Abstract
MicroRNAs (miRNAs) have been implicated in various cellular processes. They are thought to function primarily as inhibitors of gene activity by attenuating translation or promoting mRNA degradation. A typical miRNA gene produces a predominant ∼21-nucleotide (nt) RNA (the miRNA) along with a less abundant miRNA* product. We sought to identify miRNAs from the simple chordate Ciona intestinalis through comprehensive sequencing of small RNA libraries created from different developmental stages. Unexpectedly, half of the identified miRNA loci encode up to four distinct, stable small RNAs. The additional RNAs, miRNA-offset RNAs (moRs), are generated from sequences immediately adjacent to the predicted ∼60-nt pre-miRNA. moRs seem to be produced by RNAse III-like processing, are ∼20 nt long and, like miRNAs, are observed at specific developmental stages. We present evidence suggesting that the biogenesis of moRs results from an intrinsic property of the miRNA processing machinery in C. intestinalis.
miRNA genes have been observed across the Eukarya1–5. A typical miRNA arises from the processing of a larger primary transcript (pri-miRNA) that is synthesized by RNA polymerase II, as seen for protein-coding genes6. The pri-miRNA transcript forms one or multiple fixed hairpin structures that are liberated by the RNase III enzyme Drosha7. The resulting ∼70-nt hairpins (pre-miRNAs) are further processed by a separate RNAse III enzyme, Dicer, which produces stable, mature miRNAs of 20–22 nt in length8–10.
Serial processing of pre-miRNAs is usually asymmetric, resulting in the production of a single, predominant miRNA arising from either the 5′ or 3′ arm of the pre-miRNA hairpin. In some cases, the opposite arm produces what is known as a miRNA* sequence that can reach appreciable steady-state levels but is less abundant than the miRNA11. The resulting miRNA and miRNA* can regulate distinct target mRNAs in a coordinated fashion12.
It has been proposed that conserved miRNA gene families provide a distinctive evolutionary signature and that the miRNA repertoire expands along with animal complexity13. To better understand the evolutionary history of miRNA genes among the chordate lineages, we performed a high-resolution study of small RNAs from the ascidian Ciona intestinalis, which belongs to the sister group of the vertebrates14. In contrast to other well-studied model organisms, C. intestinalis possesses a uniquely simplified repertoire of small RNA cofactors, consisting of single copies of Drosha, Pasha, Dicer, TRBP/PACT and Argonaute, and just two PIWI homologs11,14,15.
Here we report that numerous miRNA loci in C. intestinalis produce one or two discrete and stable ∼20-nt small RNA species from sequences immediately adjacent to the predicted pre-miRNA hairpins, in addition to conventional miRNA and miRNA* products. The biogenesis of these distinct RNAs is not explained by current models of miRNA processing. We present evidence that moRs are derived from an unanticipated activity of the C. intestinalis miRNAbiogenesis pathway.
RESULTS
Distinct small RNAs encoded by miRNA loci
We prepared small RNA (∼16–26-nt) libraries from C. intestinalis at various developmental stages, including unfertilized eggs, early embryos, late embryos and adults. High-throughput sequencing of the resulting cDNAs was performed with an Illumina 1G Genome Analyzer. Combining earlier studies with a recently described miRNA-discovery algorithm, we defined 80 miRNA loci in the C. intestinalis genome16–18. Detailed information regarding the encoded miRNAs and their potential target mRNAs is provided in Supplementary Tables 1–4 online and at the following website: http://flybuzz.berkeley.edu/cgi-bin/CionaMicroRNAs.cgi.
Half of these genes encode a single major product (the miRNA), along with a less abundant miRNA* sequence, as is typically seen in other organisms19,20. For example, the C. intestinalis (Ci) miR-125 gene (ortholog of the prototypic lin-4 miRNA in Caenorhabditis elegans) encodes a predominant miRNA that is stably expressed at all developmental stages examined21 (Supplementary Fig. 1 online). Ci-miR-125 is most highly expressed in adults, and at the adult stage a single clone of miR-125* is also detected.
