Summary
The SH2 domain of the C-terminal Src kinase [Csk] contains a unique disulfide bond not present in other known SH2 domains. To investigate whether this unusual disulfide bond could serve a novel function, the effects of disulfide bond formation on catalytic activity of the full-length protein and on the structure of the SH2 domain were investigated. The kinase activity of full-length Csk decreases by an order of magnitude upon formation of the disulfide bond in the distal SH2 domain. NMR spectra of the fully oxidized and fully reduced SH2 domains exhibit similar chemical shift patterns and are indicative of similar, well-defined tertiary structures. The solvent-accessible disulfide bond in the isolated SH2 domain is highly stable and far from the small lobe of the kinase domain. However, reduction of this bond results in chemical shift changes of resonances that map to a cluster of residues that extend from the disulfide bond across the molecule to a surface that is in direct contact with the small-lobe of the kinase domain in the intact molecule. Normal mode analyses and molecular dynamics calculations suggest that disulfide bond formation has large effects on residues within the kinase domain, most notably within the active-site cleft. Overall, the data indicate that reversible cross-linking of two cysteines in the SH2 domain greatly impacts catalytic function and domain-domain communication in Csk.
Keywords: Csk, cysteine, disulfide, kinase, NMR, phosphorylation
The Src family of tyrosine kinases [SFKs]1 are modular enzymes responsible for controlling various aspects of cellular growth and differentiation.[1] SH3 and SH2 domains in the SFKs flank the kinase domain and repress catalytic activity through intrasteric constraints. Most notably, the SH2 domain interacts tightly with a phosphotyrosine in the C-terminus of the kinase domain, inducing a closed conformation that poorly phosphorylates protein substrates.[2] Dephosphorylation of this tail segment potently activates the kinase domain by removing these interactions and inducing large conformational changes.[3] The phosphorylation state of the C-terminus of SFKs is controlled by Csk [C-terminal Src kinase], a ubiquitously expressed tyrosine kinase also containing SH2 and SH3 domains.[4] Although Csk is constitutively active and does not require phosphorylation for activity, cellular function is controlled through adaptor proteins. In T Cells, Cbp, a large transmembrane protein, tethers Csk to the plasma membrane via contacts between a phosphotyrosine in Cbp (pTyr-314) and the SH2 domain of Csk.[5, 6] Csk can then phosphorylate and down-regulate SFKs. In addition to Cbp, additional adaptor proteins such as caveolin-1, paxillin, and IRS-1 co-localize Csk to the membrane using the same strategy so that Csk can be recruited to other locations for alternate functions in different cells.[7]
The central kinase domain in Csk shares strong homology with other tyrosine kinases and possesses a smaller nucleotide binding lobe and a larger substrate binding lobe as well as an active-site cleft consisting of a catalytic loop (residues 312–319), an activation loop (residues 332–349) and a glycine-rich loop (residues 202–207) (Fig 1A). The SH2 and SH3 domains in Csk make direct contacts with only the nucleotide binding lobe through linker segments separating the domains (SH3-SH2 & SH2-kinase).[8] The interaction of the SH2 domain with the kinase domain is critical for proper function since deleting this domain reduces catalytic activity by two orders of magnitude.[9, 10] Furthermore, it appears that this functional enhancement can be guided largely through a narrow surface in direct contact with the small lobe of the kinase domain since mutation of a single residue in the SH2-kinase linker (Phe-183) can mimic the effects of complete SH2 domain deletion.[11] How the neighboring SH2 domain enhances catalysis is still unclear. A highly dynamic relationship between the SH2 and kinase domains is suggested by the X-ray structure[8] and has been supported in solution using backbone amide exchange and normal mode analyses.[12] These studies suggest that activation may be facilitated by cantilever motions between the two domains that induce and relieve stress in key regions of the kinase domain[8, 13] and has been supported in solution using backbone amide exchange and normal mode analyses.[12]
Fig. 1. Distal disulfide bond formation regulates Csk activity.
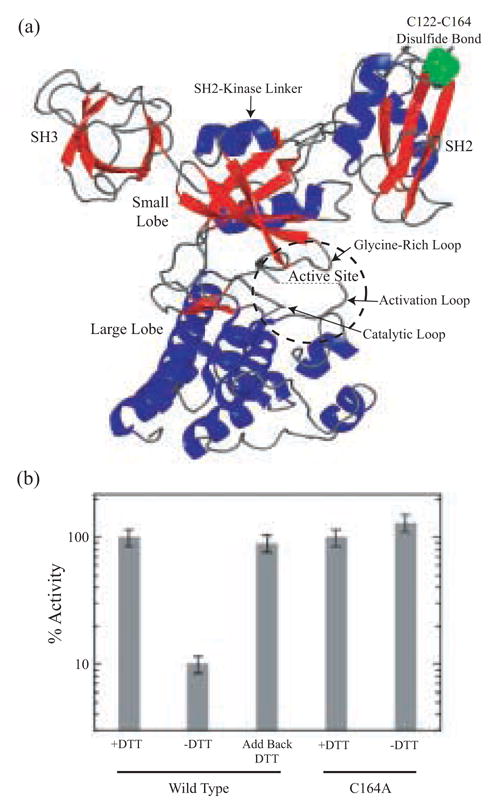
(A) Crystal structure of full-length Csk showing the SH3 and SH2 domains, ATP-binding lobe (A-lobe), peptide-binding lobe (P-lobe), the active-site cleft and the C122-C164 disulfide bond. (B) Time-dependent phosphorylation of poly(Glu4Tyr) by wild-type Csk and Csk-C164A was monitored using 32P kinetic assays. The activity of wild-type Csk was measured in the presence and absence of DTT (1 mM).
