Abstract
During apoptosis and under conditions of cellular stress, several signaling pathways promote inhibition of cap-dependent translation while allowing continued translation of specific messenger RNAs encoding regulatory and stress-response proteins. We report here that the apoptotic regulator Reaper inhibits protein synthesis by binding directly to the 40S ribosomal subunit. This interaction does not affect either ribosomal association of initiation factors or formation of 43S or 48S complexes. Rather, it interferes with late initiation events upstream of 60S subunit joining, apparently modulating start-codon recognition during scanning. CrPV IRES–driven translation, involving direct ribosomal recruitment to the start site, is relatively insensitive to Reaper. Thus, Reaper is the first known cellular ribosomal binding factor with the potential to allow selective translation of mRNAs initiating at alternative start codons or from certain IRES elements. This function of Reaper may modulate gene expression programs to affect cell fate.
Rapid changes in cellular gene expression are often brought about by regulation at the level of protein synthesis from existing mRNA transcripts. Such alterations are particularly important under conditions of cellular stress and apoptosis, and during certain stages of mitosis1. Cellular stresses such as viral infection or nutrient deprivation lead to an almost immediate shutdown of general translation accompanied by cleavage or covalent modification of one or more of the eukaryotic initiation factors (eIFs)2–5. However, this attenuation of translation is typically accompanied by a selective increase in the translation of specific regulatory proteins6–9 whose sustained expression can affect cell fate1,10. How particular mRNAs are selectively translated during periods of global translation inhibition is not well understood.
Translation of the vast majority of cellular mRNAs is initiated via recruitment of the small (40S) ribosomal subunit to the m7G cap at the 5′ end of the mRNA. The 40S subunit is thought to scan linearly along the mRNA until it engages the first AUG codon located within the proper context11. The interaction between the 40S subunit and the mRNA and the subsequent translation initiation steps are chaperoned by eIF regulatory proteins1,2,5–7. In canonical cap-dependent translation, initiation factors have many roles: they prevent premature subunit association, recruit the initiator transfer RNA to the 40S subunit to form a 43S complex, recruit the 43S complex to the mRNA, unwind the mRNA during ribosomal scanning and facilitate correct start-codon recognition12–16. When the function of particular initiation factors is compromised by cellular stressors, cap-dependent mRNAs are incapable of sustaining protein synthesis5,17.
The ability of specific mRNAs to be translated in a cap-independent manner relies primarily on their ability to directly bind the ribosome and position it at the start codon. This can be accomplished through internal ribosomal entry site (IRES) sequences in the mRNA, which, by directly recruiting the ribosome, bypass requirements for compromised initiation factors7,12. Therefore, depending on the mechanism of translation suppression, the expression from certain IRESs can be maintained under conditions in which protein expression from cap-dependent mRNAs is inhibited18.
A number of regulatory proteins have been identified that promote initiation from IRESs, but their mechanism of action is not currently known19–22. Other proteins, such as the ribosome inhibitory proteins (RIPs), directly bind the ribosome and irreversibly cleave ribosomal RNA, leading to inhibition of all translation23. Thus far, no cellular regulatory protein able to directly bind the ribosome and lead to the selective expression of specific cap-independent messages has been reported.
We and others have shown previously that Reaper, a potent apoptotic inducer, can inhibit general protein synthesis24–27. This inhibition of protein synthesis is not the result of its proapoptotic activity, as it is a genetically separable function of Reaper that does not require activation of the apoptotic program through caspase activation26. Thus, we set out to determine how Reaper protein can inhibit protein translation.
We demonstrate here that Drosophila melanogaster Reaper binds directly and specifically to the 40S subunit of the eukaryotic ribosome. This interaction does not affect early initiation events such as 43S or 48S complex formation. It acts after 48S assembly but before 60S subunit joining. Notably, Reaper’s mechanism of translation inhibition gives rise to differential inhibition of certain mRNAs. Reaper inhibits cap-dependent translation almost completely under conditions that still allow substantial cap-independent translation mediated by the CrPV IRES. Our results provide compelling evidence for a novel mechanism of translation regulation, wherein a protein regulator directly binds the 40S subunit to modulate translation initiation. This regulation has the potential to alter cell fate by rapidly changing the profile of proteins expressed.
