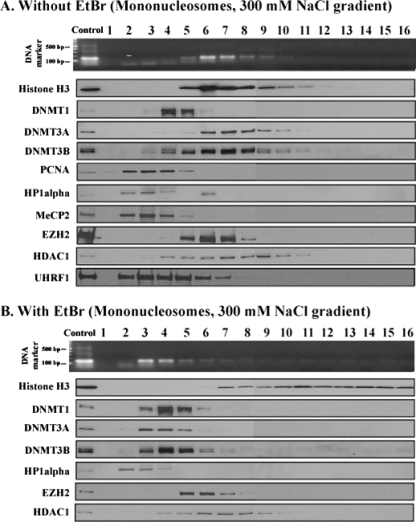
Next we prepared mononucleosomes consisting of approximately 150-bp DNA wrapped around an octamer of histones by more extensive MNase digestion of purified HCT116 nuclei and analyzed them on sucrose gradients at 300 mM NaCl. Mononucleosomes containing 150-bp DNA fragments and core histones peaked in fraction 6 (Fig. 3A). DNMT1 was mostly present in fraction 4, and UHRF1 formed a peak at fraction 3, both separated from the main nucleosomal peak.
FIG. 3.
DNMT3A/3B binding requires an intact nucleosomal structure. Mononucleosomes released by extensive digestion with MNase were resolved on a sucrose density gradient (5% to 25%) containing 300 mM NaCl. Mononucleosomes were incubated in the absence (A) or presence (B) of 300 μg/ml EtBr for 10 min at room temperature before they were loaded onto the gradients. The gradients were fractionated and analyzed as described in the legend to Fig. 2.
We tested whether DNMT1 could interact with mononucleosomes on 100 mM NaCl-containing gradients and found that it dissociated from mononucleosomes even at low ionic strength (data not shown). Since DNMT1 cosedimented with polynucleosomes on the 100 mM NaCl gradient as shown in Fig. 2A, the results together suggest that DNMT1 may interact with linker DNA. The sedimentation properties of EZH2 and HDAC1 were not altered by the increased MNase digestion (compare Fig. 2B to 3A), strongly suggesting that these proteins were not bound to nucleosomes in 300 mM NaCl. In contrast, both DNMT3A/3B localized to peaks in fraction 7, suggesting that DNMT3A/3B must be bound to mononucleosomes rather than the linker DNA and that their presence altered the sedimentation of bound nucleosomes by one fraction relative to bulk mononucleosomes. Also, the sedimentation profiles of DNMT3A/3B show a marked change when the extent of MNase digestion was altered from partial to maximum (compare Fig. 2B to 3A). MNase is a DNase that specifically targets linker DNA for digestion and has no effect on proteins, so it would not be expected to alter the distribution of DNMT3A/3B unless the enzymes were physically associated with chromatin.
EtBr disrupts nucleosomal structure by intercalating into DNA within the nucleosomes (43, 44), but it does not interfere with protein-protein interactions (37, 47). Mononucleosomes were incubated with EtBr prior to being loaded onto sucrose gradients containing 300 mM NaCl (Fig. 3B). DNA fragments of 150 bp were found mainly in fractions 3 and 4, whereas the histone proteins were detected in fractions 7 to 16, probably due to aggregation (44). The distributions of DNMT1, HP1α, EZH2, and HDAC1 were not affected by EtBr, again suggesting that they were not strongly bound to nucleosomes and were freely sedimenting, possibly as complexes with other proteins on the gradients. However, the distributions of DNMT3A/3B changed dramatically, and these enzymes were enriched mainly in factions 3 to 5, which contained the majority of the DNA but not the histone components of the nucleosome. These data show that when the histone-DNA interactions within the nucleosome are disrupted by EtBr, the DNMT3A/3B enzymes dissociate from the histone proteins and appear to cosediment with the DNA component. The data demonstrate again that the DNMT3A/3B enzymes are physically bound to nucleosomes but not directly binding to H3 and not simply cosedimenting with them on the gradients. Although we did not test whether DNMT3A/3B could maintain their interactions with EtBr-intercalated DNA, other studies have shown that these enzymes bind to DNA (10, 70) and that DNMT3A has a higher affinity for mononucleosomes than naked DNA in vitro does, whereas DNMT1 has the opposite tendency (56).