SH2 domains bind avidly to phosphotyrosine side chains of donor proteins, thereby linking signaling proteins to sites of cellular regulation.[14] SH2 domains are small (~100 aa), conserved protein modules composed of a central anti-parallel β sheet flanked by two α helices. Though not reported in the literature, examination of the crystal structure of Csk coupled with biochemical studies herein reveals that the SH2 domain in Csk possesses an additional structural refinement, a novel disulfide bond linking a cysteine at αC7 (Cys122 in Csk) with a cysteine in the BG loop (Cys164 in Csk) (Fig. 1A).[8] We sought to address whether this disulfide bond serves any structural role in the isolated Csk SH2 domain or whether its oxidation state can modulate catalytic activity in the distal kinase domain in the full-length protein. In this manuscript we describe an NMR analysis of the SH2 domain of Csk in both oxidized and reduced forms and show how chemical control of this distal structural region is correlated with two unique conformational states of the domain. We also show that the effects of oxidation state on protein conformation are not limited to the SH2 domain, despite being well removed from contact with the catalytic domain but rather extend to the active site of the neighboring kinase domain, an observation supported by activity assays and computational analyses.
Results
Regulation of Csk Activity Through Disulfide Bond Formation
We wished to determine whether reducing the Cys122-Cys164 disulfide bond in the SH2 domain of Csk would alter catalytic activity in the full-length protein. To address this issue, we performed in vitro kinetic assays to investigate the role of reducing agents in regulating Csk function. We monitored the ability of Csk to phosphorylate poly(Glu4Tyr), a general substrate for tyrosine kinases, in the presence and absence of DTT using a 32P-linked assay. Wild-type Csk was dialyzed in the absence and presence of 1 mM DTT overnight and then assayed. Activities were measured at identical enzyme concentrations (3 μM) and expressed relative to 100% for Csk in the presence of DTT (Fig. 1B). Removal of DTT from Csk results in a large reduction in catalytic activity (~10-fold). This alteration in catalytic activity is not the result of intermolecular disulfide bond formation as SDS-PAGE gels lacking reducing agents show that Csk dialyzed in the absence of DTT migrates as a monomer (data not shown). Treatment of the inactivated kinase with DTT in ‘add back’ experiments results in full recovery of the enzyme activity. Thus, the observed effects of reducing agent on catalytic activity are reversible. We measured the activity of a mutant Csk, (Csk-C164A), as a function of DTT to determine whether the loss and then recovery of activity in wild-type Csk is linked to disulfide bond oxidation state. The activity of Csk-C164A (which removes the disulfide bond) in both the presence and absence of DTT, is similar to the disulfide-reduced form of wild-type Csk (Fig. 1B). Thus, the kinetic assays suggest that catalytic activity can be regulated in a reversible manner by the oxidation state of a unique disulfide bond 40 Å removed from the active site of Csk.
Oxidation of Cysteines in Csk is Highly Specific
Using activity assays we showed that Csk can be reversibly oxidized and reduced (Fig. 1B). We next wished to determine whether oxidation results in chemical modifications to any cysteine side chains other than disulfide bond formation (e.g., sulfenic & sulfonic acids). To accomplish this we used a radiolabeled sulfhydryl-modifying agent, 14C-iodoacetamide (14C-IAM), to estimate all the available free cysteines in both the oxidized and reduced forms of Csk. In these experiments, Csk is treated with excess 14C-IAM (100 mM) at 37°C for 1 hr at pH 8 before removing all unreacted agent with exhausted dialysis. In the presence of DTT (5 mM), 100 ± 10 μM of 14C label was incorporated into 10 μM Csk. For the oxidized sample lacking any DTT, we incorporated 75 ± 10 μM 14C label into 10 μM Csk. Thus, while all 10 cysteines are labeled in reduced Csk, about 8 cysteines are labeled by 14C-IAM in the oxidized form. This result is consistent with the formation of one disulfide bond in the absence of DTT. To provide further support for these labeling results we pre-treated oxidized Csk with cold IAM (100 mM) to modify all the available cysteines. The modified protein was then reduced with DTT and then treated with 14C-IAM to modify any available cysteines not modified in the cold treatment. Under these conditions, 17 ± 7 μM 14C label was incorporated into 10 μM Csk, a value reflecting the modification of 2 cysteine side chains. Taken together, the data indicate that the reversible oxidation is specific.
Conformational Heterogeneity In the SH2 Domain of Csk
We showed that the catalytic activity of Csk is reversibly modulated by disulfide bond formation and that this activity switch is disconnected upon mutation of Cys-164. We next wanted to address the question-Is disulfide bond formation observed in the isolated SH2 domain? In an effort to address this question we investigated the conformation of the SH2 domain of Csk as a function of added reducing agent with solution NMR methods. A series of 1H-15N heteronuclear single quantum coherence (HSQC)[15] spectra were collected on a purified construct of the isolated domain expressed in bacteria. Visual inspection of the 1H-15N HSQC spectra revealed conformational heterogeneity in solution (Fig. 2A). Taken together with our analysis of the sequence-specific resonance assignments, the data indicate that 28 of the 101 residues are in slow exchange between two conformations (Fig. 2A). The sizeable difference in chemical shift values between the respective resonance sets for individual residues is fairly unusual for SH2 domains and suggests that the chemical environment of these nuclei is likely disposed in two unique structural conformations.[16, 17] The greatest chemical shift differences occur for residues directly surrounding Cys122 and Cys164 (>5 ppm in the 15N dimension and >0.3 ppm in the 1H dimension). Interestingly, the observed chemical shift heterogeneity is not limited to this local environment, but rather is extended to other regions of the structure. For example, there are significant chemical shift differences in residues 96–105, a polypeptide segment distant in primary sequence from the Cys122-Cys164 disulfide bond. Mapping the residues observed to be in slow exchange to the X-ray structure of the Csk SH2 domain[8] reveals that these heterogeneous regions form an extended surface surrounding the Cys122-Cys164 disulfide bond (Fig. 2D). Thus, the NMR data indicate that the SH2 domain of Csk equilibrates between two distinguishable conformations.
