Abstract
Nuclear receptors are multi-domain transcription factors that bind to DNA elements from which they regulate gene expression. The peroxisome proliferator-activated receptors (PPARs) form heterodimers with the retinoid X receptor (RXR), and PPAR-γ has been intensively studied as a drug target because of its link to insulin sensitization. Previous structural studies have focused on isolated DNA or ligand-binding segments, with no demonstration of how multiple domains cooperate to modulate receptor properties. Here we present structures of intact PPAR-γ and RXR-α as a heterodimer bound to DNA, ligands and coactivator peptides. PPAR-γ and RXR-α form a non-symmetric complex, allowing the ligand-binding domain (LBD) of PPAR-γ to contact multiple domains in both proteins. Three interfaces link PPAR-γ and RXR-α, including some that are DNA dependent. The PPAR-γ LBD cooperates with both DNA-binding domains (DBDs) to enhance response-element binding. The A/B segments are highly dynamic, lacking folded substructures despite their gene-activation properties.
The nuclear hormone receptors are a large family of transcription factors that directly bind and respond to ligands including steroids, thyroid hormone, retinoids, cholesterol by-products, lipids and haem1–4. These receptors contain poorly conserved A/B regions that in some cases act as potent transcriptional activators, provide sites of protein phosphorylation or form direct interactions with other receptor domains or regulatory proteins5. A central and highly conserved DBD contains two zinc-binding sites and the architectural elements capable of sequence-specific binding to DNA5,6. Hydrophobic molecules bind to the LBD, repositioning helix 12 into an active conformation that promotes the recruitment of co-regulators5,7. The nuclear receptor coactivators, including members of the steroid receptor coactivator (SRC) family, contain LXXLL motifs that dock to LBDs8,9. There have been multiple structural studies of nuclear receptors involving either the LBD or DBD fragments alone6,7,10–12. However, there has been no successful visualization of any intact nuclear receptor. Consequently, there is little information about how different domains interface to impart complex physiological and pharmacological properties.
The PPARs, like many non-steroid members of the nuclear receptor family, function as obligate heterodimers with RXR13. PPAR-α, PPAR-β/δ and PPAR-γ are encoded separately, but have overlapping tissue expression patterns, and as a group coordinate the regulation of important metabolic pathways2,14,15. These receptors control cellular processes including regulation of lipid and carbohydrate metabolism. PPAR-γ, the best-studied member of the family, is expressed in both white and brown adipocytes and regulates adipocyte differentiation, lipid storage and release16. One class of PPAR-γ ligands, the thiazolidinediones, which includes the drug rosiglitazone, are effective insulin sensitizers, and have been shown to improve glucose uptake and lower hyperglycaemia and hyperinsulinaemia17–20. The PPARs are also potential therapeutic targets for atherosclerosis, inflammation and hypertension21.
To address how different domains of PPAR-γ and RXR-α polypeptides interact within and between their polypeptides, we performed a series of crystallographic studies using wild-type intact recombinant proteins. Both receptors’ ligands, including rosiglitazone for PPAR-γ and 9-cis-retinoic acid for RXR-α, the DNA response element (PPRE) and coactivator peptides were included to examine how all the components of the complex interact. We further examined additional structures to determine if distinct ligands could modulate the multiple domain–domain interfaces. To this end we studied the crystal structures of two related complexes in which a partial agonist and a suicide inhibitor of PPAR-γ were used instead of rosiglitazone22,23.
Overall organization
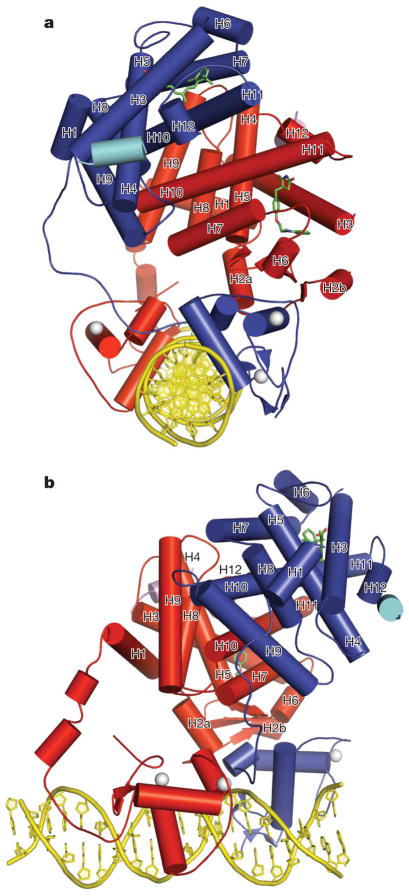
The structures we present here range in resolution from 3.1 to 3.2 Å, and were solved with single anomalous dispersion (SAD) phasing for the rosiglitazone structure, and molecular replacement for the GW9662 and BVT.13 complexes (Supplementary Table 1). Figure 1a, b shows the overall arrangement of the two receptors with respect to each other, their DNA-binding site, ligands and co-regulator (peptides) as seen in the rosiglitazone complex. As individual domains, the DBDs and LBDs of RXR-α, and the LBD of PPAR-γ, have folds highly similar to those previously reported for these isolated portions6,22,24. The structure of the PPAR-γ DBD, although previously unknown, conforms to other DBDs as expected from the high level of sequence conservation6,25. The rosiglitazone- and GW9662-containing structures belong to a different space group than the BVT.13-containing structure, and these crystal types display differences in packing. Nevertheless, the relative arrangements of the receptors, their domain–domain interactions, and the receptor interfaces with DNA and coac-tivators are similar in these crystal forms (Supplementary Fig. 1).
