Abstract
Zebrafish have emerged as a powerful model system to study leukocyte recruitment and inflammation. Here we characterize the morphology and function of inflammatory macrophages in zebrafish larvae. These macrophages can be distinguished from neutrophils by immunolabeling of L-Plastin without MPO co-expression and by an elongated morphology. Live imaging of transgenic zMPO:GFP larvae demonstrate that GFPlo macrophages migrate to wounds by extension of thin pseudopods and carry out phagocytosis of tissue debris, and FACS analysis of leukocyte markers indicates expression of CSF1R in these macrophages. These findings identify distinct functional and morphological characteristics of inflammatory macrophages in zebrafish larvae.
Keywords: Zebrafish, inflammation, macrophage, neutrophil, phagocytosis
1. Introduction
Zebrafish have become a powerful model organism to study the innate immune system [1, 2] because they are genetically tractable and amenable to small molecule screening [3, 4]. Zebrafish larvae develop an innate immune system comprised primarily of neutrophils and macrophages. Recent studies have also reported the presence of eosinophils [5] and mast cells [6]. Neutrophils can be identified by a high number of refractive granules in the cytoplasm [7], expression of myeloperoxidase (MPO, or MPX, myeloid-specific peroxidase) [8, 9], myeloperoxidase activity [9] and Sudan Black labeling [7]. Transgenic lines expressing GFP in neutrophils have been generated and have been used to gain insight into neutrophil activities within contexts of acute inflammation [10, 11], chronic inflammation [12–14], infection [15] and during hematopoietic development [5].
Studies of the leukocyte response to tissue damage and/or infection have shown that neutrophils are the first to respond, and subsequently macrophages are recruited to inflamed tissues, where their main function is phagocytosis of pathogens and tissue debris (reviewed in [16]). Multiple subtypes of the monocyte/macrophage lineage are known to exist [17], and macrophages have been shown to play a central role in host defense, with roles in both the innate and adaptive immune responses [18, 19]. Primitive macrophages have been characterized in zebrafish embryos [20], but are gradually replaced during later stages of definitive hematopoiesis [5]. Multiple genes have been suggested as markers for the monocyte/macrophage lineage in zebrafish, including L-Plastin [20] and Lysozyme C [21], however subsequent reports have indicated that these genes are expressed in other leukocytes as well [22, 23]. Colony Stimulating Factor 1 Receptor (CSF1R, also known as c-fms) shows a hematopoietic expression pattern similar to L-Plastin [24] and is co-expressed with LP but not with MPO [22, 25]. For these reasons CSF1R has become an accepted marker for zebrafish macrophages. Transgenic zebrafish expressing fluorescent proteins in primitive macrophages [26, 27] and myelo-monocytic cells (i.e., both neutrophils and macrophages) have been engineered [22, 28, 29], yet characterization of macrophages at larval developmental stages has not been well defined to date. In this study we utilize immunolabeling and time-lapse microscopy to characterize a subset of zebrafish macrophages that respond to inflammatory stimuli in zebrafish larvae.
2. Materials and Methods
2.1. Zebrafish Maintenance
All protocols using zebrafish in this study were approved by the University of Wisconsin-Madison Research Animal Resources Center. Adult AB zebrafish and embryos were maintained according to standard protocols [30] and staged as previously established [31]. Talifin wounding was performed as described [10]. Laser wounding was performed by focusing a 405nm diode laser beam at full power on a small circular area (diameter 1µm) for 1 min. All images and movies are of larvae at 3 days post-fertilization (dpf) unless specified otherwise.
2.2. Whole Mount Immunolabeling
Zebrafish larvae were fixed in 1% Formaldehyde in PBS for 2h at room temperature and immunolabeled as described [10]. Rabbit antibodies to zebrafish MPO and L-Plastin have been described [10, 13]. For MPO/L-Plastin double-immunolabeling, larvae were labeled sequentially as follows: (1) rabbit anti-zMPO, (2) FITC-conjugated goat anti-rabbit Fab fragment (Jackson), (3) rabbit anti-L-Plastin IgG conjugated to Rhodamine Red (LP-RR) using the FluoReporter Rhodamine Red-X Protein Labeling Kit (Molecular Probes), according to manufacturer’s instructions. Control experiments using rabbit anti-Pan Cadherin antibodies (Sigma) instead of anti-zMPO confirmed the LP-RR antibody binds neither a rabbit primary antibody nor the goat Fab secondary antibody (data not shown). To immunolabel GFP a rabbit anti-GFP antibody (Invitrogen) was used, followed by the LP-RR antibody. To immunolabel epidermal cells a mouse anti-p63 antibody (Novus) was used as described previously [12].
2.3. Histochemistry
Whole mount Sudan black staining of neutrophils was performed as described [7] except that a 1:20 dilution of the stain was utilized, followed by immunolabeling of L-Plastin. Cells isolated by FACS were plated on glass slides and stained using Leukocyte Peroxidase (MPO) activity kit (Sigma) according to manufacturer protocols or cells were affixed to glass slides via cytospin, air dried and stained with Wright-Giemsa (Fisher).
2.4. Image Acquisition and Analysis
Epifluorescent imaging, confocal imaging and time-lapse microscopy were performed as described [10, 12, 13]. For time-lapse movies AVI files were made using the Cinepak codec and graphics were added using ImageJ.
2.5. Fluourescence Activated Cell Sorting (FACS)
Tg(mpx:eGFP)uwm1 (zMPO:GFP) [10] larvae at 3 dpf were trypsinized (approximately 150 larvae per experiment) and sorted by FACS as described [32, 33] with minor modifications. Trypsinized cells from wild-type embryos at an identical stage were used to set the lower limit of GFP expression for each experiment.