Fig. 2. The Csk SH2 domain is in slow exchange between two conformations in solution.
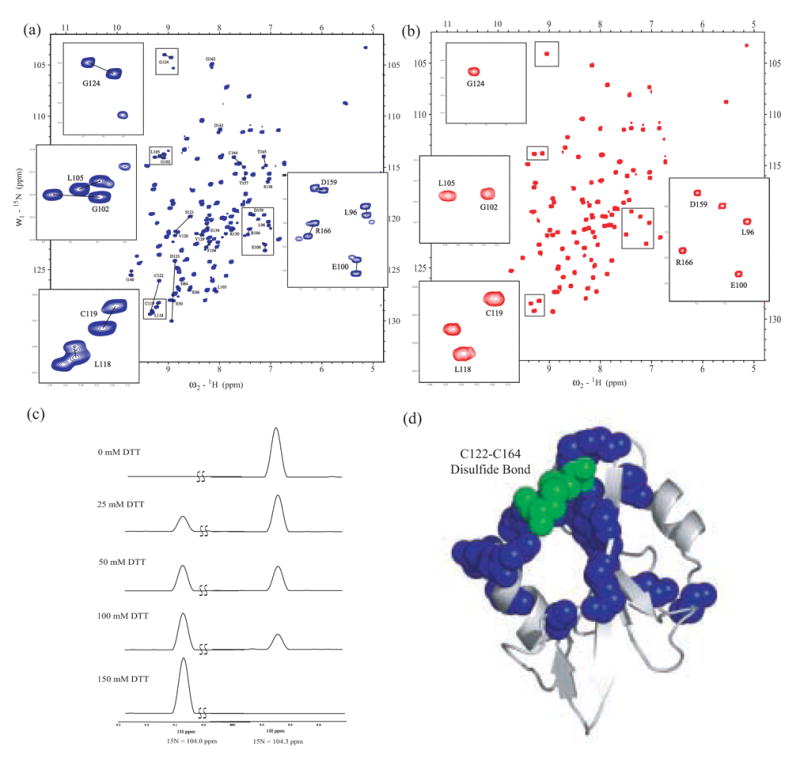
(A)1H-15N HSQC spectrum of 1.0 mM 15N-labeled Csk SH2 domain. Labeled cross-peaks correspond to residues populating double resonances. Resonances corresponding to two conformers of the same residue are connected by a black line. Boxed regions of the spectrum containing representative double resonances are expanded. (B) 1H-15N HSQC spectrum of the Csk SH2 domain (1.0 mM) in which Cys-164 is mutated to Ala. The same boxed regions of the spectrum are expanded but now show a single resonance for each residue. (C) Oxidation state of the disulfide bond in the SH2 domain specifies conformation. 1D projections of 2D 1H-15N HSQC spectra of the select region representing the dual resonances of G47. HSQC spectra of a 4 mM Csk SH2 sample in the presence of 0 mM, 25 mM, 50 mM, 100 mM and 150 mM DTT were recorded. (D) Heterogeneity maps to a contiguous surface. Residues populating dual resonances in the 1H-15N HSQC spectrum of wild-type Csk SH2 domain are mapped onto the X-ray crystal structure of the Csk SH2 domain and highlighted with blue spheres. Heterogeneity is localized to the region surrounding the disulfide bond, shown with green spheres.
Heterogeneity is Linked to Oxidation State of the Disulfide Bond
In an effort to establish the origin of the observed heterogeneity we performed structural studies of the wild-type SH2 domain as a function of DTT. 1H-15N HSQC spectra of the wild-type Csk SH2 domain (4.0 mM) were collected in the presence of varying concentrations of DTT. In the absence of DTT, the spectra indicate that the Csk SH2 domain populates a single conformation. However, addition of intermediate DTT concentrations (25–100 mM) induces a mixture of forms before a single distinguishable form prevails at high reducing agent concentration (150 mM). The effects on one representative residue (G47) are shown in Fig. 2C. At high DTT concentrations, the disulfide bond in the Csk SH2 domain sample was confirmed as reduced through Cα and Cβ chemical shift analysis (data not shown). Thus, the heterogeneity observed in the initial spectra (Fig. 2A) is likely due to a mixture of oxidized and reduced forms of the SH2 domain.
Since the conformational heterogeneity is greatest near the Cys122-Cys164 pair in the SH2 domain, we wondered whether removal of one of the cysteines could mimic a single, reduced form of the domain and simplify the observed NMR spectra. To test this possibility, the Cys-to-Ala mutation was also made in the isolated SH2 domain (SH2-C164A) and the mutant protein purified to homogeneity. The HSQC spectrum of C164A is simplified with each residue now populating only one resonance (Fig. 2B). The HSQC spectrum of the fully reduced form (150 mM DTT) of the wild-type SH2 domain precisely overlaps the spectrum of C164A (data not shown), suggesting that mutation fully mimics the reduced form of the protein. Thus, removal of the disulfide bond through either reducing agent or mutation locks the SH2 domain into only one distinguishable conformation.