As DNMT3A and DNMT3B can form heterocomplexes (34), we also tested whether the nucleosomal binding of one enzyme requires the presence of the other. We performed the same experiment using a DNMT3B-deficient HCT116 cell line (HCT116 DKO) (53) and found that DNMT3A cosedimented with mononucleosomes in the absence of DNMT3B, indicating that the interaction between DNMT3A and DNMT3B are not required for their nucleosomal binding (data not shown). PCNA, HP1α, and MeCP2 sedimented separately from both mononucleosomes and DNMT3A/3B. EZH2 and HDAC1 behaved very similarly to how they sedimented on the polynucleosome gradient (Fig. 2B). These results show that these nonhistone proteins are not involved in the stable complex between DMT3A/3B and nucleosomes.
Stable complex formation of DNMTs with chromatin after 5-aza-CdR treatment.
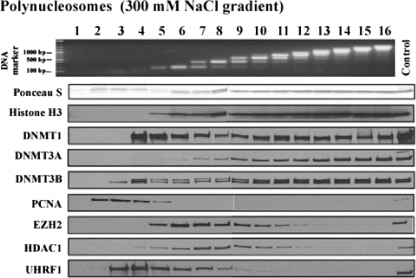
Incorporation of 5-aza-2′-deoxycytidine (5-aza-CdR) into replicating DNA leads to the formation of stable complexes between DNA and DNMTs by trapping them onto the DNA via the formation of covalent bonds which subsequently leads to gradual genomic demethylation (14, 18, 45, 58). It is important to note here that 24 h of treatment of cells with 5-aza-CdR mainly results in accumulation of high levels of hemimethylated DNA but does not cause complete genomic demethylation (46). The observation of relatively weak binding of DNMT1 to chromatin led us to examine the effects of drug treatment on the interactions of the enzyme with nucleosomes. The cells were cultured in the presence of 5-aza-CdR for 24 h before nucleus preparation, and the nucleosomes were resolved on a 300 mM NaCl-containing gradient after a partial MNase digestion (Fig. 4). DNMT1 cosedimented with nucleosomes on the high-salt gradient, indicating that it formed a complex with nucleosomal structures after 5-aza-CdR treatment due to its trapping onto the DNA by the incorporated drug (Fig. 4). UHRF1 was not trapped on nucleosomes. Thus, even though it may flip 5-methylcytosine out of double-stranded DNA (3, 4, 27), it is not required for the anchoring of DNMT1 to 5-azacytosine-containing DNA.
FIG. 4.
DNMT1 forms a stable complex in the linker DNA region with 5-aza-2′-deoxycytidine. HCT116 cells were cultured in the presence of 1 μM 5-aza-CdR for 24 h, followed by nucleus preparation. Polynucleosomes released from the nuclei digested with MNase were resolved through a sucrose density gradient (5% to 25%) containing 300 mM NaCl. DNA preparation and Western blotting were performed as described in the legend to Fig. 2.
DNMT3A/3B enzymes were still stably associated with nucleosomes possibly because of their inherent binding affinities and additionally due to their trapping by the incorporated drug (Fig. 4). However, DNMT3B, but not DNMT3A, was also detected in fraction 4 after drug treatment, suggesting that DNMT3B may have been trapped on the linker DNA region which was subsequently digested and released by MNase digestion. This observation is compatible with observations that mouse DNMT3B, but not DNMT3A, is processive in its activity (24, 25) which might give the enzyme the capacity to methylate a wider region during replication. On the other hand, it is also possible that an altered chromatin structure associated with 5-aza-CdR treatment may cause the release of some DNMT3B from the nucleosomes. The distributions of PCNA, EZH2, and HDAC1 were very similar to those observed in gradients without drug treatment (Fig. 2B), indicating that 5-azacytosine did not affect their sedimentation.
The N-terminal region of DNMT3B is essential for the strong nucleosomal association.
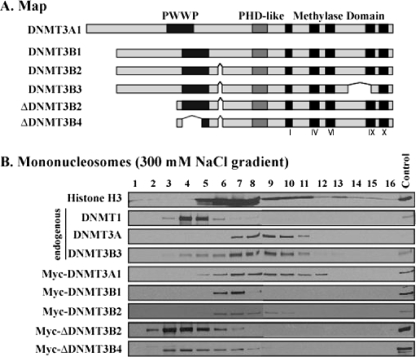
DNMT enzymes have highly conserved C-terminal catalytic methylase domains and unique N-terminal regions. DNMT3A/3B contain PWWP and PHD-like domains in their N-terminal regions (Fig. 5A) which might have a role in anchoring the enzymes to the nucleosomes. We expressed various Myc-tagged fusion proteins of DNMT3A/3B enzymes in human 293T cells and tested the distribution of DNMT3A/3B in mononucleosomal digests in 300 mM NaCl sucrose gradients (Fig. 5B). The endogenous DNMT1 and DNMT3A/3B showed distribution patterns similar to those of HCT116 cells, indicating that the strong nucleosomal association of DNMT3A/3B takes place in both cell types and is not due to potential cell-type-specific interactions. Full-length Myc-tagged DNMT3A/3B also showed that strong association with mononucleosomes; however, neither ΔDNMT3B2 nor ΔDNMT3B4 was strongly anchored to mononucleosomes, indicating that the N-terminal region of DNMT3B may play an essential role in its strong nucleosomal binding.