2.6. RNA Isolation and Reverse Transcriptase PCR (RT-PCR)
RNA was isolated using the RNeasy Mini Kit, including on-column RNAse-free DNAseI digestion (Qiagen). RT-PCR was performed as described [34] with minor modifications; 40 cycles and an anneal temperature of 55°C was used for all genes except MMP9 (50°C). Primers sequences and references are shown in Fig. S2. All primer sets were designed to span at least one intron such that amplification from contaminating genomic DNA could be distinguished by band size from amplification from spliced mRNA.
3. Results
3.1. Immunolabeling of Larval Zebrafish Leukocytes
Immunofluorescent co-labeling of zebrafish MPO and L-Plastin (LP) revealed two populations of leukocytes that express LP within the Caudal Hematopoietic Tissue (CHT) at 2–3 dpf: LP+ cells that express MPO (neutrophils) (Fig. 1A–C and Fig. S1D–F, arrowheads) and LP+ cells that do not express MPO (Fig. 1B–C and Fig.S1E–F, arrows). Earlier in development at 28 hours post fertilization (hpf), we observed some MPO+ cells that express little to no LP (Fig. S1A–C, blue arrowheads) within the posterior blood island (PBI), while most MPO+ cells also express LP (Fig. S1A–C, white arrowheads). At 2–3 dpf LP+MPO− cells are also seen in the head, eyes and over the yolk sac (Fig. S1G–I). These findings are consistent with in situ hybridization for LP [20], and since CSF1R has been shown to be co-expressed in hematopoietic cells with LP but not MPO [22], these findings suggest that the LP+MPO− cells represent macrophages.
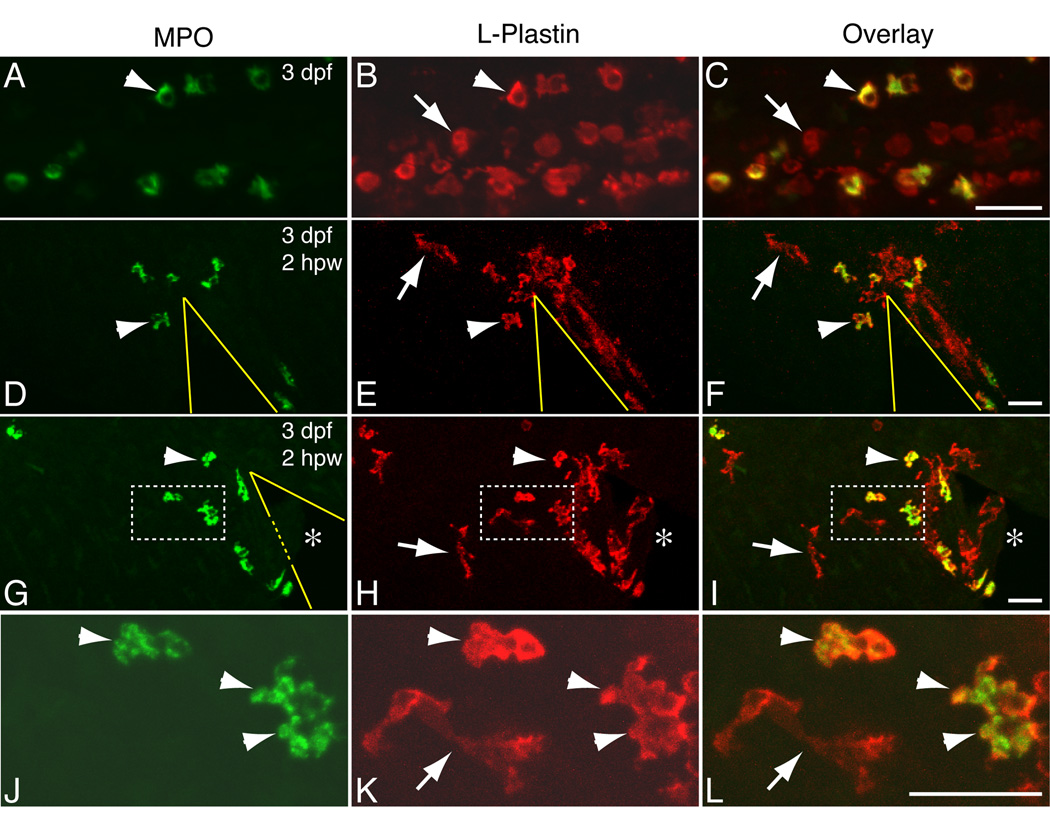
Figure 1. Confocal imaging of immunolabeled zebrafish larval leukocytes.
(A–L) Wild-type larvae at 3 dpf were sequentially immunolabeled using a rabbit antibody to MPO and a FITC-conjugated anti-rabbit Fab fragment (A,D,G,J), followed by a Rhodamine Red-conjugated rabbit antibody to L-Plastin (B,E,H,K); overlapping signals are yellow in overlay images (C,F,I,L). Arrowheads mark LP+/MPO+ neutrophils, arrows mark LP+/MPO− macrophages. (A–C) Caudal Hematopoietic Tissue (CHT). (D–L) Leukocytes responding to wounds (wedges outlined in yellow) in the tailfin, at 2 hours post-wound (hpw). Note how LP+/MPO− macrophages have accumulated along the edge of a wound (E–F) and infiltrated a small fragment of wounded tissue (marked by an * in G–I). (J–L) Higher magnification views of cells within boxed areas in G–I, note the elongated morphology of LP+/MPO− macrophages and how the granular MPO label does not overlap with the LP signal within co-expressing neutrophils. Scale bars = 25 µm.