Removal of the Disulfide Bond Affects Distal Surfaces on the SH2 Domain
Molecular Dynamics (MD) calculations probe the mechanical effects of chemical perturbation. Here we used CHARMM to conduct MD simulations of the isolated Csk SH2 domain, with explicit solvent, in order to explore the functional effects of the Cys122-Cys164 disulfide bond. Simulations of the structure with and without the disulfide bond were conducted and the resultant residue mobilities (B-factors) and RMSD values from the structure of the SH2 domain of Csk (1K9A.pdb:B) were calculated. The resultant B-factors describe the changes in the amplitudes of motions that specific residues experience, while the RMSD values describe the average displacement of specific residues from the unperturbed structure. Taken together, the theoretical B-factors and RMSD values are used to assess the physical effects of a specific chemical perturbation on the structure and dynamics of the isolated SH2 domain. Significant mechanical effects were detected upon removal of the disulfide bond, including increased mobilities and decreased fluctuations in various regions encompassing the SH2 domain (Fig. 3A). Interestingly, residues distal to the disulfide bridge show the most significant increase in B factor (residues 135 & 148–153) and form part of the docking site of the SH2 domain onto the small lobe of the kinase domain. Furthermore, the mean displacement of residues from the starting structure (RMSDs) indicates that residues 135–136 & 145–151 display the most significant changes in average position from the starting structure (Fig. 3B). Similarly, these residues within the SH2 domain are distal to the disulfide bridge and form contacts with the small lobe of the kinase domain and may provide a mechanism for the transmission of information to the kinase core.
Fig. 3. All atom MD Analysis of the SH2 Domain.
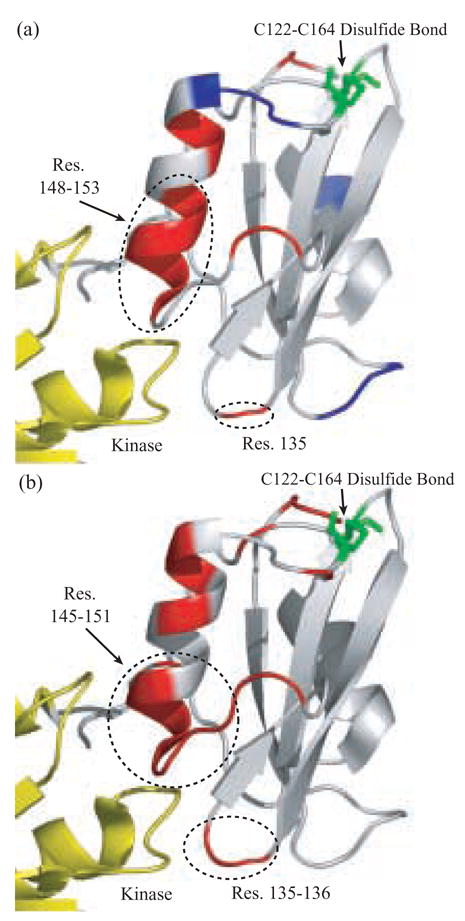
(A) Csk SH2 domain colored by change in residue B-factors upon removal of the Cys122-CysS164 disulfide bond. Residues 100–101, 135, 143–144, 148–153 and 156–157 show a positive change in B-factor and are highlighted in red. Residues 93–94, 111–113 and 159–161 show a negative change in B-factor and are highlighted in blue. (B) SH2 domain colored by change in RMSD from active conformation upon removal of disulfide bond. Residues 82, 99–101, 135–136, 143–151, 155, 157–158, 162 and 166 show an increase in RMSD and are shown in red and white indicates no change in the respective parameter upon perturbation.
Communication Between the Disulfide Bond and the Active Site of Csk
In many forms of engineering, frequency analysis is a valuable tool to learn how a device will respond to external stress. Similarly, frequency analysis of macromolecular machines, such as proteins, can illustrate the response to stress from the cellular environment. Here we employ normal mode (frequency) analysis (NMA) to study the effects of oxidative stress on the mechanical properties of Csk.
We showed previously that regions of high strain energy develop in and around the SH2-kinase linker owing to cantilever motions of the SH2 domain relative to the kinase domain.[11] Since the SH2 domain is critical for maintaining efficient catalysis in Csk,[10, 18] we wondered whether the oxidation state of the SH2 domain could influence strain energies within the Csk structure and, thus, provide theoretical grounds for understanding the effects of reducing agents on catalytic activity. To address this issue, we compared the normal modes and associated strain energies for Csk, both with and without disulfide bond formation between Cys122 and Cys164. Since crystallographic studies indicate that Csk populates two forms distinguished by the orientation of the SH2 domain relative to the kinase domain,[8] NMA were calculated for the “active” and “inactive” conformations of Csk (1K9A.pdb:B and 1K9A.pdb:C). The normalized projection of the two normal modes (compared for each conformation) for the Cα atoms is greater than 0.99999999, indicating that the path for global dynamic changes is being conserved. However, the strain energy is significantly affected by the removal of the Cys122-Cys164 disulfide bond (Fig. 4). Sizeable reductions in strain energy of residues within the glycine-rich loop (residues 205–207), the catalytic loop (residues 312–315 & 318–319) and the activation loop (residues 332–337 & 347–350) indicate that the active site is strongly influenced by changes in domain-domain communication between the kinase and SH2 domains upon disulfide reduction. Furthermore, control experiments where we removed the interactions between Lys203 and Lys225 (randomly chosen) had a negligible effect on the motion or strain energy, despite the proximity to the active site. Thus, NMA data indicate that the oxidation state of the disulfide in the SH2 domain can profoundly impact strain energies 40 Å distal to the site of chemical perturbation in both the “active” and “inactive” forms of Csk.
Fig. 4. Communication between the disulfide bond in the SH2 domain and the active site of Csk.