FIG. 5.
The N-terminal region of DNMT3B is necessary for strong nucleosomal binding. (A) Map of DNMT3A/3B isoforms showing the PWWP and PHD-like domains located in the N-terminal regions and the catalytic methylase domains in the C-terminal region. (B) Mononucleosomes from 293T cells transfected with expression vectors of various Myc-DNMT3A/3B deletion proteins were subjected to sucrose gradients containing 300 mM NaCl. Endogenous and exogenous enzymes on the gradient were detected by Western blot analysis using specific antibodies against endogenous protein and anti-Myc antibody. Gradients were analyzed as described in the legend to Fig. 2.
DNMT3A/B are enriched in methylated CGIs and repetitive DNA elements.
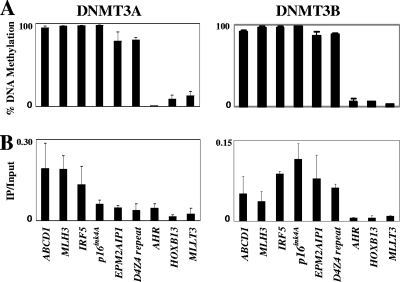
Previously, DNMT3A/3B were shown to be enriched on CpG islands when they are methylated (61), and the continued methylation of CGIs is relatively insensitive to DNMT1 depletion, suggesting roles for DNMT3A/3B in maintaining CGI methylation (17). These observations led us to examine whether DNMT3A/3B might bind preferentially to nucleosomes associated with methylated CGIs. With 293T cell lines transfected with various myc-tagged DNMT3A/3B (myc-DNMT3A/3B) derivatives available, we first determined the DNA methylation levels in various CGI regions and selected six highly methylated regions, including D4Z4 repeats and three unmethylated ones, for analysis (Fig. 6A and B). We next used ChIP with anti-myc antibodies and found that DNMT3A was enriched at the methylated ABCD1, MLH3, and IRF5 regions; however, it was less enriched in the other three methylated regions, CDKN2A, EPM2AIP1, and D4Z4, and was only marginally detectable in the three unmethylated regions, AHR, HOXB13, and MLLT3 (Fig. 6A and B). In contrast, DNMT3B was enriched in all of the six highly methylated regions, including its known target, D4Z4 (19, 69). These results indicate that all six highly methylated CGIs are enriched for DNMT3B and that DNMT3A occupies a subset of methylated CGIs, shared by DNMT3B. These data are consistent with the previous findings of Schlesinger et al. (61), who also found DNMT3A to be enriched on six different methylated CGIs and DNMT3B occupying a subset of these methylated CGIs bound by DNMT3A.
FIG. 6.
DNMT3A/3B are enriched in highly DNA methylated CpG islands. (A) The levels of DNA methylation in nine different CpG islands in 293T cells transfected with myc-DNMT3A1 or DNMT3B1 constructs were determined by the methylation-sensitive single-nucleotide primer extension assay. (B) ChIP assays were performed with anti-DNMT3A antibodies for the cells with myc-DNMT3A1 or anti-Myc antibody for the cells with myc-DNMT3B1. Values are the averages of at least triplicate determinations with standard errors (error bars) indicated.