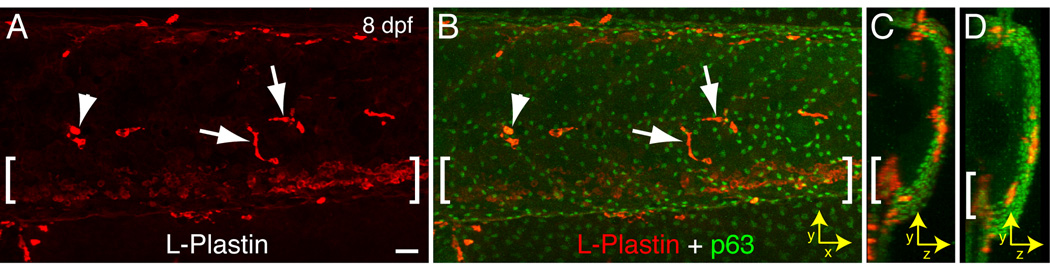
Both neutrophils and LP+MPO− cells are recruited to wounds induced in the tailfin, but display distinct morphological features that further characterize each cell type (Fig.1D–L). As previously reported [10] neutrophils recruited to a wound take on a compact, highly polarized morphology. In contrast LP+MPO– cells display an elongated morphology with no obvious polarization (Fig. 1D–I, arrows). Furthermore, LP+MPO− cells tend to line up along the edge of a wound, covering much of the exposed area of the wound (Fig. 1E–F). At higher magnification the MPO immunolabel is granular (Fig. 1J,L) and distinct from the LP label (Fig. 1K–L), providing evidence for the specificity of the two antibodies. The lipophilic dye Sudan Black has been shown to label zebrafish neutrophils [7], and double-labeling using this dye and the LP antibody confirmed the presence of a population of LP+, non-neutrophil cells that are recruited to inflamed areas (data not shown). At later developmental stages (8 dpf) LP+MPO− cells with an elongated morphology are found throughout the trunk of wild-type larvae (Fig. 2A–B, arrows). These leukocytes are found almost exclusively within the p63-expressing epidermis (Fig. 2C–D) and tend to accumulate along the dorsal margin and lateral line. The recruitment of elongated LP+MPO− cells to wounds and their placement within the epidermis indicates that these cells represent a population of tissue macrophages that respond to inflammatory stimuli.
Figure 2. L-Plastin-labeled leukocytes in the epidermis of zebrafish larvae.
Wild-type larva at 8 dpf immunolabeled with antibodies to L-Plastin (A) and the epithelial cell marker p63 (B); the CHT is denoted by brackets. (A–C) Overlay images of LP and p63 in the xy plane (A–B; anterior is to the left, dorsal side points up) and the yz plane (C; dorsal side points up) of the same embryo; in (D) a second larva labeled as in A–C is shown in the yz plane. Note the localization of LP+ cells within the p63-labeled epithelium. For A–B arrowheads mark neutrophils and arrows mark macrophages identified by morphology. Scale bar = 25 µm.
3.2. Expression of Hematopoietic Markers in Transgenic zMPO:GFP Larvae
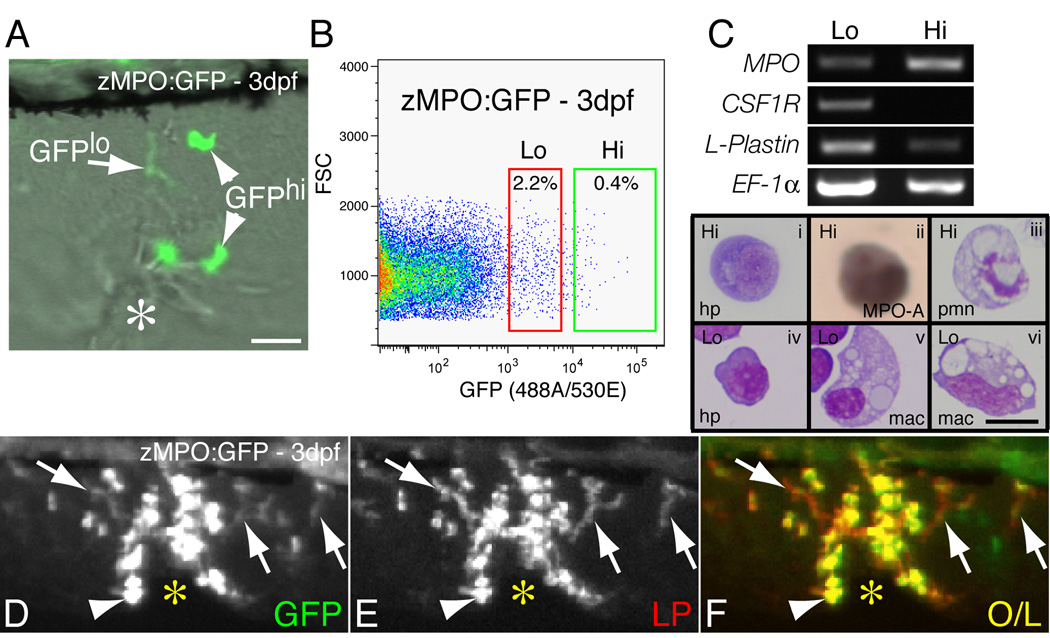
In a recent study [10] we characterized a transgenic zebrafish line (zMPO:GFP) that expresses GFP at a high level in neutrophils (GFPhi cells) and a low level in a second migratory cell type (GFPlo cells) that resembles the elongated cells seen by LP immunolabeling (Fig. 3A). In support of this interpretation, co-immunolabeling of zMPO:GFP larvae at 3 dpf using an antibody to GFP showed that both the GFPhi neutrophils (Fig. 3D–F, arrowheads), and GFPlo elongated cells (Fig. 3D–F, arrows) express L-plastin. Cells expressing GFP at a low level above background (GFP-Lo) and at a higher level (GFP-Hi) were collected by FACS of dissociated 3 dpf zMPO:GFP larvae (Fig. 3B) and analyzed by RT-PCR and histochemical analysis. Note that hereafter superscript “lo” or “hi” following “GFP” (e.g., GFPhi) will refer to leukocytes observed by microscopy, while “-Lo” or “-Hi” refers to FACS fractions. RT-PCR analysis demonstrated that LP and the neutrophil marker MPO were expressed in both fractions (Fig. 3C), with reproducible enrichment of MPO in the GFP-Hi fraction (Fig. S2). In contrast the macrophage marker CSF1R was only expressed in the GFP-Lo fraction.