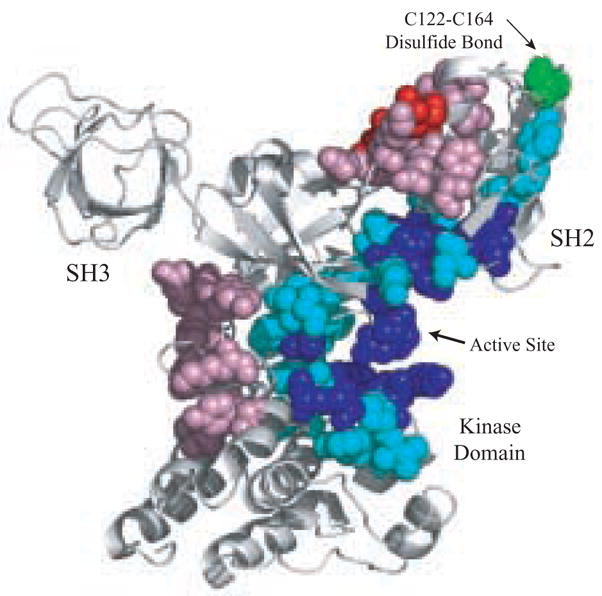
Csk is colored based on changes in strain energy upon removal of Cys122-Cys164 bond. Blue indicates a large decrease in strain, cyan indicates a moderate decrease in strain, red indicates a large increase in strain, and pink indicate a moderate increase in strain. White indicates no significant change in strain energy upon removal of the disulfide bond.
Discussion
Crystallographic studies indicate that Csk can populate two forms distinguished by ‘up’ (inactive) or ‘down’ (active) orientations of the SH2 domain relative to the kinase domain.[8] The latter orientation brings active-site residues into proper position for catalysis whereas the former orientation disrupts these residues owing to a slight rotation of the small lobe of the kinase domain. Prior backbone amide exchange and NMA studies suggested that this phenomenon may also occur in solution, thereby, constituting a unique regulatory mechanism where cantilever movements of the SH2 domain could facilitate substrate phosphorylation.[11] We now show evidence from NMR studies that the free Csk SH2 domain can exist in two distinct, low energy conformations in solution and that the population of these two forms is directly linked to the oxidation state of a novel disulfide bond within the domain. While the precise changes in SH2 domain conformation upon disulfide bond formation awaits more detailed structural investigations, the NMR data indicate that many of the impacted residues map near and some far from the bonded cysteines (Fig. 2D) and that the chemical linking of the side chains of these two residues can have effects on multiple surfaces of the SH2 domain. Such findings are consistent with the cantilever model for Csk regulation and further suggest that the relative motions between the two domains may be initiated, in part, by inherent flexibility within the SH2 domain itself.
How changes in SH2 domain orientation in Csk might influence substrate processing is a perplexing problem. While the SH2 domain ‘up’ conformer is considered catalytically competent, both forms could be required for ATP and substrate binding. For example, while the ‘open’ form of the catalytic subunit of PKA generated by lobe rotations in the kinase domain is also considered noncatalytic, it may be important for entry of ligands into the active site.[19, 20] Whether such motions play a similar role in Csk is not clear. We have observed that the binding of ligands to the SH2 domain increases the phosphoryl transfer rate and causes large changes in H-D exchange kinetics and strain energy in the active site.[11] These effects may be conveyed to the active site by changes in the amplitude and/or frequency of the cantilever motions of the SH2 domain. By analogy, it is conceivable that disulfide bond formation in the SH2 domain may lead to activity decreases by altering the population and/or interconversion rates of the active & inactive conformers. MD simulations of the isolated SH2 domain have shown that removal of the disulfide bond can substantially change the mobility of residues that pack against the small lobe of the kinase domain in the intact protein (Fig 3). Comparative NMA analysis of the full-length protein indicate that changes are not limited to the SH2 domain but extend into the active site upon disruption of the disulfide bond. The most sensitive region in the active site to disulfide bond removal is the glycine-rich loop of Csk (residues 202–207, Fig. 4) which contains three highly conserved glycines that greatly control catalytic activity.[21, 22] We also observed that activation of Csk through phosphopeptide binding to the SH2 domain decreases strain energies in this glycine-rich loop.[11] Likewise, in this study, activation of Csk through disulfide bond removal decreases strain in this loop. In addition, our current NMA suggests that removal of the disulfide bond lowers the activation barrier and facilitates the interconversion of the “active” and “inactive” states. This interesting correspondence suggests that there may be a direct correlation between changes in strain energies within certain active-site sequences, ground-state fluctuations and catalytic activation.