Unexpectedly, the remaining half of C. intestinalis miRNA loci encode previously uncharacterized small RNAs, in addition to conventional miRNA and miRNA* products. This new class of RNAs arises from sequences located adjacent to the predicted pre-miRNA stem-loop, and we hereafter refer to them as ‘moRs’, for miRNA-offset RNAs. Only small RNAs with 5′ monophosphates and free 3′ hydroxyl groups can be cloned by the method used in this study (see Methods), although they could contain modifications on the 2′ oxygen, as seen for Piwi-interacting RNAs (piRNAs) and some miRNA species22. Most moR sequences are 19–20 nt in length, whereas C. intestinalis miRNAs range in size between 19 nt and 22 nt (Supplementary Fig. 2a online). Overall, moRs are considerably less abundant than miRNAs, but just ∼50% less abundant than miRNA* sequences (1,552 total reads and 3,353 total reads, respectively) (Supplementary Table 4b). In general, moRs show greater 5′ heterogeneity than miRNA or miRNA* sequences (Supplementary Fig. 2b). However, several abundantly expressed moRs, such as 5′ moRs 124-1 and 219, contain a rigid 5′-terminal nucleotide identity and show developmental regulation, suggesting that particular moRs may be under selective pressure, as has been suggested for the 5′ ends of miRNAs23.
It is possible that the C. intestinalis miRNA loci encoding moRs contain unique structural features, as compared to those that do not24. Global comparisons of base-pairing probabilities across the extended pre-miRNA loci in C. intestinalis revealed only modest structural differences between the two classes of miRNA loci (Supplementary Fig. 3 online). Overall, C. intestinalis miRNA loci maintain a similar base-pairing probability trace as those seen in Drosophila melanogaster, suggesting that C. intestinalis miRNA genes lack an intrinsic, species-specific structure. Similarly, there is no obvious difference in the size of the loop sequences in pre-miRs that produce moRs and those that do not (∼13 nt and ∼15 nt, respectively; Supplementary Fig. 4a online). In addition, we analyzed sequence motifs for all small RNAs cloned in this study. Whereas C. intestinalis miRNAs retained the expected 5′-uracil bias, no obvious motifs were apparent in the moRs25 (Supplementary Fig. 4b). Thus, it is currently unclear why they arise from particular miRNA loci.
Defining characteristics of moRs
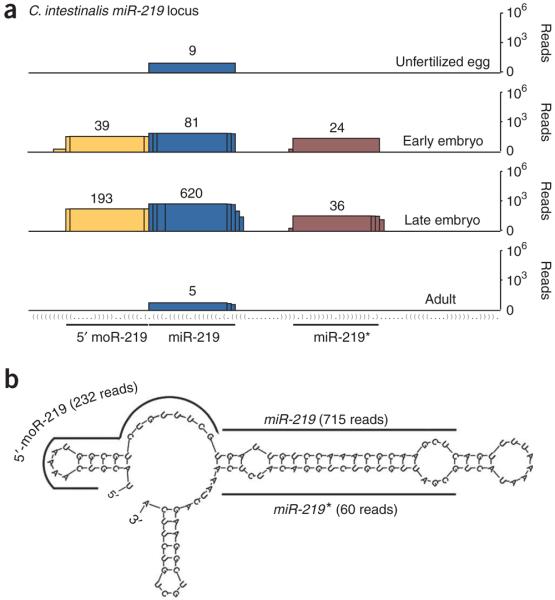
The C. intestinalis miR-219 gene (Ci-miR-219) encodes a predicted 57-nt pre-miRNA hairpin that is processed to produce miR-219 and miR-219*. In addition, a 5′ moR product (5′-moR-219) arises from sequences located immediately adjacent to miR-219 (Fig. 1a,b). The predominant miR-219 and miR-219* sequences are each ∼21 nt in length, whereas the 5′-moR-219 sequence is 20 nt (Fig. 1b).
Figure 1.
Developmental expression of small RNAs encoded by the C. intestinalis miR-219 locus. (a) Graphical depiction of small RNAs that map to the miR-219 locus at four developmental time points, indicated to the right. The histograms represent overlapping Illumina sequencing reads (numbered above stack) centered at each position (miRNA, blue; miRNA*, burgundy; 5′-moR, yellow). The y axis is plotted on a log scale. The secondary structure of the locus is presented in parenthetical format. (b) Locations of miRNA, miRNA* and moR sequences on the predicted secondary structure surrounding the pre–miR-219 hairpin. mFold was used to predict pre-miRNA secondary structure here and in the following figures45,46.