Figure 3. Hematopoeitic gene expression in zMPO:GFP larvae.
(A) Still image (GFP overlaid onto DIC) from a time-lapse movie of zMPO:GFP (3 dpf) leukocytes responding to a wound (*) in the ventral tailfin; GFPlo leukocytes and GFPhi neutrophils are marked. (B) Flow cytometry of zMPO:GFP larvae at 3 dpf; GFP-Lo and GFP-Hi fractions are marked with boxes; cells from wild-type larvae were used to set the lowest level of GFP expression. (C) RT-PCR of hematopoietic markers (MPO, CSF1R and L-Plastin; EF-1α is used as a loading control) from GFP-Lo and -Hi fractions marked in B; cells sorted from either fraction and stained by Wright-Giemsa (i,iii–vi) or MPO activity (ii, MPO-A) are also shown. Hematopoietic precursor (hp); polymorphonuclear neutrophil (pmn); macrophage (mac). (D–F) Immunolabeling of zMPO:GFP (3 dpf) larva, 2h following a wound (*) in the caudal tailfin; (D) GFP antibody, (E) L-Plastin, (F) overlay. Note elongated LP+ cells that also express a low level of GFP (arrows); arrowheads mark GFPhi neutrophils, which also express LP. Scale bar = 10 µm.
Genes expressed by non-hematopoietic cells that also express the zMPO:GFP transgene [10] were observed primarily in the GFP-Lo fraction and to a very small extent in the GFP-Hi fraction (Fig. S2), indicating the presence of contaminating non-immune cells. This was supported by histochemical analysis that indicated the presence of non-hematopoietic cells primarily in the GFP-Lo fractions (data not shown). Both the GFP-Hi and GFP-Lo fractions contained hematopoietic cells consisting predominantly of precursors (Fig. 3Ci,iv) that we were unable to adequately distinguish any further by morphology alone. However, the majority of cells in the GFP-Hi fraction stained positive for neutrophil-specific MPO activity (Fig. 3Cii) while MPO-positive cells were relatively rare in the GFP-Lo fraction (data not shown). Furthermore, cells with more mature neutrophil and macrophage morphologies were identified in the GFP-Hi (Fig. 3Ciii) and GFP-Lo (Fig. 3Cv,vi) fractions, respectively. Taken together these RT-PCR and histochemical results indicate that the GFP-Hi fraction is enriched with neutrophils (which also express LysozymeC, MMP9 and MMP13, see Fig. S2) at varying stages of development, while the GFP-Lo fraction contains (among other hematopoietic precursors) macrophages, which likely represent the GFPlo cells observed by microscopy (Fig. 3A).
3.3. Migratory Characteristics and Phagocytic Behavior of GFPlo Macrophages
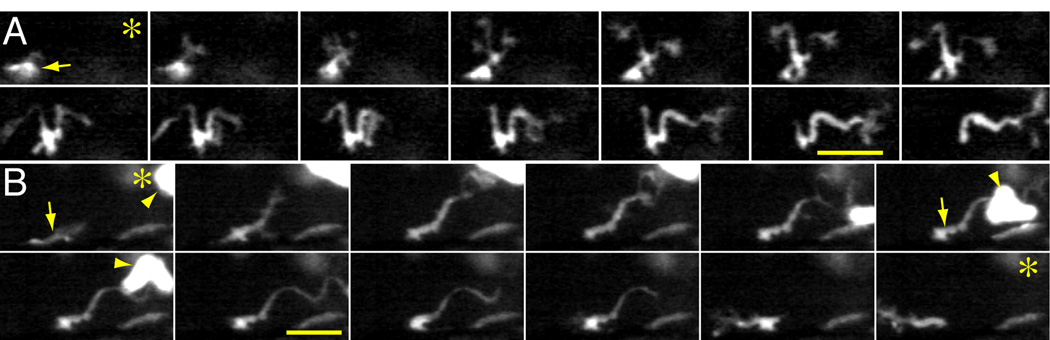
In response to tissue wounding GFPlo cells were observed to extend one or more thin pseudopods and then migrate by streaming the remaining cell contents into one of these pseudopods (Fig. 4A and Movie S1). This process can be repeated several times with sequential pseudopod selection (Movie S2), creating a stepwise migration that is distinct from the more gliding movement of GFPhi neutrophils [10]. This mode of migration by extension of thin pseudopods is typically observed, seen in 11 out of 11 GFPlo cells from one time-lapse movie (which includes the cells in Movie S1 and Movie S2), and 8 out of 8 cells in another. GFPlo cells were also observed to extend and maintain a pseudopod toward a wound, followed by retraction of the pseudopod (Fig. 4B and Movie S3). Similar to LP− immunolabeled macrophages, GFPlo cells line up along the edge of wounds, where they extend pseudopods and remain relatively stationary (Movie S4), often remaining after GFPhi neutrophils have left the wound (observed by 8 out of 8 GFPlo cells in Movie S4) by retrograde migration [10].
Figure 4. Migratory characteristics of GFPlo macrophages in transgenic zMPO:GFP larvae.