The observed correlation of changes in strain energy that accompanies catalysis is reminiscent of the “Elastomeric Rack Mechanism of Enzyme Catalysis” first proposed by Eyring.[23] In this model, while ordering of the active site structure is necessary to bind the substrate, the large relative size of the enzyme to the active site suggests that the useful function of the enzyme active-site periphery is to facilitate the population of multiple nearly isoenergetic conformations in solution. These conformations may constitute “open and closed” or “active and inactive” molecules that interconvert during catalytic cycling. The enzyme facilitates catalysis by transmitting stress into the substrate and increasing its reactivity, and “it seems necessary to assume that, in many cases, these stresses must be transmitted mechanically.” In this light, the conformational changes in Csk could represent an almost muscular action that facilitates catalysis not unlike a “boa constrictor-like action within the biologically active giant molecules (enzyme) that accompanies their conformational changes. Any victim (substrate) caught in the writhing coils of a boa constrictor would do anything to get out, including reacting chemically.”[23]
While the effects of disulfide bond reduction on strain energies within the active site are correlated with activity increases (Figs. 1B & 4), this chemical modification has other effects on Csk as well. For example, decreases in strain energy in the active site are accompanied by increases in strain energies in other regions of the kinase domain (Fig. 4). Removal of the disulfide bond increases strain near the SH2-kinase linker (residues 72–81) and the hinge region connecting the small and large lobes within the kinase domain (residues 267–272). This could be the result of cantilever motions that rock the SH2 domain counterclockwise about the SH2-kinase linker, increasing strain at the lobe hinge and subsequently relieving strain at the active site. In this manner, mechanical stresses are finely tuned in Csk with increases in strain at some regions being carefully balanced by decreases at other regions. This communication is reminiscent of studies on dihydrofolate reductase where a network of coupled motions work cohesively to support substrate binding and catalysis.[24]
The presence of a novel disulfide bond in the SH2 domain of Csk raises some very compelling questions regarding biological control of activity. While it is normally assumed that disulfide bonds will be unstable in the reducing environment of the cell, we have detected this disulfide in the presence of intermediate levels of DTT (Figs. 1A & 2A). Whether this bond is present in the cell under normal conditions is still unknown. We propose that given the close apposition of both cysteines in the structure, a portion of the Csk molecules may be oxidized in certain cell types. While Csk is a cytosolic enzyme it is normally localized to lipid rafts in T cells via the Cbp adaptor protein.[25] Localization of this type could further shield the disulfide from oxidation. Another exciting possibility is that the disulfide bond may be highly sensitive to oxidative conditions such as those anticipated in inflammatory cells where superoxides are generated upon cell activation.[26, 27] Whether such inducible conditions could chemically modify the disulfide bond is unknown but raises compelling questions regarding the regulation of Csk under certain oxidative stress conditions.
Materials and Methods
Materials
Dithiothreitol (DTT) and the poly(Glu4Tyr) peptide were obtained from Sigma. Adenosine triphosphate (ATP), 3-(N-morpholino) propanesulphonic acid (MOPS), magnesium chloride, potassium chloride, Kodak imaging film (Biomax MR) and liquid scintillant were obtained from Fisher Scientific. [γ-32P]ATP was obtained from NEN Products. 14C-iodoacetamide was obtained from MP Biomedicals. [15N] ammonium sulfate, [13C] glucose and D2O were obtained from Isotec and Cambridge Isotope Laboratories
Generation of Csk Constructs
(His)6-tagged full-length human Csk [28] was used as the template to generate the C164A point mutation introduced by the Quick-Change (Stratagene) method. The gene segment encoding the Csk SH2 Domain, residues 78–178, was amplified using the polymerase chain reaction (PCR). The amplified fragment was cloned into the pET-21a(+) vector at the BamH1–Xho1 sites. All constructs were confirmed by DNA sequencing.
Protein Expression and Purification
Full-length Csk enzymes were expressed in Escherichia coli strain BL21(DE3) as described[29] and purified by Ni2+ affinity chromatography as described.[28] The purified full-length enzymes were dialyzed against 10 mM Tris-Cl (pH 8.0), 100 mM KCl with 10% glycerol and then concentrated to approximately 50 μM and stored at −80°C. The Csk SH2 Domain constructs were expressed in E. coli strain BL21(DE3) as described[30] in modified M9 minimal medium and were purified by Ni2+ affinity chromatography as described.[28] The modified M9 medium contained [15N] ammonium sulfate (2 g/L) and/or [13C] glucose (2 g/L) for uniformly 15N labeled and 15N/13C labeled protein, respectively. The purified Csk SH2 constructs were dialyzed against 10 mM phosphate buffer (pH 7.0), 50 mM NaCl with 0.5 mM DTT and then concentrated to approximately 1–4 mM for NMR experiments. Various DTT concentrations were added to a Csk SH2 domain sample with no DTT present in the buffer for NMR experiments. All protein samples contained 5% D2O (v/v) for NMR experiments. Protein concentrations were determined by the method of Gill & von Hippel[31] and purity was assessed by SDS-PAGE.
NMR Spectroscopy
All NMR spectra were acquired at 25°C on a Bruker DMX 500-MHz spectrometer. A gradient-enhanced HSQC experiment with minimal water saturation[15] was used for all 1H-15N correlation experiments. Sequence-specific assignments of the polypeptide backbone resonances of the Csk SH2 domain were obtained by standard triple resonance techniques.[32] The programs NMRPipe[33] and Sparky[34] were used for spectral processing and data analysis, respectively.
Autographic Assay for poly(Glu4Tyr) phosphorylation
Time-dependent phosphorylation of poly(Glu4Tyr) (5 mg/mL) by wild-type or mutant Csk enzymes (3 μM) was carried out in 100 mM MOPS (pH 7.0), 100 mM KCl, 10 mM free Mg2+, [γ-32P]ATP (600–1000 cpm pmol−1) in the presence and absence of DTT (1 mM) at 23°C. Reactions were quenched with 10 uL of SDS/PAGE loading buffer. A fraction of the total quenched reaction mixture (20 uL) was separated using a 10% SDS-PAGE gel. Bands corresponding to the phosphorylated poly(Glu4Tyr) peptide were excised from the dried SDS/PAGE gel and quantified on the 32P channel in liquid scintillant. The activity of the enzymes was attained by measuring the first minute of the reaction and establishing the initial velocity of the reaction.