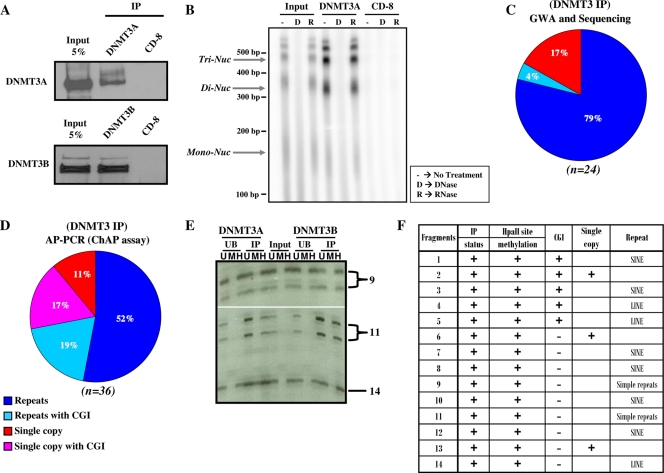
To gain a more global perspective, we next performed a native ChIP assay, followed by direct sequencing of the immunoprecipitated DNA. Nucleosomes isolated from MNase-digested nuclei were immunoprecipitated using DNMT3A/3B-specific antibodies. The specificities of the DNMT3A/3B antibodies were confirmed by Western blot analysis of the immunoprecipitated proteins. Both DNMT3A/3B antibodies showed specific enrichment of their target proteins compared to the CD8 control antibody (Fig. 7A). Furthermore, nucleic acids pulled down by the DNMT3A antibody were analyzed to ascertain whether nuclear RNA plays a role in the binding of DNMT3s to nucleosomes. We found that the DNMT3A antibody specifically pulled down DNA and not RNA, showing that RNA is not involved in DNMT3A binding to nucleosomes. Also, the DNA pulled down by DNMT3A antibody showed enrichment of nucleosomal DNA, indicating direct physical binding of DNMT3s to the nucleosomes. The CD8 control antibody showed no enrichment of either DNA or RNA and hence could not be used to provide a control for the evaluation of background in the native ChIP experiment (Fig. 7B). DNA pulled down by DNMT3A/3B antibodies was purified and used for whole-genome amplification. Sequence analysis of 24 DNA clones obtained from this amplified DNA sample showed that 83% of the clones contained SINE and LINE repeats of which 4% contained CGIs (Fig. 7C). The human genome is comprised of approximately 50% repetitive DNA (6, 64). Thus, the proportion of repeat sequences found in the amplified DNA sample is more than 1.5-fold higher than expected, suggesting that DNMT3A/3B preferentially associate with repetitive DNA elements.
FIG. 7.
DNMT3A/3B preferentially associate with methylated repetitive DNA elements and CpG islands, but not with nuclear RNA. (A) Western blot analysis was performed to analyze the proteins immunoprecipitated using DNMT3A, DNMT3B, and control CD8 antibodies. DNMT3A/3B antibodies specifically pulled down DNMT3A/3B proteins. (B) Nucleic acids (DNA and/or RNA, if any) pulled down by DNMT3A and CD8 antibodies from pooled polynucleosomal fractions from a partially digested sucrose gradient were 32P end labeled, treated with DNase or RNase or not treated, and then analyzed on a denaturing polyacrylamide/urea electrophoresis gel. The positions of trinucleosomes (Tri-Nuc), dinucleosomes (Di-Nuc), and mononucleosomes (Mono-Nuc) and molecular size markers are indicated to the left of the gel. (C) Distribution of different sequence classes in samples obtained by DNMT3A/3B IP. Sequence analysis of DNA fragments isolated by genome-wide amplification (GWA) of DNA precipitated by a mixture of DNMT3 antibodies shows preferential association of DNMT3A/3B with repetitive elements, such as SINES, LINES, etc. (D) Selective amplification of the DNA in the IP material through AP-PCR using GC-rich primers (ChAP assay [ChIP assay coupled with an arbitrarily primed PCR]) also shows the presence of repeats and single-copy CGIs. (E) Native ChIP assays were performed with DNMT3A/3B-specific antibodies followed by DNA purification from input, unbound (UB), and immunoprecipitated (IP) samples. The DNA samples were then digested with buffer alone (U), MspI (M), and HpaII (H) and subsequently used for an AP-PCR. The PCR products were resolved on a sequencing polyacrylamide gel, and part of the gel is shown as an example. The two strands of the same fragment sometimes resolved as separate bands on the gel due to the slight difference in their molecular weights. Fourteen informative bands were then extracted from the gel and sequenced. The numbers to the right of the gel denote the corresponding fragment number in panel F. (F) Properties of DNA fragments containing HpaII sites that were isolated from the sequencing polyacrylamide gel.
To confirm that DNMT3A/3B may also specifically bind to CGI regions, we adopted a nondirect approach and performed a methyl-sensitive AP-PCR experiment for DNA fragments enriched in the DNMT3A/3B-bound fraction. We used GC-rich random primers to preferentially amplify GC-rich CGI regions. This is an effective way to assess the methylation status of a number of different CGI sequences at the same time, since it selects CGI sequences in an independent manner (41). Sequence analysis of 36 DNA fragments obtained from the AP-PCR-amplified DNA sample showed that 71% of the sequenced fragments were repeats and 36% were CGIs (Fig. 7D). Fourteen IP-positive informative bands containing HpaII sites were identified, and we found that all 14 identified fragments were resistant to HpaII digestion, i.e., they all were methylated at internal HpaII (CCGG) sites (Fig. 7E and F). These data suggest that DNMT3A/3B associate with repeat elements, which is in agreement with previously published data (10, 11, 33). Taken together, these data strongly suggest that DNMT3A/3B preferentially bind to methylated genomic targets.