Like most miRNAs, nearly all 5′-moR-219 clones maintain an invariant 5′ end (223 of 232 total reads)23. Each of the three small RNAs observed at the miR-219 locus showed developmental regulation (Fig. 1a). Only miR-219 was detected in unfertilized eggs and adults (Fig. 1a), whereas both miR-219* and 5′-moR-219 were seen during embryogenesis.
In some cases, two distinct moRs are produced from a single miRNA gene, in addition to miRNA and miRNA* sequences (Fig. 2). The Ci-miR-124 locus encodes a pri-miRNA containing two tandem, but slightly different, ∼58-nt pre-miRNAs (Fig. 2b). The resulting miRNAs, miR-124-1 and miR-124-2, are identical, and the sequence shows peak expression, as evidenced by increased read counts (see miR-133 example below), in advanced-stage embryos (Fig. 2a). Both pre-miRNAs produce 5′ and 3′ moRs during embryogenesis (Fig. 2a). We observed the 3′ moR from the pre–miR-124-2 hairpin in both early embryos and late embryos, but the 3′ moR from the pre-miR-124-1 hairpin was detected only in early embryos. Moreover, 5′-moR-124 RNAs are considerably more abundant than the 3′-moR-124 RNAs, a result that is typical of the moRs and reminiscent of the processing of miRNA and miRNA* sequences, as well as processing of pri-miRNA 5′ and 3′ arms by Drosha26–28.
Figure 2.
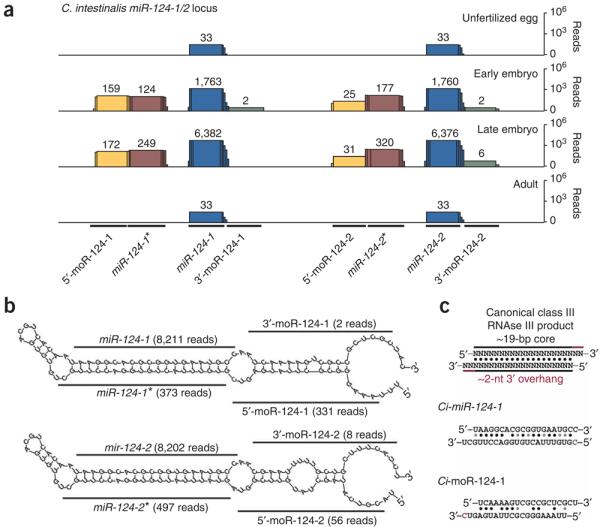
Coincident expression of 5′ and 3′ moR sequences from the C. intestinalis miR-124 locus. (a) Sequencing reads at each position of the miR-124 cluster are shown (miRNA, blue; miRNA*, burgundy; 5′-moR, yellow; 3′-miRNA, green). (b) miRNA and moR sequences aligned with sequence surrounding the predicted pre–miR-124-1 and pre–miR-124-2 stem-loop structures. A red ‘C’ in the pre–miR-124-1 structure indicates a shared base between multiple 5′-moR and miR-124-1* clones. (c) Standard class III RNAse III product is shown (above), depicting an ∼19-nt core of matched RNA bases, along with an ∼2-nt 3′ overhang. Aligned sequences are shown in the context of the predicted secondary structure of the pri-miRNA for miR-124-1 (top) and miR-124-1* (bottom), as well as 5′-moR-124-1 (bottom) and 3′-moR-124-1 (top). A shared base between loci is marked as a red “C”.
Notably, alignment of coincident 5′ and 3′ moR sequences from numerous miRNA loci suggests that they arise from RNAse III processing (∼21-nt duplexed RNAs with ∼2-nt 3′ overhangs)29 (Fig. 2c and Supplementary Fig. 5 online).
Despite the high prevalence of moRs associated with miRNA loci in C. intestinalis, we found that, overall, moR sequences are poorly conserved as compared to miRNAs between C. intestinalis and a related ascidian species, Ciona savignyi, and moRs are even less conserved than miRNA* sequences (Supplementary Fig. 4c). However, it has been noted that well-conserved small RNAs are expressed at higher levels than those lacking conservation19,30. This is true for most miRNAs when comparing C. intestinalis to C. savignyi (Supplementary Fig. 4c). Similarly, abundant moRs are also better conserved than those found at low copy number. Nonetheless, the general lack of conservation raises the possibility that moRs may represent unstable processing intermediates during the biogenesis of miRNAs. Such intermediates might be produced through a generic RNA-degradation mechanism that leaves behind spurious and variably sized small RNAs. However, as with miRNAs, the high copy number and near uniformity of clones at each locus suggests that moRs are produced mainly as ∼20-nt RNAs. To further address this point, we used northern assays to directly examine the expression and size distribution of miRNA and moRs in C. intestinalis embryos (see below).