Sequential images from time-lapse movies of GFPlo macrophages; note that brightness and contrast have been increased to view cellular details. (A; from Movie S1) A GFPlo macrophage (arrow) extends two pseudopods, then “follows” the pseudopod nearest to a wound (out of frame, oriented by an *) by moving the cell body into this pseudopod; 1 min separates each frame. (B; from Movie S3) A GFPlo macrophage (arrow) extends a pseudopod toward a wound (out of frame, oriented by an *) and maintains the pseudopod in this position for several minutes (3 min separates each frame) before retracting it and migrating in the opposite direction; a GFPhi neutrophil is marked with arrowheads. Scale bars = 25 µm.
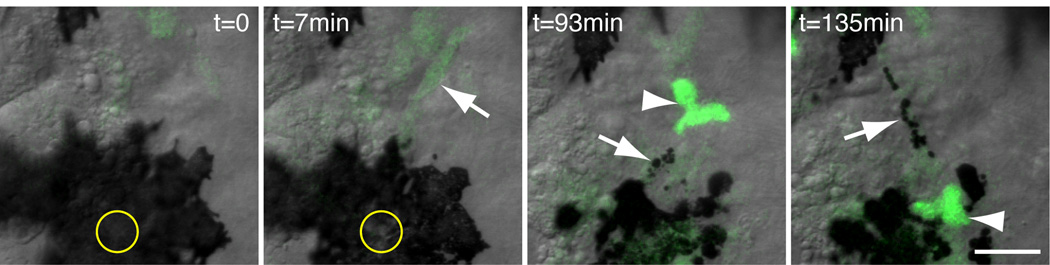
However, retrograde migration away from a wound and toward the vasculature is also exhibited by GFPlo macrophages (Movie S3). GFPlo macrophages were also observed to phagocytose pigment following laser-mediated wounding of a melanocyte (Fig. 5, Movie S5), providing functional evidence that they are macrophages [7]. Importantly, these macrophages are observable by time-lapse phase-contrast microscopy and can be distinguished from neutrophils by the morphological and migratory characteristics described in this study.
Figure 5. Phagocytosis of Cell Debris by GFPlo macrophages.
A melanocyte over the yolk sac is ruptured by laser wounding (circle denotes targeted area, t=0 is pre-wound), resulting in the recruitment of GFPlo macrophages (arrows), which phagocytose pigment from the ruptured cell; a GFPhi neutrophil (arrowheads) also responds. Images are from Movie S5. Scale bar = 20 µm.
4. Discussion
Collectively the data in this report characterize a class of leukocytes in zebrafish larvae whose morphology and functional properties indicate that they are a population of macrophages that are distinct from primitive macrophages [20] and neutrophils. Prior to more detailed characterization we propose to call these macrophages “inflammatory macrophages”, based on their involvement in inflammatory processes in larvae (Fig.1 D–I); note that this is distinct from “inflammatory monocytes” described previously [35]. Several different subsets of the macrophage/monocyte lineage have been characterized in higher eukaryotes [17] including specialized tissue-specific subsets [18]. Microglial cells, derived from LP+ precursors, have been described in zebrafish larvae and reside in the central nervous system [36]; however no other macrophage subsets have been defined in zebrafish to date. We expect that as development progresses the degree of macrophage heterogeneity will concurrently increase in zebrafish larvae. By using our zMPO:GFP transgenic fish, further macrophage markers based on tissue residency could be defined by dissection of larvae [37], followed by FACS and RT-PCR (Fig. 3B–C). The positioning of inflammatory macrophages throughout the epidermis in older larvae (Fig. 2) suggests that some may be resident tissue macrophages, while their morphology and migratory characteristics (Fig. 4) resemble that of dendritic cells [38].
To date there have been few studies of macrophages during larval stages of zebrafish development, and these have been hindered somewhat by discrepancies in expression of hematopoietic markers. Multiple labs have produced transgenic fish that express fluorescent proteins in macrophages [28, 29] by use of promoter fragments from the Lysozyme C gene, which was previously thought to be macrophage-specific [21]. However, these transgenic lines label both neutrophils and macrophages, and multiple reports indicate that Lysozyme C is also expressed in neutrophils [22, 23], consistent with our findings (Fig. S2). In the zMPO:GFP transgenic line [10], neutrophils and inflammatory macrophages can be readily distinguished based on GFP expression (Fig. 3A). Since we see no MPO immunolabel in LP+ macrophages at high magnification (Fig. 1J–L), the low amount of GFP expressed by macrophages in the zMPO:GFP line may be due to elements in the transgenic construct rather than a reflection of low levels of MPO activity observed in macrophages in other species [39]. The expression of MPO in the macrophage-enriched GFP-Lo fraction (Fig. 3C) is most likely from neutrophil precursors [9]. Few hematopoietic markers seem to be specific to macrophages, which hinders further characterization of this lineage. A role for MMP13 in macrophage migration has been shown [28], however we found that neutrophils also express MMP13 (Fig. S2). Use of CSF1R as a marker for macrophages is generally accepted but is somewhat limited due to expression in neural crest cells [40]. Outside of marker genes the only described methods to identify macrophages require phagocytosis of fluorescent bacteria [41] or the dye Neutral Red [24]. This manuscript therefore provides important evidence that enables the identification of this subset of macrophages on the basis of morphology, in addition to the expression of cellular markers.