Figure 1. Overall structure of the PPAR-γ–RXR-α complex on PPRE.
a, b, Orthogonal views are shown in which RXR-α is blue and PPAR-γ is red. The ligands rosliglitazone and 9-cis-retinoic acid are shown in green, the Zn(II) ions are white, and the coactivator LXXLL peptides are in light blue and purple.
The hinge regions of nuclear receptors had not been previously seen beyond the short carboxy (C)-terminal extensions (CTEs) of some DBDs26. In PPAR-γ, the CTE forms a significant DNA interaction and is followed by two helical segments that reach the LBD (Fig. 1a, b). The RXR-α CTE forms one of the dimer contacts with PPAR-γ DBD, but is otherwise devoid of secondary structure and is more flexible. This flexibility may be an adaptive feature that allows RXR-α to be more promiscuous as a heterodimeric partner for numerous nuclear receptors, allowing their complexes to bind to direct repeats with multiple spacer sizes and half-site geometries. The PPAR-γ protein has its LBD and DBD closely positioned. In contrast, the RXR-α LBD and DBD are far apart from each other, with the space between them filled by the PPAR-γ LBD.
The PPAR-γ LBD has been the most exploitable drug site within the heterodimer, as ligands that bind to RXR-α are likely to inadvertently act on other RXR-α complexes. The PPAR-γ LBD is located prominently as the centrepiece of the complex, around which all other domains are positioned (Fig. 1). This arrangement leads to the consideration of whether some currently known or yet to be identified PPAR-γ ligands could impact other receptor domains. In contrast to the highly connected PPAR-γ LBD, the RXR-α LBD forms no contacts within the complex outside the PPAR-γ LBD. The two receptor polypeptides are asymmetrically positioned with respect to each other, whether viewed as intact polypeptides or as individual domains (Figs 1 and 2b).
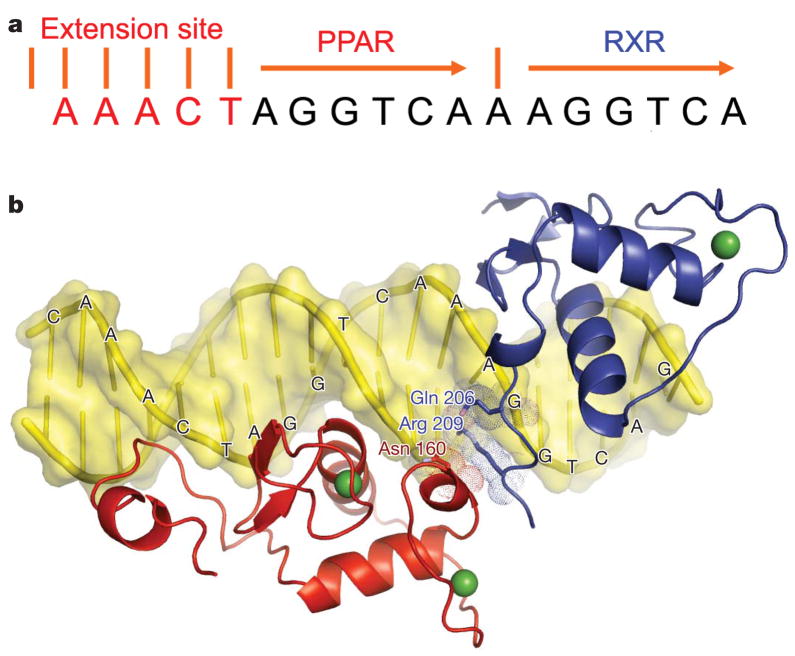
Figure 2. DNA-binding features of the complex.
a, The sequence of PPRE element containing direct repeats of the canonical AGGTCA half-site separated by one base pair (DR1) together with the upstream sequence AAACT specificity element. b, The DBDs and their C-terminal extensions are shown for PPAR-γ (red) and RXR-α (blue). These domains form a polar, DNA-dependent head-to-tail interaction through side chains that converge over the spacer element.
Binding to PPRE DNA
The PPRE element we used contains two copies of the consensus half-site AGGTCA, separated by a single base pair, forming the so-called DR1 element (Fig. 2A)27. This element contains an additional AAACT motif positioned 5′ to the DR127. The two half-sites are distinguishable by their 5′ and 3′ positioning on the DR1 (Fig. 2a, b). PPAR-γ and RXR-α display half-site selective binding that results in a polar arrangement of their DBDs, with PPAR-γ residing upstream of RXR-α. This polarity is in agreement with a previous biochemical study, and is seen in all three crystal structures described here27.
A major determinant of the polarity is the PPAR-γ CTE, which interacts significantly with the 5′ flanking sequence (described below). A second and more subtle determinant of the polarity is the requirement that both receptors form productive interactions between their DBDs. PPAR-γ and RXR-α DBDs interact within the minor groove of the spacer (Fig. 2b). The DNA dependence of DBD–DBD contacts is clear as the two proteins bury a modest 30 Å surface area between them. This size is negligible compared with standard dimer interfaces formed between protein pairs. The spacer DNA minor groove shelters the highly polar side chains of the interacting residues (Asn 160 from PPAR-γ, and Arg 209 and Gln 206 of RXR-α) from solvent. One interpretation of this arrangement is that the PPRE DNA allosterically contributes to its own binding by directly bridging the two receptor DBDs. We previously described the crystallographic arrangement of RXR–RAR on DR1, based on their isolated DBDs28. The observed polarity and DBD–DBD contacts are precisely the same for PPAR–RXR, as are the residues involved in these dimer interfaces (Supplementary Fig. 2).