Direct detection of moRs as discrete small RNAs
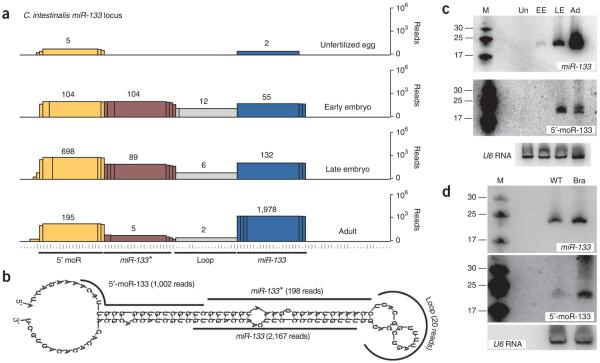
Vertebrate miR-133 genes are often part of a bicistronic pri-miRNA that also contains miR-1, and the two miRNAs work together to promote mesodermal fates31. A similar genomic linkage is seen in C. intestinalis, and previous studies have shown that the primary transcript containing miR-1 and miR-133 is selectively expressed in developing tail muscles during C. intestinalis embryogenesis32. The C. intestinalis miR-133 locus encodes separate miRNA, miRNA* and 5′ moR products (Fig. 3). miR-133 reads steadily increase during embryogenesis and reach peak levels in adults (Fig. 3a). We found that the 5′-moR-133 RNA is most abundant in late embryos and is present at an equal or higher read count than miR-133 and miR-133* at all embryonic stages examined.
Figure 3.
Direct detection of the 5′-moR-133 species. (a) Overlapping sequencing reads at each position along the miR-133 locus (miRNA, blue; miRNA*, burgundy; loop, gray; 5′-moR, yellow). (b) Alignment of sequenced reads on the predicted structure surrounding pre–miR-133. (c) Total RNA (∼30 μg per lane) was used for northern blots showing the ∼21-nt miR-133 (above) and 5′-moR-133 (middle) species throughout C. intestinalis development (M, size markers; Un, unfertilized eggs; EE, early embryos; LE, late embryos; Ad, adult animals). A northern blot for U6 RNA was used as a loading control (below). (d) As in c, comparing tailbud-stage C. intestinalis embryos that are unelectroporated (wild type, WT) or electroporated with a Ci-Brachyury enhancer:minimal Ci-miR-133 transgene (Bra). The Ci-Brachyury enhancer drives expression in the developing notochord33.
The levels of miR-133 and 5′-moR-133 detected in northern assays are in agreement with the sequencing frequencies obtained from the cDNA libraries (Fig. 3c). There is a progressive increase in the steady-state levels of miR-133 in unfertilized eggs, early embryos, late-stage embryos and adults (Fig. 3c, above). Similarly, the predicted 5′-moR-133 RNA was detected as a stable product (appearing as a doublet of ∼19–20-nt species in adults), with peak levels seen in late embryos. There was no indication of a smear or ‘ladder’ of higher- or lower-molecular-weight products, as would be expected if moRs represented incompletely degraded hairpin sequences or cleaved pri-miRNA transcripts. Moreover, ectopic expression of Ci-miR-133 directed by a Ci-Brachyury enhancer in the developing C. intestinalis notochord—the primitive chordate backbone—resulted in increased accumulation of both 5′-moR-133 and miR-133, indicating that expression of a discrete moR is correlated with that of the host miRNA transcript33 (Fig. 3d.)
Drosophila pri-miRNAs produce moRs in the Ciona tadpole
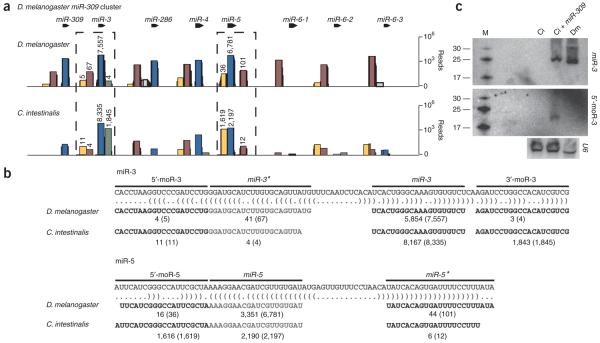
The preceding analysis suggests that moRs arise from an intrinsic property of the C. intestinalis small RNA–biogenesis machinery (see Discussion). To test this possibility, the miR-309 miRNA cluster (also known as ‘8-miR’) from D. melanogaster was selectively expressed in C. intestinalis34,35 (Fig. 4). We reasoned that the pri-miR-309 transcript would be more likely to produce detectable moRs when expressed in the C. intestinalis tadpole because it seems to produce such products, albeit rarely, in D. melanogaster (Fig. 4a).