DISCUSSION
Our current results suggest that DNMT1 associates relatively weakly with chromatin, mostly at linker or nucleosome-free DNA regions possibly assisted by factors such as PCNA and/or UHRF1 (8, 15, 51). The observation of a “loose” association of DNMT1 with chromatin may be consistent with the highly dynamic, transient nature of the interaction of Dnmt1 with the replication machinery (59, 60) and its continuous loading onto constitutive heterochromatin during G2/M phase (16). As such, DNMT1 may be capable of methylating hemimethylated sites on the linker DNA missed during the replication process, possibly with the aid of UHRF1 (3, 4, 27, 62).
The strong interaction of DNMT3A/3B enzymes with nucleosomes is very striking. This “anchoring” is manifested even at 300 mM salt and alters the sedimentation properties of mononucleosomes. Our data suggest a strong physical interaction between DNMT3A/3B and nucleosomes, since the sedimentation properties of DNMT3A/3B were strongly influenced by the extent of MNase digestion, which as a DNase, would not directly alter the sedimentation behavior of a protein. Also, the fact that ethidium bromide treatment altered the position of DNMT3A/3B on the gradient without altering the sedimentation profile of other proteins like EZH2 (which are not physically bound to nucleosomes at 300 mM salt) argues for a strong physical association of DNMT3A/3B with nucleosomes. Since this anchoring does not require the presence of proteins such as EZH2 or HP1 which have previously been proposed to mediate the binding of these enzymes to chromatin (22, 66), it suggests that these enzymes directly interact with intact nucleosomal structures. Recent studies of mammalian germ cells have shown that DNMT3A2 and DNMT3L form a complex in vitro (31) and that DNMT3L targets nucleosomes, suggesting that the DNMT3A2/DNMT3L complex may bind to nucleosomes (52). It remains a question as to whether a similar structure might exist in somatic cells, where there is no expression of DNMT3L proteins. Indeed, a single nucleosome reconstituted in vitro can accommodate up to two molecules of DNMT3A1, supporting this idea (26, 32). Separation of the DNMT3A/3B from the histone components of the nucleosome by EtBr disruption suggests that the enzymes may depend on the interaction with DNA as well as with histones for stable anchoring. While we cannot rule out the possibility that other proteins may have roles in initially recruiting DNMT3A/3B to the target regions, none of those examined were required for anchoring. DNMT3B seems to require the N-terminal region of the enzymes for stable binding to nucleosomes, since ΔDNMT3B variants were easily dissociated from mononucleosomes by 300 mM salt. Since these variants have active de novo methylating activity, this lack of tethering might play a role in the generation of spurious methylation patterns such as those found in lung cancer (67, 68).
Our understanding of the exact mechanisms by which eukaryotic cells maintain cytosine methylation patterns still relies, to a large extent, on the seminal ideas of Riggs (54) and Holiday and Pugh (29). They suggested that patterns are established de novo during embryogenesis and then faithfully copied by a “maintenance” enzyme. DNMT1 clearly can play a maintenance role in that it has a marked preference for hemimethylated DNA (7), is localized to the replication fork (38), and is assisted in its DNA binding by PCNA (15) and UHRF1 (8). It seems reasonable to hypothesize that it “reads” the pattern on the parent strand and copies it to the daughter strand during DNA replication. However, the maintenance of global DNA methylation seems to require DNMT1 in cooperation with DNMT3A and/or DNMT3B, since knockout of these enzymes leads to a loss of most of the CpG methylation (11, 40, 53).
The biochemical approaches outlined in this paper may help explain how CpG-rich regions such as repeats and CGIs are kept methylated, since they suggest that DNMT3A/3B are highly localized to a subset of nucleosomes containing methylated CpG sites. We and others previously proposed that DNMT3A and DNMT3B may fill the gaps in the unmethylated CpG sites missed by DNMT1 (11, 40). We now propose that DNMT3A/3B associated with nucleosomes containing methylated CpG sites may have a role in maintaining methylation in these regions. DNMT1 may not be able to process a large number of hemimethylated sites generated in some regions such as CpG islands and some repeats due to the rapidity of the replication process (59), leaving some unmethylated CpG sites. DNMT3A/3B may still remain bound to the DNA after the passage of the replication fork and have the opportunity for methylating those unmethylated CpG sites as well as for carrying out their de novo methylation activity, before chromatin assembly takes place on the newly synthesized DNA strands. On the other hand, ΔDNMT3B variants may be aberrantly targeted, contributing to abnormal methylation patterns observed in tumorigenesis (67, 68).