The inflammatory macrophages described in this paper were characterized on the basis of morphology and immunolabeling of two intracellular proteins expressed only in hematopoietic cells, MPO and LP. At 2–3 dpf inflammatory macrophages can be identified by LP expression without MPO and an elongated morphology (Fig. 1B–C and Fig. S1E–F, arrows). MPO expression at 2–3 dpf is confined to neutrophils, which also express LP (Fig. 1A–C and Fig. S1D–F, arrowheads) as observed in other studies at similar embryonic stages [7, 22]. Similar immunolabeling results in 2 dpf embryos have been shown at much lower resolution [41]. At ~24 hpf our results (Fig. S1A–C) differ slightly than those of previous studies of embryos at early time-points where co-expression of MPO and LP mRNA was rarely seen [8, 23]. These discrepancies may reflect differences in sensitivity between detection methods or the developmental stage of neutrophil precursors, e.g., MPO expression may precede that of LP in early neutrophils. LP+MPO− leukocytes are recruited to wounds in 2–3 dpf larvae (Fig.1 D–I and data not shown); since cells expressing CSF1R mRNA can be recruited to wounds in the tailfins of 2 dpf embryos [42], this strongly suggests that the LP+ cells responding to a wound are macrophages.
Within a developmental setting such as the CHT, both neutrophils and macrophages appear amoeboid (Fig. 1A–C), while upon migration through tissues inflammatory macrophages adopt a distinct elongated morphology (Fig. 1K). Live imaging of inflammatory macrophages in transgenic zMPO:GFP larvae provided further insight into the biology and functions of this cell type. Inflammatory macrophages migrate by extension of long pseudopods (Movie S1 and Movie S2), reminiscent of the “flowing and squeezing” model shown by leukocytes migrating in a 3-dimensional environment [43]. During migration these cells are highly deformable, and able to adopt a very thin, elongated morphology (Fig. 4A). Prolonged pseudopod extension without migration was also observed (Fig. 4B and Movie S3), likely suggesting a role in either sensing or cell signaling. The possibility that these extensions are used to communicate with other leukocytes is intriguing, but we do not have any substantial evidence of this to date.
Several qualitative observations similar to previous reports [16] provide further evidence that the LP+MPO−/GFPlo leukocytes we observe are macrophages. During the inflammatory response macrophages are known to arrive after neutrophils and are generally retained at an inflamed area longer than neutrophils. Similarly, GFPlo inflammatory macrophages tend to migrate slower and arrive later than GFPhi neutrophils at wounds in the tailfin of zebrafish larvae [10]. Inflammatory macrophages are observed to line up along the edge of the wound (Fig. 1E–F) and remain at the wound after neutrophils have left (Movie S4). In support of these observations, electron microscopy has shown that zebrafish macrophages form stable junctions with other cells at a wound, which was speculated to serve a “tethering” function [44]. Neutrophils sometimes round up at wound, remaining in this state for several minutes at a time [10]. We have not observed any instances of this by inflammatory macrophages, which remain elongated and extend pseudopods upon arresting at a wound (Movie S4).
Macrophages are known to be highly phagocytic. Phagocytosis of erythrocytes has been shown in primitive macrophages [20], and we now show that inflammatory macrophages are also highly phagocytic of cellular debris (Movie S5). Other reports have shown that larval macrophages phagocytose bacteria [7, 41]. In future studies it will be important to determine if these inflammatory macrophages possess the capacity to present antigen to lymphocytes. Transgenic zebrafish expressing fluorescent proteins in other hematopoietic cells [33, 45, 46] crossed to the zMPO:GFP transgenic will enable novel observations of interactions between immune cell types in zebrafish larvae.
Supplementary Material
Wild-type embryos were sequentially immunolabeled using a rabbit antibody to MPO and a FITC-conjugated anti-rabbit Fab fragment (A,D,G), followed by a Rhodamine Red-conjugated rabbit antibody to L-Plastin (B,E,H); overlapping signals are yellow in overlay images (C,F,I). All frames are epifluorescence images, anterior is to the left and dorsal side up; arrows mark LP+/MPO− macrophages, white arrowheads mark LP+/MPO+ neutrophils, blue arrowheads mark LP−/MPO+ cells. (A–C) 28 hpf, Posterior Blood Island. (D–F) 2 dpf, Caudal Hematopoietic Tissue. (G–I) 2 dpf, head and yolk sac (composite images at different focal planes).
RT-PCR of cells from GFP-Lo and -Hi fractions, as in Fig. 3B but from a different preparation of cells and RNA. Genes are indicated to the left, cell type expression is indicated in parentheses; EF-1α is used as a loading control. Primer sequences are to the right of each gene, followed by references for each primer sequence; primers to CSF1R and Lysozyme C were designed in this study, therefore references detailing the expression patterns of these genes are listed in blue.
A GFPlo macrophage (starting in the lower left corner) extends two pseudopods, then “follows” the pseudopod nearest to a wound (out of frame, in the upper right corner) by moving the cell body into this pseudopod; a non-hematopoietic GFP+ cell is present in the upper right corner of the frame. Compression is 6 frames per second, with 1 min actual time between each frame.
A GFPlo macrophage sequentially extends multiple pseudopods during migration toward a wound (out of frame, in the upper left corner). Compression is 6 frames per second, with 1 min actual time between each frame.
A GFPlo macrophage (marked “1”) extends a pseudopod toward a wound (the edge of which is marked with a yellow line) and maintains the pseudopod in this position for several minutes before retracting it and migrating in the opposite direction; a GFPhi neutrophil can be seen migrating in the upper right corner of the frame. Later, a second GFPlo macrophage (marked “2”) migrates away from the wound and back to the vasculature (at the bottom of the frame). Compression is 6 frames per second, with 1 min actual time between each frame.
Following 2h of wound response, GFPlo macrophages (numbered 1–8) are lined along a wound (the edge of which is marked with a yellow line) and remain there after GFPhi neutrophils (marked with arrowheads) have left the wound. The vasculature is at the bottom of the frame, and a non-hematopoietic GFP+ cell is indicated (nh). Compression is 6 frames per second, with 1 min actual time between each frame.
A melanocyte over the yolk sac is ruptured by a laser-induced wound, resulting in the recruitment of GFPlo macrophages that phagocytose the pigment from the dead melanocyte. GFPhi neutrophils (note the large difference in GFP intensity) are also recruited but do not exhibit phagocytosis. Elapsed time is indicated in the lower left corner.