Figure 4.
Ectopic expression of Drosophila pri-miRNAs can induce moR production in C. intestinalis embryos. (a) Small RNAs were cloned from 2–4-hour-old D. melanogaster Toll10b mutant embryos (above), which contain only mesodermal cell types, or tailbud-stage C. intestinalis embryos expressing the entire D. melanogaster pri–miR-309 cluster (below), and were subjected to Illumina sequencing. The resulting sequencing reads are shown at each position along the D. melanogastermiR-309 locus (miRNA, blue; miRNA*, burgundy; 5′-moR, yellow; intervening loop, gray). (b) The most abundant reads overlapping the respective regions of the miR-3 (above) or miR-5 (below) loci are shown. The number of clones matching the exact sequence depicted is shown in comparison to the overall number of clones overlapping that segment (in parentheses). (c) Northern blots showing miR-3 (above) and 5′-moR-3 (middle) in C. intestinalis and D. melanogaster embryos. For each well, ∼50 μg total RNA was analyzed from tailbud-stage C. intestinalis embryo that were unelectroporated (Ci), similarly staged C. intestinalis embryos electroporated with D. melanogastermiR-309 expression plasmids (Ci + miR-309), or 2–4-hour-old Toll10b embryos. Below is shown a northern blot in which a cross-reactive probe for U6 RNA was used as a loading control.
We separately placed the entire miR-309 cluster under the control of three different tissue-specific enhancers from C. intestinalis that direct expression in the notochord, epidermis and mesenchyme, respectively33,36. All three transgenes were coelectroporated into fertilized eggs, and the embryos were allowed to develop to the tailbud stage (after neurogenesis). Total RNA was extracted from these embryos and subjected to high-throughput sequencing or used for northern assays.
Drosophila melanogaster moRs are produced at high steady-state levels in C. intestinalis, and here we focused on the miR-3 and miR-5 genes within the miR-309 cluster. We detected only four 3′-moR-3 RNA reads in the D. melanogster embryo, whereas in C. intestinalis we observed nearly 2,000 copies (Fig. 4a,b). There is also a marked increase in the levels of the 5′-moR-5 RNA produced in C. intestinalis as compared with those in D. melanogaster. Nearly all copies of this moR RNA contain homogenous 5′ and 3′ termini (1,616 of 1,629 cloned copies are identical; Fig. 4b). In contrast, miR-3 was cloned at high frequency in D. melanogaster and C. intestinalis. Using northern assays, we identified similar levels of miR-3 in C. intestinalis and D. melanogaster embryos, a result that is consistent with the similar number of reads detected by sequence analysis. However, using a specific 5′-moR-3 hybridization probe, we detected a discrete band, without any obvious intermediate products, only in C. intestinalis embryos ectopically expressing the miR-309 cluster (Fig. 4c ).
There is no obvious correlation between the efficiency of moR biogenesis and the size of the loop sequence in the pre-miRNAs or conservation of other features. For example, the pre-miRNAs encoding miR-3 and miR-5 contain loops of 13 nt and 18 nt, respectively, but nonetheless produce similar yields of moRs. These experiments clearly demonstrate that the stable expression of moRs is an intrinsic feature of the C. intestinalis small RNA–processing machinery.
DISCUSSION
We have presented a high-resolution analysis of small RNAs during the development of the simple chordate, C. intestinalis. In the course of documenting 80 C. intestinalis miRNA genes, a distinct species of small RNAs was found to arise from sequences immediately 5′ and 3′ of the expected miRNA and miRNA* products. We have termed these small RNAs moRs (miRNA-offset RNAs).
moRs arise from ∼50% of the detected miRNA loci in C. intestinalis. However, there is no obvious sequence or structural difference between those miRNA loci that produce moRs and those that do not. This observation raises the possibility that moRs might reflect an intrinsic property of the small RNA–biogenesis machinery in C. intestinalis (see below). It is currently unclear why this machinery fails to produce moRs from half of the C. intestinalis miRNA genes and why there is differential accumulation of individual moRs during C. intestinalis development.