Cysteine alkylation using 14C-iodoacetemide
The number of free cysteines was measured using a sulfhydryl-modifying agent, 14C-iodoacetamide (14C-IAM). Full-length Csk (10 uM) dialyzed in the presence or absence of 5 mM DTT was treated with 14C-IAM (100 mM) at 37° in the dark for 1 hour in a buffer containing 6 M GdnHCl, 10 mM Tris-HCl (pH 8), 100 mM KCl and 10% glycerol. Unreacted 14C-IAM was then removed from each sample using dialysis and the incorporation of 14C into the Csk samples were quantified using the 14C channel in liquid scintillant. This protocol was also performed for samples lacking Csk and these CPMs were subtracted from the samples containing Csk. To determine how may cysteines are involved in disulfide bond formation, a sequential modification-reduction protocol was employed.[35] Csk dialyzed in the absence of DTT was reacted with unlabeled IAM (100 mM) using the same conditions as that for the labeled reagent. After this time, DTT (50 mM) was added to reduce any disulfide bonds present. After incubation for 1 hour at 37°, 14C-IAM was added to alkylate irreversibly all newly reduced cysteines.
All Atom Calculations
Molecular dynamics was performed on the SH2 domain (residues 78–178) of the active form of Csk (chain B in 1K9A.pdb)[8] using CHARMM[36] with the potential function parameter set 22. 1.5 ns runs were performed with and without a disulfide bond connecting Cys122 and Cys164. Both simulations were run with constant pressure (1 ATM) and temperature (300K). Explicit water was modeled with the TIP3P potential [37] in an orthorhombic cell of linear dimensions 60, 53 and 52 angstroms with periodic boundary conditions. For non-bonded terms, we used a cutoff of 14 angstroms, a shifting function for van der Waals interactions and a force-switching function for electrostatics. The SHAKE algorithm[38] was used to constrain covalent bonds involving hydrogen atoms. The Verlet algorithm was used with an integration step of 2 fs. RMSD of the Cα atoms was computed for residues 83–173 using PROFIT.[39] Contacts of Structural Units (CSU) were derived with CSU software.[40]
Normal Mode Analysis
Normal mode analysis was conducted on the active conformation of Csk using CHARMM[36] using the Tirion Potential,[41] in conjunction with the RTB method.[42] In NMA the dynamic properties of a biological system are approximated as a set of independent harmonic oscillators (i.e. normal modes). Two sets of normal modes were determined, one set corresponding to all atoms within 8 Angstroms interacting, including interactions between Cys122 and Cys164, and one set without interactions between Cys122 and Cys164. Strain energy was calculated as described,[43] and the difference in strain was determined.
Acknowledgments
This work was supported by NIH grants GM 68168 to J.A.A. and DK54441 to P.A.J.
Footnotes
Abbreviations: Csk, Carboxyl terminal Src kinase; DTT; dithiothreitol; HSQC, heteronuclear single quantum coherence; IAM, iodoacetamide; MD, molecular dynamics; NMA, normal mode analysis; polyGlu4Tyr, random heteropolymer of glutamic acid and tyrosine; SH2, Src homology 2 domain; SH3, Src homology 3 domain; SFK, Src family kinase; Src, tyrosine kinase from Rous Sarcoma virus.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 2.Sicheri F, Kuriyan J. Structures of Src-family tyrosine kinases. Curr Opin Struct Biol. 1997;7(6):777–85. doi: 10.1016/s0959-440x(97)80146-7. [DOI] [PubMed] [Google Scholar]
- 3.Cowan-Jacob SW, et al. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure (Camb) 2005;13(6):861–71. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Okada M, et al. CSK: a protein-tyrosine kinase involved in regulation of src family kinases. J Biol Chem. 1991;266(36):24249–52. [PubMed] [Google Scholar]
- 5.Brdicka T, et al. Phosphoprotein associated with glycosphingolipid-enriched microdomains (PAG), a novel ubiquitously expressed transmembrane adaptor protein, binds the protein tyrosine kinase csk and is involved in regulation of T cell activation. J Exp Med. 2000;191(9):1591–604. doi: 10.1084/jem.191.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawabuchi M, et al. Transmembrane phosphoprotein Cbp regulates the activities of Src-family tyrosine kinases. Nature. 2000;404(6781):999–1003. doi: 10.1038/35010121. [DOI] [PubMed] [Google Scholar]
- 7.Chong YP, Mulhern TD, Cheng HC. C-terminal Src kinase (CSK) and CSK-homologous kinase (CHK)--endogenous negative regulators of Src-family protein kinases. Growth Factors. 2005;23(3):233–44. doi: 10.1080/08977190500178877. [DOI] [PubMed] [Google Scholar]
- 8.Ogawa A, et al. Structure of the carboxyl-terminal Src kinase, Csk. Journal of Biological Chemistry. 2002;277(17):14351–14354. doi: 10.1074/jbc.C200086200. [DOI] [PubMed] [Google Scholar]
- 9.Sun G, Budde RJ. Mutations in the N-terminal regulatory region reduce the catalytic activity of Csk, but do not affect its recognition of Src. Arch Biochem Biophys. 1999;367(2):167–72. doi: 10.1006/abbi.1999.1253. [DOI] [PubMed] [Google Scholar]
- 10.Sondhi D, Cole PA. Domain interactions in protein tyrosine kinase Csk. Biochemistry. 1999;38(34):11147–11155. doi: 10.1021/bi990827+. [DOI] [PubMed] [Google Scholar]
- 11.Wong L, et al. Dynamic coupling between the SH2 domain and active site of the COOH terminal Src kinase, Csk. Journal of Molecular Biology. 2004;341(1):93–106. doi: 10.1016/j.jmb.2004.05.060. [DOI] [PubMed] [Google Scholar]