Both DBDs have α-helices directly in register with the AGGTCA sequences (Supplementary Fig. 3). Hydrogen-bonding contacts are made between the DBD amino acids and the major groove features of the half-sites. PPAR-γ makes more base and phosphate backbone interactions than RXR-α (Supplementary Figs 3 and 4). The PPAR-γ hinge region (CTE portion) makes an extensive DNA interaction as well, binding to the upstream AAACT element (Supplementary Fig. 4). This interaction is similar to that of the Rev–Erb CTE with its AACT-binding sequence upstream of its DR2-binding site26,29. Both PPAR and Rev–Erb deeply embed the tetrapeptide RFGR from their CTEs into this DNA minor groove (Supplementary Fig. 4). DNA sequences are not clearly distinguishable from their minor groove side, yet in the case of PPREs the AAACT sequence is well conserved27. We showed previously that this conserved sequence provides the DNA deformability and minor groove distortions that allow recognition by RFGR sequences29.
Cooperation between receptor domains
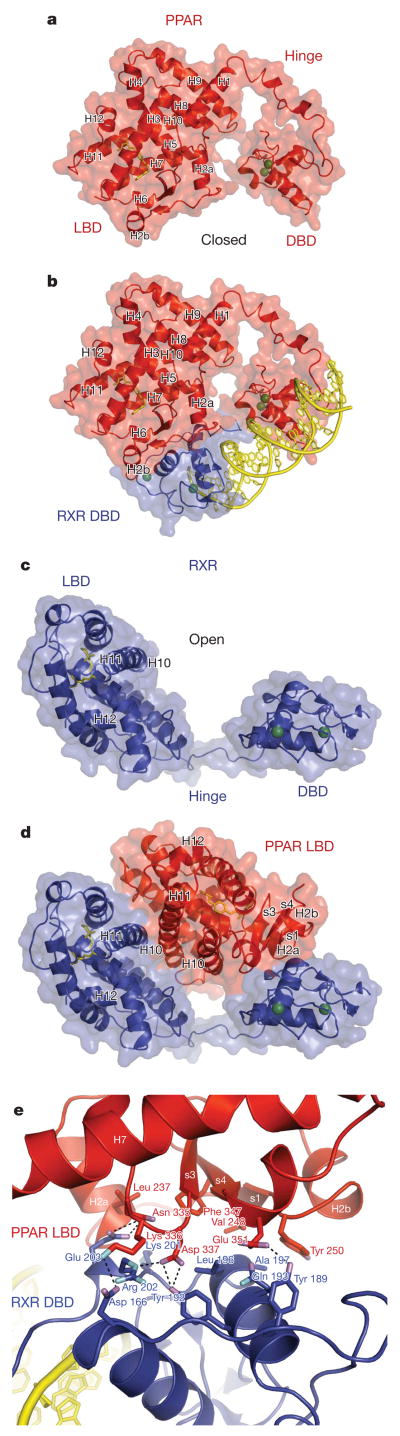
The DBD–DBD interface is only one of the three distinct hetero-dimerization surfaces between the receptors. Additionally, a dimer interface is formed by the two LBDs (Fig. 1a, b)24. This interface is DNA independent and buries 2,160 Å of solvent-accessible surface, using helices 7, 9 and 10 of each receptor. A third, previously unknown, heterodimerization interface is also observed between the PPAR-γ LBD and the DBD CTE region of RXR-α. The DNA dependence of this interaction is suggested in Fig. 3b, e. Electron density of all three of the heterodimerization interfaces is shown in Supplementary Figures 5–7.
Figure 3. Domain–domain interactions involving the PPAR-γ LBD.
a, The PPAR-γ LBD is closely positioned to the PPAR-γ DBD. b, The PPAR-γ LBD (red) interfaces with both receptor DBDs to enhance their DNA association. RXR DBD is in blue. c, The LBD and DBD of RXR-α, do not interact. d, PPAR-γ LBD bisects both RXR-α domains. The proximity of the PPAR-γ ligand to both RXR-α domains can be seen. e, The side-chain interactions between PPAR-γ LBD and RXR-α DBD in the rosiglitazone complex. The PPAR-γ LBD residue Phe 347 lies at the centre of the interface.
Figures 3c, d demonstrate how both the DBD and LBD of RXR-α are intersected by the PPAR-γ LBD. A feature unique to the LBD of PPARs is their β-strand elements S1, S2, S3 and S4, which face the RXR-α DBD (Figs 3b, d, e). The linchpin of this interaction appears to be PPAR-γ LBD’s Phe 347, which resides in strand S4 and forms hydrophobic interactions with RXR-α DBD (Fig. 3e). As the amino acids shown in Fig. 3e are conserved among both PPARs and RXRs, other PPAR family members are expected to form the same interface.