Acknowledgments
We thank A. Riggs for generously providing DNMT3A/3B expression vectors. We also thank T. Miranda for great discussions and suggestions.
This work was supported by grant CA082422 from the National Cancer Institute.
Footnotes
Published ahead of print on 20 July 2009.
REFERENCES
- 1.Aapola, U., K. Kawasaki, H. S. Scott, J. Ollila, M. Vihinen, M. Heino, A. Shintani, K. Kawasaki, S. Minoshima, K. Krohn, S. E. Antonarakis, N. Shimizu, J. Kudoh, and P. Peterson. 2000. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics 65:293-298. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, A., I. Suetake, J. Miyagawa, T. Fujio, T. Chijiwa, H. Sasaki, and S. Tajima. 2001. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 29:3506-3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arita, K., M. Ariyoshi, H. Tochio, Y. Nakamura, and M. Shirakawa. 2008. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature 455:818-821. [DOI] [PubMed] [Google Scholar]
- 4.Avvakumov, G. V., J. R. Walker, S. Xue, Y. Li, S. Duan, C. Bronner, C. H. Arrowsmith, and S. Dhe-Paganon. 2008. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 455:822-825. [DOI] [PubMed] [Google Scholar]
- 5.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein, B. E., A. Meissner, and E. S. Lander. 2007. The mammalian epigenome. Cell 128:669-681. [DOI] [PubMed] [Google Scholar]
- 7.Bestor, T. H., and V. M. Ingram. 1983. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl. Acad. Sci. USA 80:5559-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bostick, M., J. K. Kim, P. O. Esteve, A. Clark, S. Pradhan, and S. E. Jacobsen. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760-1764. [DOI] [PubMed] [Google Scholar]
- 9.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. [DOI] [PubMed] [Google Scholar]
- 10.Chen, T., N. Tsujimoto, and E. Li. 2004. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 24:9048-9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, T., Y. Ueda, J. E. Dodge, Z. Wang, and E. Li. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, T., Y. Ueda, S. Xie, and E. Li. 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277:38746-38754. [DOI] [PubMed] [Google Scholar]
- 13.Chen, Z. X., J. R. Mann, C. L. Hsieh, A. D. Riggs, and F. Chedin. 2005. Physical and functional interactions between the human DNMT3L protein and members of the de novo methyltransferase family. J. Cell. Biochem. 95:902-917. [DOI] [PubMed] [Google Scholar]
- 14.Christman, J. K., N. Schneiderman, and G. Acs. 1985. Formation of highly stable complexes between 5-azacytosine-substituted DNA and specific non-histone nuclear proteins. Implications for 5-azacytidine-mediated effects on DNA methylation and gene expression. J. Biol. Chem. 260:4059-4068. [PubMed] [Google Scholar]
- 15.Chuang, L. S., H. I. Ian, T. W. Koh, H. H. Ng, G. Xu, and B. F. Li. 1997. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277:1996-2000. [DOI] [PubMed] [Google Scholar]
- 16.Easwaran, H. P., L. Schermelleh, H. Leonhardt, and M. C. Cardoso. 2004. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 5:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egger, G., S. Jeong, S. G. Escobar, C. C. Cortez, T. W. Li, Y. Saito, C. B. Yoo, P. A. Jones, and G. Liang. 2006. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl. Acad. Sci. USA 103:14080-14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egger, G., G. Liang, A. Aparicio, and P. A. Jones. 2004. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457-463. [DOI] [PubMed] [Google Scholar]
- 19.Ehrlich, M. 2003. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin. Immunol. 109:17-28. [DOI] [PubMed] [Google Scholar]
- 20.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 21.Fuks, F., W. A. Burgers, N. Godin, M. Kasai, and T. Kouzarides. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20:2536-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gal-Yam, E. N., S. Jeong, A. Tanay, E. Egger, A. S. Lee, and P. A. Jones. 2006. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gowher, H., and A. Jeltsch. 2001. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpA (sic) sites. J. Mol. Biol. 309:1201-1208. [DOI] [PubMed] [Google Scholar]