Acknowledgements
We gratefully acknowledge Andrea Gallagher for zebrafish maintenance and lab support, and the University of Wisconsin-Madison Flow Cytometry Facilities for expert guidance and support. This work was supported by an Arthritis Foundation post-doctoral fellowship (J.R.M.) and NIH grants to A.H. (R01 GM074827) and the University of Wisconsin-Madison Department of Hematology (M.E.D.).
Abbreviations
- CSF1R
Colony Stimulating Factor 1 Receptor
- dpf
days post-fertilization
- FACS
Fluorescence Activated Cell Sorting
- GFP
green fluorescent protein
- hpf
hours post-fertilization
- LP
L-Plastin
- MMP
Matrix metalloproteinase
- MPO
Myeloperoxidase
- PBS
Phosphate Buffered Saline
- RFP
red fluorescent protein
- RR
Rhodamine Red
- RT-PCR
Reverse Transcriptase Polymerase Chain Reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carradice D, Lieschke GJ. Zebrafish in hematology: sushi or science? Blood. 2008;111(7):3331–3342. doi: 10.1182/blood-2007-10-052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Jong JL, Zon LI. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu Rev Genet. 2005;39:481–501. doi: 10.1146/annurev.genet.39.073003.095931. [DOI] [PubMed] [Google Scholar]
- 3.Murphey RD, Zon LI. Small molecule screening in the zebrafish. Methods. 2006;39(3):255–261. doi: 10.1016/j.ymeth.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat Rev Genet. 2001;2(12):956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134(23):4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobson JT, Seibert J, Teh EM, Da'as S, Fraser RB, Paw BH, Lin TJ, Berman JN. Carboxypeptidase A5 identifies a novel mast cell lineage in the zebrafish providing new insight into mast cell fate determination. Blood. 2008;112(7):2969–2972. doi: 10.1182/blood-2008-03-145011. [DOI] [PubMed] [Google Scholar]
- 7.Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, Mordelet E, Zapata A, Shinomiya H, Herbomel P. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2008;111(1):132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- 8.Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, Look AT. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98(3):643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- 9.Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98(10):3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- 10.Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J Leukoc Biol. 2006;80(6):1281–1288. doi: 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- 11.Renshaw SA, Loynes CA, Trushell DM, Elworthy S, Ingham PW, Whyte MK. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 12.Walters KB, Dodd ME, Mathias JR, Gallagher AJ, Bennin DA, Rhodes J, Kanki JP, Look AT, Grinblat Y, Huttenlocher A. Muscle degeneration and leukocyte infiltration caused by mutation of zebrafish fad24. Dev Dyn. 2009;238(1):86–99. doi: 10.1002/dvdy.21821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathias JR, Dodd ME, Walters KB, Rhodes J, Kanki JP, Look AT, Huttenlocher A. Live imaging of chronic inflammation caused by mutation of zebrafish Hai1. J Cell Sci. 2007;120(Pt 19):3372–3383. doi: 10.1242/jcs.009159. [DOI] [PubMed] [Google Scholar]
- 14.Dodd ME, Hatzold J, Mathias JR, Walters KB, Bennin DA, Rhodes J, Kanki JP, Look AT, Hammerschmidt M, Huttenlocher A. The ENTH domain protein Clint1 is required for epidermal homeostasis in zebrafish. Development. 2009 doi: 10.1242/dev.038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brannon MK, Davis JM, Mathias JR, Hall CJ, Emerson JC, Crosier PS, Huttenlocher A, Ramakrishnan L, Moskowitz SM. Pseudomonas aeruginosa Type III secretion system interacts with phagocytes to modulate systemic infection of zebrafish embryos. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dale DC, Boxer L, Liles WC. The phagocytes: neutrophils and monocytes. Blood. 2008;112(4):935–945. doi: 10.1182/blood-2007-12-077917. [DOI] [PubMed] [Google Scholar]
- 17.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5(12):953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 18.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon S. The macrophage: past, present and future. Eur J Immunol. 2007;37 Suppl 1:S9–S17. doi: 10.1002/eji.200737638. [DOI] [PubMed] [Google Scholar]
- 20.Herbomel P, Thisse B, Thisse C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development. 1999;126(17):3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Wen Z. Cloning and expression pattern of the lysozyme C gene in zebrafish. Mech Dev. 2002;113(1):69–72. doi: 10.1016/s0925-4773(01)00658-x. [DOI] [PubMed] [Google Scholar]
- 22.Meijer AH, van der Sar AM, Cunha C, Lamers GE, Laplante MA, Kikuta H, Bitter W, Becker TS, Spaink HP. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol. 2008;32(1):36–49. doi: 10.1016/j.dci.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Su F, Juarez MA, Cooke CL, Lapointe L, Shavit JA, Yamaoka JS, Lyons SE. Differential regulation of primitive myelopoiesis in the zebrafish by Spi-1/Pu.1 and C/ebp1. Zebrafish. 2007;4(3):187–199. doi: 10.1089/zeb.2007.0505. [DOI] [PubMed] [Google Scholar]
- 24.Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238(2):274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- 25.Clay H, Ramakrishnan L. Multiplex fluorescent in situ hybridization in zebrafish embryos using tyramide signal amplification. Zebrafish. 2005;2(2):105–111. doi: 10.1089/zeb.2005.2.105. [DOI] [PubMed] [Google Scholar]
- 26.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol. 2002;248(2):307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 27.Redd MJ, Kelly G, Dunn G, Way M, Martin P. Imaging macrophage chemotaxis in vivo: Studies of microtubule function in zebrafish wound inflammation. Cell Motil Cytoskeleton. 2006 doi: 10.1002/cm.20133. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Bai XT, Zhu KY, Jin Y, Deng M, Le HY, Fu YF, Chen Y, Zhu J, Look AT, Kanki J, Chen Z, Chen SJ, Liu TX. In vivo interstitial migration of primitive macrophages mediated by JNK-matrix metalloproteinase 13 signaling in response to acute injury. J Immunol. 2008;181(3):2155–2164. doi: 10.4049/jimmunol.181.3.2155. [DOI] [PubMed] [Google Scholar]