The PPAR-γ LBD contacts both receptor DBDs to stabilize their DR1 binding (Fig. 3b). The DNA-binding properties of intact nuclear receptors are typically of higher affinity than those of their isolated DBDs30. The current structure shows how regions outside the DBD can contribute to DNA recognition and affinity by interacting with DBDs. The PPAR-γ LBD also brings its own positively charged surface near the negatively charged DNA phosphodiester backbone. An example is Lys 240 from the PPAR-γ LBD, which has its basic side chain positioned in close proximity to the DNA backbone, indirectly contributing to favourable electrostatic interactions with DNA.
To test the idea that the PPAR-γ LBD interface with RXR-α DBD is contributing to the DNA binding of the complex, we made a point mutation at a key hydrophobic residue in PPAR-γ LBD, Phe 347. This residue is near the surface of the LBD, and distant from both the ligand-binding pocket and coactivator docking sites (Fig. 3e). Our DNA binding assays show that the Phe347Ala-mutant PPAR-γ negatively affects PPRE binding (Supplementary Figs 8 and 9). The weakened ability to bind DNA is further manifested in the receptors’ inability to activate transcription in response to rosiglitazone (Supplementary Fig. 10). These data demonstrate that a residue positioned on the surface of a receptor LBD can strongly influence DNA binding, confirming the ability of the PPAR-γ LBD, through domain–domain interactions, to affect the receptors’ DNA-binding properties.
A comparison of the ligand-binding pockets of the intact receptors with previous structurally defined LBDs alone did not reveal any unexpected differences. Functional PPREs almost exclusively consist of DR1 sites31. Other RXR heterodimers, which can use multiple response elements, are likely to show different types of domain–domain interaction, and possibly different modes of ligand binding, when bound to their different target DNAs. For example, the RAR–RXR complex displays a distinctly different behaviour for ligand binding and gene activation on DR5 versus DR1 elements32.
Effect of PPAR-γ ligands on receptor organization
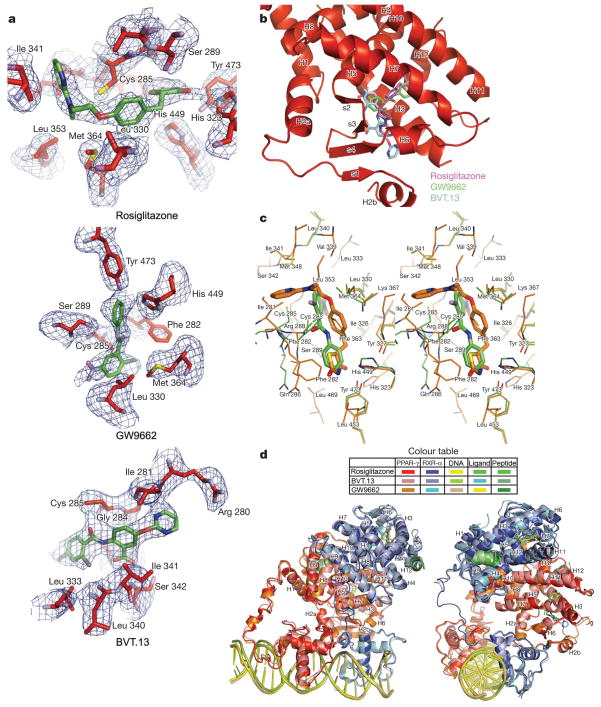
We considered whether different ligands could affect the domain–domain interactions. We selected two other PPAR-γ ligands that we believed would have distinct properties from rosiglitazone. The first was the antagonist GW9662, which can act as a suicide inhibitor of PPAR-γ23. A second ligand we used was BVT.13, which in its bound position was expected to have a significant reach into the unique β-strand region of the PPAR-γ LBD, the region strongly coupled to the RXR-α DBD22. In all three crystal structures, we confirmed by electron density that these ligands were occupying the pocket (Fig. 4a).
Figure 4. Effect of different ligands on the PPAR-γ–RXR-α complex.
a, Electron density (Fo −-Fc) omit maps around the PPAR-γ ligands rosiglitazone, BVT.13 and GW9662 from the their respective complexes. b, The three PPAR-γ ligands as a group occupy distinct portions of the receptor’s Y-shaped pocket. c, Comparison of the binding mode of rosiglitazone with GW9662 within the PPAR-γ pocket. GW9662 does not form a covalent bond with Cys 285. d, Superposition of crystal structures obtained with the different PPAR-γ ligands showing minor variations in overall domain organization. The colours are indicated for each complex.
As a group, rosiglitazone, GW9662 and BVT.13 gave rise to a ‘Y’shaped pocket (Fig. 4b). GW9662, which has been shown biochemically to be capable of forming a covalent bond with a cysteine in the PPAR-γ LBD pocket, resides in the pocket without having made the covalent bond in the intact PPAR-γ (Fig. 4c). Nevertheless, the positioning of the nitro-aryl chloride portion of GW9662 shows this ligand is poised to form a covalent adduct, given the proximity of its displaceable chloride group to the cysteine sulphur. The covalent bond could in principle form in a small population of PPAR-γ receptors that were not captured in our crystallization condition. BVT.13 has a longer reach and is inserted more directionally towards the β-strand segments of the PPAR-γ LBD (Fig. 4a, b). A previous study showed that this ligand could also dramatically influence the dynamics of the β-strand elements of PPAR-γ LBD22. With both the GW9662 and BVT.13 ligands, the overall conformation of the two receptors, including all of the domain–domain, receptor–DNA, and receptor–coactivator interactions, were not found significantly altered from those seen in the rosiglitazone complex (Fig. 4d).