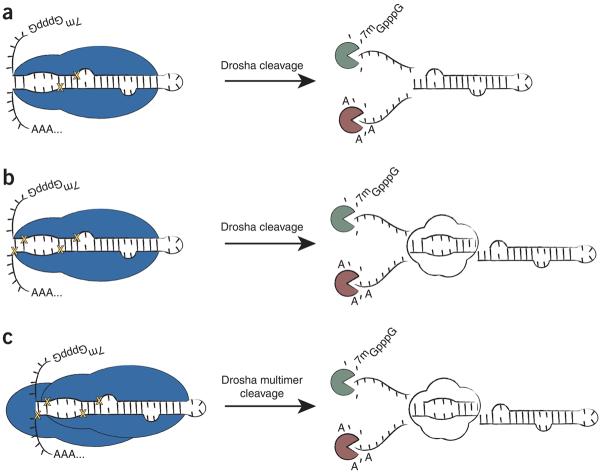
Putative moR products are seen in D. melanogaster and mouse embryonic stem cells, although they are extremely rare19,37. It was suggested that they might arise as by-products from exonuclease digestion of pri-miRNAs. According to this view, the pre-miRNA stem-loop would be excised from the pri-miRNA by Drosha, followed by decapping and 5′-3′ degradation, leaving behind fortuitously cloned ∼21-mers near the base of the pre-miRNA (summarized in Fig. 5). We have presented evidence suggesting that this mechanism probably does not apply to the biogenesis of C. intestinalis moRs. These products are far more abundant in C. intestinalis as compared with D. melanogaster and mouse. Moreover, the most abundant moRs contain homogenous 5′ and 3′ termini, and northern assays did not detect intermediate cleavage products (a smear or ladder), as would be expected from such processive degradation (Figs. 3c,d and 4c).
Figure 5.
A speculative model for the biogenesis of moRs. (a) Previous analysis of D. melanogaster and mouse small RNAs suggested that pre-miRNA-proximal sequences (analogous to moRs) were by-products of exonucleolytic degradation following excision of the pre-miRNA hairpin by Drosha (Drosha is represented in blue and yellow crosses indicate where Drosha cuts). (b) moR production may result via excision of an ∼20-nt, imperfectly paired duplex RNA at the immediate base of the pre-miRNA stem-loop, following two concurrent or sequential cuts by a single Drosha molecule. (c) Alternatively, a multimeric complex containing at least two Drosha molecules could associate with a substrate pri-miRNA. Here each Drosha molecule would cleave the pri-miRNA at a distinct position, liberating the pre-miRNA, as well as the ∼20-nt moR duplex.
In C. intestinalis, distinct 5′ and 3′ moRs arise from sequences located between the bicistronic Ci-miR-124-1/2 pre-miRNAs and from an ectopically expressed D. melanogaster pri-miRNA cluster. It is difficult to reconcile the proposed exonucleolytic degradation model with the occurrence of such moRs, because this intervening region should be equally accessible to 5′-3′ and 3′-5′ exonucleases38,39. Once again, such processing would be expected to produce a range of small RNAs rather than the discrete products that are actually observed.
Altogether, the simplest explanation for the biogenesis of moRs is that they arise during Drosha processing of the pri-miRNA transcript. Drosha is a class II RNAse III enzyme containing two tandem RNAse III domains28,40. Following intramolecular dimerization of these domains, the enzyme cleaves the pri-miRNA substrate at a single site (two total phosphodiester bonds), releasing a 5′ and a 3′ product in addition to the pre-miRNA. Analysis of coincident 5′ and 3′ moRs from numerous miRNA loci (such as those arising near miR-124-1) suggests that they may be paired in a manner similar to products generated through a bona fide RNAse III-like mechanism. That is, the duplexed RNAs contain ∼2-nt 3′ overhangs, as seen for Dicer products29.
For a lone C. intestinalis Drosha molecule to produce moRs, the single processing center must cut in a processive fashion at two sites along the pri-miRNA substrate, which is inconsistent with the prevailing model for Drosha activity28. Interactions among Drosha molecules could reconcile this apparent discrepancy. Such a mechanism is suggested by the recent demonstration of multimerized human Drosha complexes28. Notably, mouse embryonic stem cells lacking Dicer show enriched levels of moR-like sequences, which are lost upon disruption of Drosha activity37.