- 29.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nusslein-Volhard C, Dahm R. A Practical Approach. New York, New York: Oxford University Press Inc.; 2002. Zebrafish. [Google Scholar]
- 31.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 32.Covassin L, Amigo JD, Suzuki K, Teplyuk V, Straubhaar J, Lawson ND. Global analysis of hematopoietic and vascular endothelial gene expression by tissue specific microarray profiling in zebrafish. Dev Biol. 2006;299(2):551–562. doi: 10.1016/j.ydbio.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4(12):1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 34.Hsu K, Traver D, Kutok JL, Hagen A, Liu TX, Paw BH, Rhodes J, Berman JN, Zon LI, Kanki JP, Look AT. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood. 2004;104(5):1291–1297. doi: 10.1182/blood-2003-09-3105. [DOI] [PubMed] [Google Scholar]
- 35.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19(1):71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 36.Peri F, Nusslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133(5):916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 37.Bertrand JY, Kim AD, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 2008;135(10):1853–1862. doi: 10.1242/dev.015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427(6970):154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 39.Kaplow LS. Simplified Myeloperoxidase Stain Using Benzidine Dihydrochloride. Blood. 1965;26:215–219. [PubMed] [Google Scholar]
- 40.Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a subpopulation of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127(14):3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 41.Clay H, Davis JM, Beery D, Huttenlocher A, Lyons SE, Ramakrishnan L. Dichotomous role of the macrophage in early Mycobacterium marinum infection of the zebrafish. Cell Host Microbe. 2007;2(1):29–39. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crowhurst MO, Layton JE, Lieschke GJ. Developmental biology of zebrafish myeloid cells. Int J Dev Biol. 2002;46(4):483–492. [PubMed] [Google Scholar]
- 43.Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, Hirsch K, Keller M, Forster R, Critchley DR, Fassler R, Sixt M. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453(7191):51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- 44.Cvejic A, Hall C, Bak-Maier M, Flores MV, Crosier P, Redd MJ, Martin P. Analysis of WASp function during the wound inflammatory response--live-imaging studies in zebrafish larvae. J Cell Sci. 2008;121(Pt 19):3196–3206. doi: 10.1242/jcs.032235. [DOI] [PubMed] [Google Scholar]
- 45.Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP, Zon LI, Look AT, Trede NS. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci U S A. 2004;101(19):7369–7374. doi: 10.1073/pnas.0402248101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin HF, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, Handin RI. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106(12):3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild-type embryos were sequentially immunolabeled using a rabbit antibody to MPO and a FITC-conjugated anti-rabbit Fab fragment (A,D,G), followed by a Rhodamine Red-conjugated rabbit antibody to L-Plastin (B,E,H); overlapping signals are yellow in overlay images (C,F,I). All frames are epifluorescence images, anterior is to the left and dorsal side up; arrows mark LP+/MPO− macrophages, white arrowheads mark LP+/MPO+ neutrophils, blue arrowheads mark LP−/MPO+ cells. (A–C) 28 hpf, Posterior Blood Island. (D–F) 2 dpf, Caudal Hematopoietic Tissue. (G–I) 2 dpf, head and yolk sac (composite images at different focal planes).
RT-PCR of cells from GFP-Lo and -Hi fractions, as in Fig. 3B but from a different preparation of cells and RNA. Genes are indicated to the left, cell type expression is indicated in parentheses; EF-1α is used as a loading control. Primer sequences are to the right of each gene, followed by references for each primer sequence; primers to CSF1R and Lysozyme C were designed in this study, therefore references detailing the expression patterns of these genes are listed in blue.
A GFPlo macrophage (starting in the lower left corner) extends two pseudopods, then “follows” the pseudopod nearest to a wound (out of frame, in the upper right corner) by moving the cell body into this pseudopod; a non-hematopoietic GFP+ cell is present in the upper right corner of the frame. Compression is 6 frames per second, with 1 min actual time between each frame.
A GFPlo macrophage sequentially extends multiple pseudopods during migration toward a wound (out of frame, in the upper left corner). Compression is 6 frames per second, with 1 min actual time between each frame.
A GFPlo macrophage (marked “1”) extends a pseudopod toward a wound (the edge of which is marked with a yellow line) and maintains the pseudopod in this position for several minutes before retracting it and migrating in the opposite direction; a GFPhi neutrophil can be seen migrating in the upper right corner of the frame. Later, a second GFPlo macrophage (marked “2”) migrates away from the wound and back to the vasculature (at the bottom of the frame). Compression is 6 frames per second, with 1 min actual time between each frame.
Following 2h of wound response, GFPlo macrophages (numbered 1–8) are lined along a wound (the edge of which is marked with a yellow line) and remain there after GFPhi neutrophils (marked with arrowheads) have left the wound. The vasculature is at the bottom of the frame, and a non-hematopoietic GFP+ cell is indicated (nh). Compression is 6 frames per second, with 1 min actual time between each frame.
A melanocyte over the yolk sac is ruptured by a laser-induced wound, resulting in the recruitment of GFPlo macrophages that phagocytose the pigment from the dead melanocyte. GFPhi neutrophils (note the large difference in GFP intensity) are also recruited but do not exhibit phagocytosis. Elapsed time is indicated in the lower left corner.