Dynamic properties of PPAR-γ
The A/B segment of PPAR-γ is a potent transcriptional activator, contains a phosphorylation site and can interact with other proteins33–36. However, the A/B segments could not be visualized in any of the three complexes, presumably because of their high mobility. We examined their dynamic properties using amide hydrogen/deuterium exchange mass spectrometry (H/D-Ex)37,38. The H/D exchange rate is significantly slower in structured regions than unstructured portions. The H/D exchange levels of the intact PPAR-γ with no ligands, with rosiglitazone or with GW9662 were measured. The H/D exchange levels of a third complex were also measured which simultaneously contained rosiglitazone, RXR-α, 9-cis-retinoic acid and PPRE DR1, similar to our crystallographic samples but lacking peptide elements. The H/D-Ex analysis allowed us to judge the relative dynamic properties of segments along the PPAR-γ protein.
Figure 5a shows that DNA protected the amino-acid region 170–193 from exchange, the portion of PPAR-γ that inserts into the minor groove of the 5′ DR1 extension. The amino-acid region 434–442 in helix 10 and region 333–340 also showed slower exchange compared with the PPAR-γ protein without RXR-α. The amino-acid region 434–442 is involved in the LBD–LBD heterodimerization and region 333–340 interacts with RXR-α DBD (see residues 335–337 in Fig. 3e). Importantly, in all samples, the most rapidly exchanging region was the A/B portion. Sequence alignments further demonstrate that these receptors lack significant stretches of hydrophobic residues and/or a meaningful level of amino-acid conservation in their A/B regions to suggest the existence of a folded motif (Supplementary Figs 11 and 12).
Figure 5. The dynamic features of the receptors.
H/D-Ex data for intact PPAR-γ. From top, in a ligand-free state, with rosiglitazone, with GW9662, and with all of rosiglitazone, RXR-α, 9-cis-retinoic acid and PPRE DNA (RXR, etc.). Each horizontal colour block represents an analysed peptic fragment. The deuteration level of each peptide at each time point is colour-coded as in the insert. Each block contains five time points (from top, 15, 50, 150, 500, and 1,500 s).
Coactivator interactions
Both RXR-α and PPAR-γ assume the so-called active conformation on their LBD in all three structures. This conformation consists of both receptors having their helix 12 appropriately positioned by ligands to facilitate the docking of coactivator LXXLL motifs (Fig. 1a, b). In the context of the entire complex, the location of the coactivator peptides is distantly isolated from all of the other protein–protein interaction sites (Fig. 1). This positioning suggests no direct modulation of coactivator binding from other receptor domains. The PPRE is also very distant from the coactivator-binding site. Therefore, to recruit promoter-specific co-regulators, PPAR binding sites may be required to cooperate with other specific promoter elements that are positioned nearby.
A so-called phantom ligand effect has also been described in some RXR complexes, where an RXR ligand can cause dissociation of co-repressors and recruitment of coactivators at the non-covalently bound subunit of the heterodimer39. The current work does not provide a direct explanation for the phantom ligand effect or for receptor permissivity; instead, it shows two individual LXXLL motifs interacting independently at the PPAR-γ and RXR-α LBDs. In the case of RXR–RAR heterodimers, it has been shown that synergy between their ligands, resulting in enhanced transcription, can be created by the increased interaction efficiency of a single p160 protein40.
Discussion
The importance of the PPARs as a nuclear receptor subfamily is underscored by their validated roles in human disease and the proven beneficial role of some PPAR ligands for patients with type 2 diabetes. As many other non-steroid receptors also form heterodimers with RXR, the current structural analysis suggests the possibility of many different types of domain–domain interaction both within a single polypeptide and through the heterodimeric partners. The PPAR-γ–RXR-α polypeptides reside in a polar arrangement set by the 5′ extension of the DR1 and the lone spacer base pair. PPAR-γ occupies most of the DR1 including its specific 5′element (Supplementary Fig. 13). The DNA element largely dictates the type of domain–domain interactions in the complex, by predisposing both receptors to a unique polarity and spatial arrangement.
The observed architecture of the DBD and LBD receptor domains, in the context of the current structures, does not differ significantly from the same domains crystallized in isolation12,24,41. However, the domain–domain interactions involving intact nuclear receptors are defined fully here for the first time. The PPAR-γ LBD lies at the centre of the organized complex and is juxtaposed against every ordered domain from both proteins. Three distinct heterodimerization interfaces are formed between PPAR-γ and RXR-α. The interconnected nature of both receptors is intimately linked to their mode of binding to the PPRE DNA. Moreover, the PPAR-γ LBD assists both DBDs in their binding to DNA. This observation is supported by the finding that a single point mutation in the PPAR-γ LBD can severely impact DNA binding by the heterodimer. The structures also allow for a better understanding of PPAR-γ missense mutations associated clinically with partial lipodystrophy, insulin resistance and colon cancer (Supplementary Table 2).