- 25.Gowher, H., and A. Jeltsch. 2002. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J. Biol. Chem. 277:20409-20414. [DOI] [PubMed] [Google Scholar]
- 26.Gowher, H., C. J. Stockdale, R. Goyal, H. Ferreira, T. Owen-Hughes, and A. Jeltsch. 2005. De novo methylation of nucleosomal DNA by the mammalian Dnmt1 and Dnmt3A DNA methyltransferases. Biochemistry 44:9899-9904. [DOI] [PubMed] [Google Scholar]
- 27.Hashimoto, H., J. R. Horton, X. Zhang, M. Bostick, S. E. Jacobsen, and X. Cheng. 2008. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 455:826-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermann, A., H. Gowher, and A. Jeltsch. 2004. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 61:2571-2587. [DOI] [PubMed] [Google Scholar]
- 29.Holliday, R., and J. E. Pugh. 1975. DNA modification mechanisms and gene activity during development. Science 187:226-232. [PubMed] [Google Scholar]
- 30.Jair, K. W., K. E. Bachman, H. Suzuki, A. H. Ting, I. Rhee, R. W. Yen, S. B. Baylin, and K. E. Schuebel. 2006. De novo CpG island methylation in human cancer cells. Cancer Res. 66:682-692. [DOI] [PubMed] [Google Scholar]
- 31.Jia, D., R. Z. Jurkowska, X. Zhang, A. Jeltsch, and X. Cheng. 2007. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449:248-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurkowska, R. Z., N. Anspach, C. Urbanke, D. Jia, R. Reinhardt, W. Nellen, X. Cheng, and A. Jeltsch. 2008. Formation of nucleoprotein filaments by mammalian DNA methyltransferase Dnmt3a in complex with regulator Dnmt3L. Nucleic Acids Res. 36:6656-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato, Y., M. Kaneda, K. Hata, K. Kumaki, M. Hisano, Y. Kohara, M. Okano, E. Li, M. Nozaki, and H. Sasaki. 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 16:2272-2280. [DOI] [PubMed] [Google Scholar]
- 34.Kim, G. D., J. Ni, N. Kelesoglu, R. J. Roberts, and S. Pradhan. 2002. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 21:4183-4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura, H., and K. Shiota. 2003. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J. Biol. Chem. 278:4806-4812. [DOI] [PubMed] [Google Scholar]
- 36.Klose, R. J., and A. P. Bird. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31:89-97. [DOI] [PubMed] [Google Scholar]
- 37.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 38.Leonhardt, H., A. W. Page, H. U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865-873. [DOI] [PubMed] [Google Scholar]
- 39.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 40.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22:480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang, G., J. C. Lin, V. Wei, C. Yoo, J. C. Cheng, C. T. Nguyen, D. J. Weisenberger, G. Egger, D. Takai, F. A. Gonzales, and P. A. Jones. 2004. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. USA 101:7357-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, J. C., S. Jeong, G. Liang, D. Takai, M. Fatemi, Y. C. Tsai, G. Egger, E. N. Gal-Yam, and P. A. Jones. 2007. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell 12:432-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMurray, C. T., E. W. Small, and K. E. van Holde. 1991. Binding of ethidium to the nucleosome core particle. 2. Internal and external binding modes. Biochemistry 30:5644-5652. [DOI] [PubMed] [Google Scholar]
- 44.McMurray, C. T., and K. E. van Holde. 1986. Binding of ethidium bromide causes dissociation of the nucleosome core particle. Proc. Natl. Acad. Sci. USA 83:8472-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michalowsky, L. A., and P. A. Jones. 1987. Differential nuclear protein binding to 5-azacytosine-containing DNA as a potential mechanism for 5-aza-2′-deoxycytidine resistance. Mol. Cell. Biol. 7:3076-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen, C. T., D. J. Weisenberger, M. Velicescu, F. A. Gonzales, J. C. Lin, G. Liang, and P. A. Jones. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 62:6456-6461. [PubMed] [Google Scholar]
- 47.Nielsen, A. L., M. Oulad-Abdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7:729-739. [DOI] [PubMed] [Google Scholar]
- 48.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 49.Okano, M., S. Xie, and E. Li. 1998. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 19:219-220. [DOI] [PubMed] [Google Scholar]