It is possible that Drosha produces ‘double cuts’ in most or all organisms, not just C. intestinalis. However, the resulting moR RNAs may be subject to rapid degradation by an unknown pathway. Ciona intestinalis might have a modified version of this degradation pathway to produce high steady-state levels of moRs. Future studies will explore the mechanistic details of moR biogenesis and function in C. intestinalis development.
METHODS
Small RNA cloning and detection
We collected adult C. intestinalis animals from Half Moon Bay, California, and maintained them in an artificial seawater tank. We carried out fertilization, dechorionation and electroporations as previously described33. Total RNA was extracted from unfertilized eggs, cleavage stage, tadpole-stage embryos and adults using the miRVana miRNA Isolation Kit (Ambion). Small RNA cloning was carried out as previously described41. Basically, from ∼30 μg of total RNA, only 17–25-nt RNAs were size selected via 15% denaturing PAGE. The 3′‘modban-1’ adaptor (IDT) was ligated to the RNAs from this fraction with RNA ligase (Ambion) in ATP-free reaction buffer41, and appropriately ligated RNAs were size selected via 15% denaturing PAGE. The modified RNAs were subsequently ligated to a 5′ linker (Solexa linker) in the presence of RNA ligase and in reaction buffer with ATP. The resulting RNA library was reverse transcribed to a cDNA library with SuperScript II (Invitrogen). cDNA was amplified using Illumina sequencing–specific primers, and the resulting libraries were sequenced on an Illumina 1G Genome Analyzer. In parallel, small RNAs were extracted using TRIZOL (Invitrogen), cloned and sequenced, as above, from staged, 2–4-hour-old D. melanogaster Toll10b embryos34. Northern blotting assays were performed as described previously42.
We cloned the D. melanogaster miR-309 cluster by amplifying the locus from yw genomic DNA using pfuUltra High Fidelity polymerase (Stratagene) and the TOPO TA cloning system (Invitrogen).
Ci-Brachyury, Ci-FoxF and Ci-Twist enhancers were used to drive transgene expression in the C. intestinalis notochord, epidermis and mesenchyme, respectively33,36.
Primers used for amplification of the Ci-Twist enhancer were Ci-Twist-F (forward), 5′-ACCACAGCTTCTATTATATA-3′, and Ci-Twist-R (reverse), 5′-CATCGTGTGTTGATTGATTT-3′.
Probe sequences for the Ci-miR-133 northern assay were Ci-miR-133 (5′-CAGCTGGTTGAAGGGGACCAAA-3′), Ci-5′-moR-133 (5′-GACCGACACCCGCAATGTTT-3′) and Ci-U6 (5′-GTCATCCTTGCGCAGGGGCCATGCTA ATCTTCTCTGTATCGTTCC-3′).
The C. intestinalis miR-133 amplification primers were Ci-miR-133-F (forward), 5′-CGTTTTATACGGTTATATACAGG-3′, and Ci-miR-133-R (reverse), 5′-TATTTCCGACTACTGAGCG-3′.
The Drosophila miR-309 cluster amplification primers were Dme-8miR-F (forward), 5′-TGCAGACAAATGACGAATTGA-3′, and Dme-8miR-R (reverse), 5′-CCGACCCTTTCAGGTAACAA-3′.
The probe sequences for the Drosophila miR-3 northern assay were Dme-miR-3, 5′-TGAGACACACTTTGCCCAGTGAT-3′ and Dme-5′-moR-3, 5′-CAGGATCGGGACCTTAGGTG-3′.
Data analysis
The standard Illumina pipeline (GAPipeline-0.3.0) was used to extract sequenced reads. Nucleotide positions 1 to 26 were aligned to the C. intestinalis (JGI version 1.0) or D. melanogaster (version 4.3) genomes using ELAND, and for the calculation of position-specific error rates18,43. Supplementary Figure 6a online shows the average error rate, defined as the estimated probability of a base call being incorrect as a function of nucleotide position for each of the four lanes (libraries) studied. The error rate model for the Illumina pipeline was calibrated on the basis of uniquely aligned reads to the genome, and then applied to all reads. The average error rate (averaged over all reads) rises sharply beyond the twenty-first base, consistent with an assumption that the reads should be dominated by miRNA sequences of roughly 21 nt, as subsequent unaligned bases of the 3′ adaptor would be scored as low quality. Reads were trimmed so as to optimize the total nucleotide quality in a dynamic programming approach that produced trimmed reads such that the maximum acceptable error rate over the trimmed sequence is less than 10% (QPHRED = 10), the total quality of the read is optimized globally over all start and stop positions, and the resulting length is greater than or equal to 17 nt44.