We found that the A/B domains are intrinsically flexible as they were not observable in the crystal structure. These receptor portions are also not conserved in sequence. The intrinsic flexibility in this receptor portion of PPAR-γ was independently verified by H/D-Ex. Examination of several different crystal structures of PPAR-γ–RXR-α complexes indicated that three distinct PPAR-γ ligands did not substantially impact the ordering of the A/B regions or alter the domain–domain interactions within the complexes. A summary of all the critical sites of interactions on PPAR-γ, RXR-α and the PPRE is presented in Supplementary Figures 11–13. We expect that the other PPARs could form a similar arrangement with RXRs on DR1 PPREs, as the major sites of domain–domain interactions are well conserved in amino-acid sequence.
Owing to their direct involvement in many disease pathways, most of the nuclear receptors have been the subject of compound screening assays to find selective small-molecule modulators that bind to the LBD pockets. For practical reasons, such efforts at drug discovery have mainly relied on isolated LBDs, not intact receptor homo- or heterodimers on DNA. Moreover, the major readout for compound screening assays has been co-regulator recruitment properties. These studies have been guided by the assumption that different nuclear receptor ligands can impart their gene-selective actions strictly through their differential impact on co-regulator affinities. Given that co-regulator availability differs in cell types, the altered affinities are thought to give rise to selective modulator properties in these ligands.
The current structural examination shows that the LBD of a receptor can be tightly coupled to other receptor segments both within its own polypeptide and with the heterodimeric partner. The LBD can also have an important function in modulating DNA binding depending on the manner in which the complex is organized. It remains plausible that some nuclear receptor ligands could provide their selective gene modulation properties by imparting a graded set of response element affinities, and not through coactivator affinities alone. More sophisticated small-molecule screening and crystallization efforts are needed that use fully assembled and intact nuclear receptors to test this concept properly.
METHODS SUMMARY
Expression, purification and crystallization
The hRXR-α (NP_002948) and hPPAR-γ (NP_619726) proteins were expressed using pET46 Ek/LIC vectors in BL21(DE3) Escherichia coli cells (Novagen). Cells were induced with 1.0 mM IPTG at 17 °C overnight, collected and lysed in 20 mM Tris, pH 7.5, 500 mM NaCl, 30 mM imidazole and 10% glycerol. Proteins were purified on His-Bind resin (Novagen) and on SP-Sepharose columns (GE Healthcare). Ligands, coactivator-peptide (EKHKILHRLLQDSY) and DNA were added. The oligonu-cleotide strands 5′-GCAAACTAGGTCAAAGGTCAG-3′ and 5′-CTGACCTTTGACCTAGTTTGC-3′ were prepared as described previously26. Crystals were grown with 15–18% PEG 3350, 25 mM MgCl2, 100 mM NH4CL, 5 mM dithiothreitol, 0.1 M MES, pH 6.5, at 4 °C.
Data collection and structure determination
Diffraction data were collected at 100 K at the Argonne National Laboratory beamline SER-CAT 20ID and processed with HKL-3000 (ref. 42). The rosiglitazone-containing structure was solved using phases obtained from a SAD experiment. Both Refmac5 (CCP4) and CNS 1.2 were used for initial structure refinement, during which restraints were placed on DNA geometry43–45. The rosiglitazone structure was used to solve the BVT.13 and GW9662 structures by molecular replacement using PHASER46.
Hydrogen/deuterium exchange experiments
An exchange reaction was initiated by mixing 10 μl of protein stock solution (with or without ligand and/or DNA) with 10 μl of D2O. After incubation at 4 ± 1 °C for a predetermined time (15–1,500 s), the H/D-exchange reaction was quenched by shifting to pH 2.5 with 30 μl of 8 M urea and 1 M TCEP at 1 °C. The quenched solution was immediately pumped over a pepsin column (104 μl bed volume)38. Peptide fragments were separated by a C18 column. Mass spectrometric analyses were performed with a Thermo Fisher LCQ instrument, and the centroids of probe peptide isotopic envelopes were determined using a program developed in-house47. The corrections for back-exchange were made using methods described previously48.
METHODS
Expression, purification and crystallization
The hRXR-α (NP_002948) and hPPAR-γ (NP_619726) were expressed as His-tagged proteins from pET46 Ek/LIC vector in BL21(DE3) E. coli cells (Novagen). The seleno-methionyl derivative of hRXR-α was prepared by expression in B834(DE3) E. coli grown in minimal medium supplemented with Seleno-Met (Sigma). Cells were induced with 1.0 mM IPTG at 17 °C overnight, harvested and lysed in 20 mM Tris, pH 7.5, 500 mM NaCl, 30 mM imidazole and 10% glycerol. Each protein was purified on His-Bind resins (Novagen) and subsequently on SP-Sepharose columns (GE Healthcare), and their concentrations estimated based on calculated extinction values. 9-cis-retinoic acid (Sigma) and rosiglitazone (Cayman Chemicals) were added at 7× molar excess to RXR and PPAR, respectively. Synthetic coac-tivator peptide (EKHKILHRLLQDSY) was added in 2× molar ratio to each receptor. RXR and PPAR were then combined in 1:1 molar ratio and subsequently mixed with DNA oligonucleotide (DR1) at 1.5× molar ratio. The oligonucleotide strands containing the essential segments 5′-GCAAAC-TAGGTCAAAGGTCAG-3′ and 5′-CTGACCTTTGACCTAGTTTGC-3′ were purified and annealed as described previously26. The heterodimeric complex was purified on Superdex 200 (GE Healthcare) gel filtration column. The complex was concentrated to 3.0 mg ml−1 and crystals were grown using the hanging drop method against a reservoir of 15–18% PEG 3350, 25 mM MgCl2, 100 mM NH4CL, 5 mM dithiothreitol, 0.1 M MES, pH 6.5, at 4 °C. Crystals were streaked through a cryosolvent solution containing the reservoir solution supplemented with 34% glycerol and flash frozen in liquid nitrogen.