- 50.Okuwaki, M., and A. Verreault. 2004. Maintenance DNA methylation of nucleosome core particles. J. Biol. Chem. 279:2904-2912. [DOI] [PubMed] [Google Scholar]
- 51.Ooi, S. K., and T. H. Bestor. 2008. Cytosine methylation: remaining faithful. Curr. Biol. 18:R174-R176. [DOI] [PubMed] [Google Scholar]
- 52.Ooi, S. K., C. Qiu, E. Bernstein, K. Li, D. Jia, Z. Yang, H. Erdjument-Bromage, P. Tempst, S. P. Lin, C. D. Allis, X. Cheng, and T. H. Bestor. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552-556. [DOI] [PubMed] [Google Scholar]
- 54.Riggs, A. D. 1975. X inactivation, differentiation, and DNA methylation. Cytogenet. Cell Genet. 14:9-25. [DOI] [PubMed] [Google Scholar]
- 55.Riggs, A. D., and Z. Xiong. 2004. Methylation and epigenetic fidelity. Proc. Natl. Acad. Sci. USA 101:4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robertson, A. K., T. M. Geiman, U. T. Sankpal, G. L. Hager, and K. D. Robertson. 2004. Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem. Biophys. Res. Commun. 322:110-118. [DOI] [PubMed] [Google Scholar]
- 57.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 58.Santi, D. V., A. Norment, and C. E. Garrett. 1984. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc. Natl. Acad. Sci. USA 81:6993-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schermelleh, L., A. Haemmer, F. Spada, N. Rosing, D. Meilinger, U. Rothbauer, M. C. Cardoso, and H. Leonhardt. 2007. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 35:4301-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schermelleh, L., F. Spada, H. P. Easwaran, K. Zolghadr, J. B. Margot, M. C. Cardoso, and H. Leonhardt. 2005. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Methods 2:751-756. [DOI] [PubMed] [Google Scholar]
- 61.Schlesinger, Y., R. Straussman, I. Keshet, S. Farkash, M. Hecht, J. Zimmerman, E. Eden, Z. Yakhini, E. Ben-Shushan, B. E. Reubinoff, Y. Bergman, I. Simon, and H. Cedar. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39:232-236. [DOI] [PubMed] [Google Scholar]
- 62.Sharif, J., M. Muto, S. Takebayashi, I. Suetake, A. Iwamatsu, T. A. Endo, J. Shinga, Y. Mizutani-Koseki, T. Toyoda, K. Okamura, S. Tajima, K. Mitsuya, M. Okano, and H. Koseki. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450:908-912. [DOI] [PubMed] [Google Scholar]
- 63.Smallwood, A., P. O. Esteve, S. Pradhan, and M. Carey. 2007. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 21:1169-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smit, A. F., and A. D. Riggs. 1995. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 23:98-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takeshima, H., I. Suetake, H. Shimahara, K. Ura, S. Tate, and S. Tajima. 2006. Distinct DNA methylation activity of Dnmt3a and Dnmt3b towards naked and nucleosomal DNA. J. Biochem. (Tokyo) 139:503-515. [DOI] [PubMed] [Google Scholar]
- 66.Vire, E., C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, L. Morey, A. Van Eynde, D. Bernard, J. M. Vanderwinden, M. Bollen, M. Esteller, L. Di Croce, Y. de Launoit, and F. Fuks. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871-874. [DOI] [PubMed] [Google Scholar]
- 67.Wang, J., G. Walsh, D. D. Liu, J. J. Lee, and L. Mao. 2006. Expression of ΔDNMT3B variants and its association with promoter methylation of p16 and RASSF1A in primary non-small cell lung cancer. Cancer Res. 66:8361-8366. [DOI] [PubMed] [Google Scholar]
- 68.Wang, L., J. Wang, S. Sun, M. Rodriguez, P. Yue, S. J. Jang, and L. Mao. 2006. A novel DNMT3B subfamily, DeltaDNMT3B, is the predominant form of DNMT3B in non-small cell lung cancer. Int. J. Oncol. 29:201-207. [PubMed] [Google Scholar]
- 69.Weisenberger, D. J., M. Velicescu, J. C. Cheng, F. A. Gonzales, G. Liang, and P. A. Jones. 2004. Role of the DNA methyltransferase variant DNMT3b3 in DNA methylation. Mol. Cancer Res. 2:62-72. [PubMed] [Google Scholar]
- 70.Yokochi, T., and K. D. Robertson. 2002. Preferential methylation of unmethylated DNA by mammalian de novo DNA methyltransferase Dnmt3a. J. Biol. Chem. 277:11735-11745. [DOI] [PubMed] [Google Scholar]
- 71.Yoo, C. B., S. Jeong, G. Egger, G. Liang, P. Phiasivongsa, C. Tang, S. Redkar, and P. A. Jones. 2007. Delivery of 5-aza-2′-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 67:6400-6408. [DOI] [PubMed] [Google Scholar]