- 12.Wong L, et al. Coupled motions in the SH2 and kinase domains of Csk control Src phosphorylation. J Mol Biol. 2005;351(1):131–43. doi: 10.1016/j.jmb.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 13.Lieser SA, et al. Coupling phosphoryl transfer and substrate interactions in protein kinases. Biochim Biophys Acta. 2005;1754(1–2):191–9. doi: 10.1016/j.bbapap.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Marengere LE, Pawson T. Structure and function of SH2 domains. J Cell Sci Suppl. 1994;18:97–104. doi: 10.1242/jcs.1994.supplement_18.14. [DOI] [PubMed] [Google Scholar]
- 15.Mori S, Abeygunawardana C, O’Neill Johnson M, Van Zul PCM. Improved sensitivity of HSCQ spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson. 1995;B108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 16.Mulhern TD, To C, Cheng HC. 1H, 13C and 15N chemical shift assignments of the SH2 domain of the Csk homologous kinase. J Biomol NMR. 2002;24(4):363–4. doi: 10.1023/a:1021657406480. [DOI] [PubMed] [Google Scholar]
- 17.Ojennus DD, Fleissner MR, Wuttke DS. Reconstitution of a native-like SH2 domain from disordered peptide fragments examined by multidimensional heteronuclear NMR. Protein Sci. 2001;10(11):2162–75. doi: 10.1110/ps.18701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin XF, et al. Probing the communication between the regulatory and catalytic domains of a protein tyrosine kinase, Csk. Biochemistry. 2005;44(5):1561–1567. doi: 10.1021/bi048142j. [DOI] [PubMed] [Google Scholar]
- 19.Zheng J, et al. Crystal structures of the myristylated catalytic subunit of cAMP- dependent protein kinase reveal open and closed conformations. Protein Sci. 1993;2(10):1559–73. doi: 10.1002/pro.5560021003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsigelny I, et al. 600 ps molecular dynamics reveals stable substructures and flexible hinge points in cAMP dependent protein kinase. Biopolymers. 1999;50(5):513–24. doi: 10.1002/(SICI)1097-0282(19991015)50:5<513::AID-BIP5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 21.Hirai TJ, Tsigelny I, Adams JA. Catalytic assessment of the glycine-rich loop of the v-Fps oncoprotein using site-directed mutagenesis [In Process Citation] Biochemistry. 2000;39(43):13276–84. doi: 10.1021/bi001216g. [DOI] [PubMed] [Google Scholar]
- 22.Grant BD, et al. Kinetic analyses of mutations in the glycine-rich loop of cAMP- dependent protein kinase. Biochemistry. 1998;37(21):7708–15. doi: 10.1021/bi972987w. [DOI] [PubMed] [Google Scholar]
- 23.Eyring H, Johnson FH. The elastomeric rack in biology. Proc Natl Acad Sci U S A. 1971;68(10):2341–4. doi: 10.1073/pnas.68.10.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annual Review of Biochemistry. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 25.Torgersen KM, et al. Release from Tonic Inhibition of T Cell Activation through Transient Displacement of C-terminal Src Kinase (Csk) from Lipid Rafts. J Biol Chem. 2001;276(31):29313–29318. doi: 10.1074/jbc.C100014200. [DOI] [PubMed] [Google Scholar]
- 26.Monteiro HP, Stern A. Redox modulation of tyrosine phosphorylation-dependent signal transduction pathways. Free Radical Biology and Medicine. 1996;21(3):323–333. doi: 10.1016/0891-5849(96)00051-2. [DOI] [PubMed] [Google Scholar]
- 27.Cao HM, Sanguinetti AR, Mastick CC. Oxidative stress activates both Src-kinases and their negative regulator Csk and induces phosphorylation of two targeting proteins for Csk: caveolin-1 and paxillin. Experimental Cell Research. 2004;294(1):159–171. doi: 10.1016/j.yexcr.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Lin X, Lee S, Sun G. Functions of the activation loop in Csk protein-tyrosine kinase. J Biol Chem. 2003;278(26):24072–7. doi: 10.1074/jbc.M210596200. [DOI] [PubMed] [Google Scholar]
- 29.Bougeret C, et al. Recombinant Csk Expressed in Escherichia-Coli Is Autophosphorylated on Tyrosine Residue(S) Oncogene. 1993;8(5):1241–1247. [PubMed] [Google Scholar]
- 30.Waksman G, et al. Crystal-Structure of the Phosphotyrosine Recognition Domain Sh2 of V-Src Complexed with Tyrosine-Phosphorylated Peptides. Nature. 1992;358(6388):646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 31.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182(2):319–26. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 32.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy, Principles and Practice. Academic Press, Inc; San Diego: 1996. [Google Scholar]
- 33.Delaglio F, et al. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Goddard TD, Kneller DG. SPARKY. 2000;3 [Google Scholar]
- 35.Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2(11):e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks BR, et al. Charmm - a Program for Macromolecular Energy, Minimization, and Dynamics Calculations. Journal of Computational Chemistry. 1983;4(2):187–217. [Google Scholar]
- 37.Jorgensen WL, et al. Comparison of Simple Potential Functions for Simulating Liquid Water. Journal of Chemical Physics. 1983;79(2):926–935. [Google Scholar]
- 38.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. Journal of Computational Physics. 1977;23(3):327–341. [Google Scholar]
- 39.Martin ACR. http://www.bioinf.org.uk/software/profit/
- 40.Sobolev V, et al. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15(4):327–332. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 41.Tirion MM. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Physical Review Letters. 1996;77(9):1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- 42.Tama F, et al. Building-block approach for determining low-frequency normal modes of macromolecules. Proteins. 2000;41(1):1–7. doi: 10.1002/1097-0134(20001001)41:1<1::aid-prot10>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 43.Miyashita O, Onuchic JN, Wolynes PG. Nonlinear elasticity, proteinquakes, and the energy landscapes of functional transitions in proteins. Proc Natl Acad Sci U S A. 2003;100(22):12570–5. doi: 10.1073/pnas.2135471100. [DOI] [PMC free article] [PubMed] [Google Scholar]