Data collection and structure determination
Diffraction data were collected on frozen crystals at 100 K using the Argonne National Laboratory at beamline SER-CAT 20ID. These data were integrated and scaled using HKL-3000 (ref. 42). The rosiglitazone-containing structure was initially solved, wherein phases were obtained using a SAD-phased electron-density map derived from the 3.1- Å selenomethionine–RXR-containing complex. Selenium sites were identified and refined using SHELXD, MLPHARE and DM features of HKL-3000. Both Refmac5 (CCP4) and CNS 1.2 were used for initial structure refinement, during which restraints were placed on DNA geometry43–45. Restraints were released in the later stages of refinement. The 3.1-Å refined rosiglitazone complex structure was used to solve the BVT.13 and GW9662 structures by molecular replacement using the program PHASER46. All data collection and refinement statistics are listed in Supplementary Table 1.
Hydrogen/deuterium exchange experiments
An exchange reaction was initiated by mixing 10 μl of protein stock solution (18 μM PPAR-γ ± 144 μM ligand ± DNA in 50 mM Tris, 200 mM NaCl, pH 8.0) with 10 μl of D2O. After incubation at 4 ± 1 °C for a predetermined time (15, 50, 150, 500 or 1,500 s), the H/D-exchange reaction was quenched by shifting to pH 2.5 with 30 μl of 8 M urea and 1 M TCEP at 1 °C. The quenched solution was immediately pumped over a pepsin column (104 μl bed volume) filled with porcine pepsin (Sigma) immobilized on Poros 20 AL media at 30 mg ml−1 according to the manufacturer’s instructions, with 0.05% TFA (200 μl min−1) for 3 min with contemporaneous collection of proteolytic products by a trap column (4 ml bed volume). Subsequently, the peptide fragments were eluted from the trap column and separated by C18 column (Magic C18, Michrom BioResources) with a linear gradient of 15% solvent B to 40% solvent B over 23 min (solvent A, 0.05% TFA in water; solvent B, 80% acetonitrile, 20% water, 0.01% TFA; flow rate 5–10 μl min−1). Mass spectrometric analyses were performed with an LCQ mass spectrometer (Thermo Fisher) with capillary temperature at 200 °C.
The centroids of probe peptide isotopic envelopes were measured using a program developed in-house in collaboration with Sierra Analytics47. The corrections for back-exchange were made using methods described previously48. Deuteration level (%) = [(m(P) − m(N))/(m(F) − m(N))] × 100 Deuterium incorporation (#) = [(m(P) − m(N))/(m(F) − m(N))] × MaxD where m(P), m(N) and m(F) are the centroid value of partly deuterated peptide, non-deuterated peptide and fully deuterated peptide, respectively. MaxD is the maximum deuterium incorporation calculated by subtracting the number of prolines in the third or later amino acid and two from the number of amino acids in the peptide of interest (assuming the first two amino acids cannot retain deuterons49). For PPAR-γ full length, the average deuterium recovery of a fully deuterated sample ((m(F) − m(N))/MaxD) was 66%.
Supplementary Material
Acknowledgments
We thank K. S. Molnar, S. J. Tuske and S. J. Coales for providing assistance with the H/D-Ex studies; M. Chruszcz and W. Minor for assistance with diffraction data processing and analysis; P. Rogers for analysis of mutant receptor expression; and P. Griffin for providing BVT.13.
Footnotes
Author Information Atomic coordinates and structure factors for the reported crystal structures are deposited in the Protein Data Bank under accession numbers 3DZY, 3DZU and 3E00. Reprints and permissions information is available at www.nature.com/reprints.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions V.C. expressed, purified and crystallized the samples, and with P.H. collected data and solved the structure. Y.H. performed the H/D-Ex work. S.R. provided the expression systems. Y.W. and T.P.B. performed the electrophoretic mobility shift assay and transcription reporter assays. F.R. supervised the work and wrote the manuscript.