The trimming procedure can be described formally as follows. An optimal trimming can be achieved by defining a penalty P associated with making an incorrect base call at a given nucleotide n. Using the position-specific error probability, en, one can define an expected score for a given nucleotide as sn = 1 ⋅ (1 – en)+P ⋅ en = 1 – (P +1) ⋅ en. The total expected score for a trimming of nucleotide sequence to start at position and end at position j is then given by:
One is then free to choose the penalty, such that the expected score is zero when the error rate is the maximum tolerated, so and any error rate greater than emax will produce a negative contribution to the score. A dynamic programming search then globally optimizes S(i, j) over all start and stop positions44. Further details of the data analysis rationale and methodology are available in the Supplementary Methods online. A meta-analysis of the distribution for all processed reads across a miRNA locus is presented in Supplementary Figure 7 online.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Tonkin of the Vincent J. Coates Genomics Sequencing Laboratory for assistance with high-throughput sequencing and general expertise, H. Melichar for critical reading of the manuscript and members of the Levine laboratory for discussions. B.H. is supported by an American Cancer Society Postdoctoral Fellowship. This work was funded by a grant from the US National Institutes of Health (34431) to M.L.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Accession codes. Gene Expression Omnibus: Small RNA sequencing data have been deposited with accession code GSE13625.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 5.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 6.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 7.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 9.Grishok A, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 10.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 12.Okamura K, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc. Natl. Acad. Sci. USA. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dehal P, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 15.Murphy D, Dancis B, Brown JR. The evolution of core proteins involved in microRNA biogenesis. BMC Evol. Biol. 2008;8:92. doi: 10.1186/1471-2148-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedlander MR, et al. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 17.Fu X, Adamski M, Thompson EM. Altered miRNA repertoire in the simplified chordate, Oikopleura dioica. Mol. Biol. Evol. 2008;25:1067–1080. doi: 10.1093/molbev/msn060. [DOI] [PubMed] [Google Scholar]
- 18.Prochnik SE, Rokhsar DS, Aboobaker AA. Evidence for a microRNA expansion in the bilaterian ancestor. Dev. Genes Evol. 2007;217:73–77. doi: 10.1007/s00427-006-0116-1. [DOI] [PubMed] [Google Scholar]
- 19.Ruby JG, et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark A, et al. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slack F, Ruvkun G. Temporal pattern formation by heterochronic genes. Annu. Rev. Genet. 1997;31:611–634. doi: 10.1146/annurev.genet.31.1.611. [DOI] [PubMed] [Google Scholar]
- 22.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both microRNAs and their miRNA strands in flies. Curr. Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 25.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 26.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 28.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr. Opin. Struct. Biol. 2007;17:138–145. doi: 10.1016/j.sbi.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Axtell MJ. Evolution of microRNAs and their targets: are all microRNAs biologically relevant? Biochim. Biophys. Acta. 2008;1779:725–734. doi: 10.1016/j.bbagrm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 2006;20:2728–2738. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 34.Biemar F, et al. Comprehensive identification of Drosophila dorsal-ventral patterning genes using a whole-genome tiling array. Proc. Natl. Acad. Sci. USA. 2006;103:12763–12768. doi: 10.1073/pnas.0604484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 36.Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134:3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- 37.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 39.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 41.Brennecke J, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 42.Haley B, Hendrix D, Trang V, Levine MA. simplified miRNA-based gene silencing method for Drosophila melanogaster. Dev. Biol. 2008;321:482–490. doi: 10.1016/j.ydbio.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Norden-Krichmar TM, Holtz J, Pasquinelli AE, Gaasterland T. Computational prediction and experimental validation of Ciona intestinalis microRNA genes. BMC Genomics. 2007;8:445. doi: 10.1186/1471-2164-8-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chapman J. Whole Genome Shotgun Assembly in Theory and Practice. Univ. California; Berkeley: 2004. pp. 50–51. PhD Thesis. [Google Scholar]
- 45.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of rna secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.