References
- 1.Nagy L, Schwabe JW. Mechanism of the nuclear receptor molecular switch. Trends Biochem Sci. 2004;29:317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghuram S, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nature Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin L, et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 5.Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear receptor structure: implications for function. Annu Rev Physiol. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- 6.Khorasanizadeh S, Rastinejad F. Nuclear-receptor interactions on DNA-response elements. Trends Biochem Sci. 2001;26:384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- 7.Wurtz JM, et al. A canonical structure for the ligand-binding domain of nuclear receptors. Nature Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 8.McKenna NJ, O’Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 9.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Lambert MH, Xu HE. Activation of nuclear receptors: a perspective from structural genomics. Structure. 2003;11:741–746. doi: 10.1016/s0969-2126(03)00133-3. [DOI] [PubMed] [Google Scholar]
- 11.Greschik H, Moras D. Structure-activity relationship of nuclear receptor-ligand interactions. Curr Top Med Chem. 2003;3:1573–1599. doi: 10.2174/1568026033451736. [DOI] [PubMed] [Google Scholar]
- 12.Rastinejad F. Retinoid X receptor and its partners in the nuclear receptor family. Curr Opin Struct Biol. 2001;11:33–38. doi: 10.1016/s0959-440x(00)00165-2. [DOI] [PubMed] [Google Scholar]
- 13.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 14.Chen F, Law SW, O’Malley BW. Identification of two mPPAR related receptors and evidence for the existence of five subfamily members. Biochem Biophys Res Commun. 1993;196:671–677. doi: 10.1006/bbrc.1993.2302. [DOI] [PubMed] [Google Scholar]
- 15.Lazar MA. PPARγ, 10 years later. Biochimie. 2005;87:9–13. doi: 10.1016/j.biochi.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Lehrke M, Lazar MA. The many faces of PPARγ. Cell. 2005;123:993–999. doi: 10.1016/j.cell.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 17.Lehmann JM, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ) J Biol Chem. 1995;270:12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 18.Olefsky JM, Saltiel AR. PPARγ and the treatment of insulin resistance. Trends Endocrinol Metab. 2000;11:362–368. doi: 10.1016/s1043-2760(00)00306-4. [DOI] [PubMed] [Google Scholar]
- 19.Staels B. PPAR agonists and the metabolic syndrome. Therapie. 2007;62:319–326. doi: 10.2515/therapie:2007051. [DOI] [PubMed] [Google Scholar]
- 20.Willson TM, et al. The structure-activity relationship between peroxisome proliferator-activated receptor γ agonism and the antihyperglycemic activity of thiazolidinediones. J Med Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- 21.Berger J, Wagner JA. Physiological and therapeutic roles of peroxisome proliferator-activated receptors. Diabetes Technol Ther. 2002;4:163–174. doi: 10.1089/15209150260007381. [DOI] [PubMed] [Google Scholar]
- 22.Bruning JB, et al. Partial agonists activate PPARγ using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. doi: 10.1016/j.str.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Leesnitzer LM, et al. Functional consequences of cysteine modification in the ligand binding sites of peroxisome proliferator activated receptors by GW9662. Biochemistry. 2002;41:6640–6650. doi: 10.1021/bi0159581. [DOI] [PubMed] [Google Scholar]
- 24.Gampe RT, Jr, et al. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Mol Cell. 2000;5:545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 25.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 26.Zhao Q, Khorasanizadeh S, Miyoshi Y, Lazar MA, Rastinejad F. Structural elements of an orphan nuclear receptor-DNA complex. Mol Cell. 1998;1:849–861. doi: 10.1016/s1097-2765(00)80084-2. [DOI] [PubMed] [Google Scholar]
- 27.Ijpenberg A, Jeannin E, Wahli W, Desvergne B. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. J Biol Chem. 1997;272:20108–20117. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- 28.Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J. 2000;19:1045–1054. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sierk ML, Zhao Q, Rastinejad F. DNA deformability as a recognition feature in the reverb response element. Biochemistry. 2001;40:12833–12843. doi: 10.1021/bi011086r. [DOI] [PubMed] [Google Scholar]
- 30.Ozers MS, et al. Equilibrium binding of estrogen receptor with DNA using fluorescence anisotropy. J Biol Chem. 1997;272:30405–30411. doi: 10.1074/jbc.272.48.30405. [DOI] [PubMed] [Google Scholar]
- 31.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 32.Kurokawa R, et al. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 33.Werman A, et al. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor γ (PPARγ). Differential activity of PPARγ-1 and -2 isoforms and influence of insulin. J Biol Chem. 1997;272:20230–20235. doi: 10.1074/jbc.272.32.20230. [DOI] [PubMed] [Google Scholar]
- 34.Adams M, Reginato MJ, Shao D, Lazar MA, Chatterjee VK. Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. J Biol Chem. 1997;272:5128–5132. doi: 10.1074/jbc.272.8.5128. [DOI] [PubMed] [Google Scholar]
- 35.Castillo G, et al. An adipogenic cofactor bound by the differentiation domain of PPARγ. EMBO J. 1999;18:3676–3687. doi: 10.1093/emboj/18.13.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao D, et al. Interdomain communication regulating ligand binding by PPAR-γ. Nature. 1998;396:377–380. doi: 10.1038/24634. [DOI] [PubMed] [Google Scholar]
- 37.Maier CS, Deinzer ML. Protein conformations, interactions, and H/D exchange. Methods Enzymol. 2005;402:312–360. doi: 10.1016/S0076-6879(05)02010-0. [DOI] [PubMed] [Google Scholar]
- 38.Hamuro Y, et al. Rapid analysis of protein structure and dynamics by hydrogen/deuterium exchange mass spectrometry. J Biomol Tech. 2003;14:171–182. [PMC free article] [PubMed] [Google Scholar]
- 39.Schulman IG, Li C, Schwabe JW, Evans RM. The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes Dev. 1997;11:299–308. doi: 10.1101/gad.11.3.299. [DOI] [PubMed] [Google Scholar]
- 40.Germain P, Iyer J, Zechel C, Gronemeyer H. Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature. 2002;415:187–192. doi: 10.1038/415187a. [DOI] [PubMed] [Google Scholar]
- 41.Nolte RT, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 42.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project. Number 4. The CCP 4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nature Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamuro Y, et al. Hydrogen/deuterium-exchange (H/D-Ex) of PPARγ LBD in the presence of various modulators. Protein Sci. 2006;15:1–10. doi: 10.1110/ps.062103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.