RESULTS
Reaper binds the ribosome
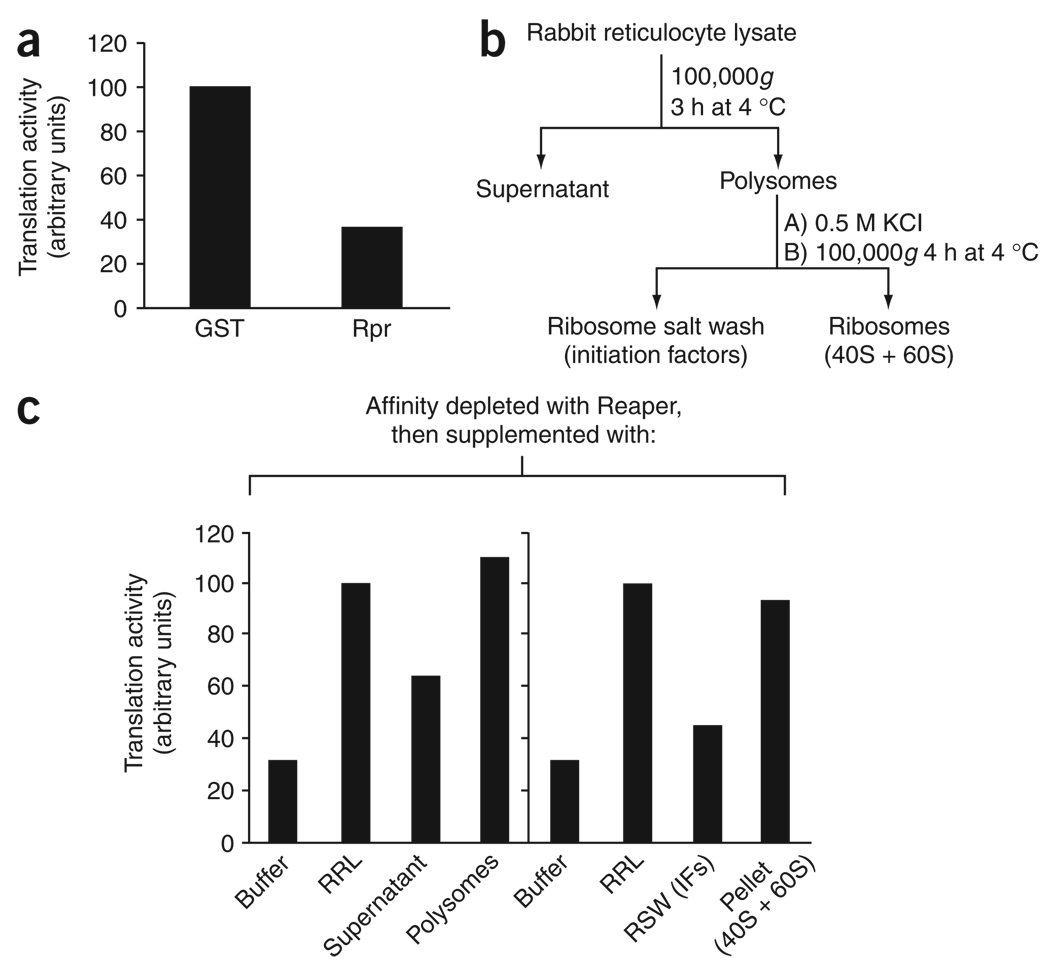
As reported previously, synthetic peptides encoding either the full-length, 65-residue Reaper protein or Reaper lacking its first 15 residues (Reaper 16–65) inhibit general translation upon addition to rabbit reticulocyte lysate (RRL) or Xenopus laevis egg extract translation systems26. To determine the molecular mechanism by which Reaper modulates protein translation, we wished to identify factors required for this inhibition in the RRL28,29. Given the absence of any obvious catalytic domains in Reaper, we reasoned that Reaper might bind directly to translational regulators, and we therefore set out to determine whether RRL depleted of Reaper-interacting factors could retain the ability to translate. For this purpose, the fragment containing residues 16–65 of Reaper (Reaper16–65) was synthesized as an N-terminally biotinylated peptide. The Reaper peptide was then coupled to streptavidin sepharose and incubated with RRL containing endogenous mRNAs. Protein-bound beads were retrieved by centrifugation, and the RRL depleted of Reaper-bound factors was assayed for its ability to support protein translation of endogenous mRNAs. Depletion of Reaper-bound factors markedly compromised the translational activity of the RRL (Fig. 1a). These data suggest that Reaper might interact physically with an essential component of the translation machinery.
Figure 1.
Ribosomes restore Reaper’s inhibitory activity. (a) Translation of endogenous RRL mRNAs in the presence or absence of Reaper (Rpr). Biotinylated Reaper peptide or GST protein coupled to streptavidin beads was used to affinity-deplete untreated RRL (containing endogenous RRL mRNA). Translation was assayed and quantitated as described in Methods. (b) Schematic representation of the fractionation protocol used to purify Reaper’s translational inhibitory activity (protocol adapted from refs. 28,29). (c) Translation of endogenous mRNAs as in a, except that RRL was supplemented before the translation reaction with either buffer, fresh RRL, supernatant or polysome pellet (containing polyribosomes and associated initiation factors) (left chart), or buffer, fresh RRL, ribosomal salt wash (RSW) containing initiation factors (IFs) or a ribosomal pellet (right chart). Activity is normalized to that of the affinity-depleted RRL rescued with fresh RRL.
To determine which translational components might be relevant to Reaper’s inhibitory activity, we fractionated RRL by centrifugation and determined whether the resulting supernatant, containing the bulk of the initiation factors, or the pellet, containing ribosomes, could restore translation to the RRL depleted of Reaper-interacting factors (Fig. 1b)28,29. To our surprise, we found that the ribosome-containing fraction was able to complement the depleted extracts. Furthermore, washing of the ribosomes with 0.5 M KCl to remove associated factors did not compromise the ability of the ribosomal fraction to restore translation (Fig. 1c). The ribosomal fraction was unable to enhance translation of the control extracts depleted with glutathione S-transferase (GST) protein, which ruled out the possibility that complementation of the depleted extracts resulted from simple addition of active ribosomes (Supplementary Fig. 1 online). Together, these data suggest that Reaper inhibited translation through effects on the ribosomes.
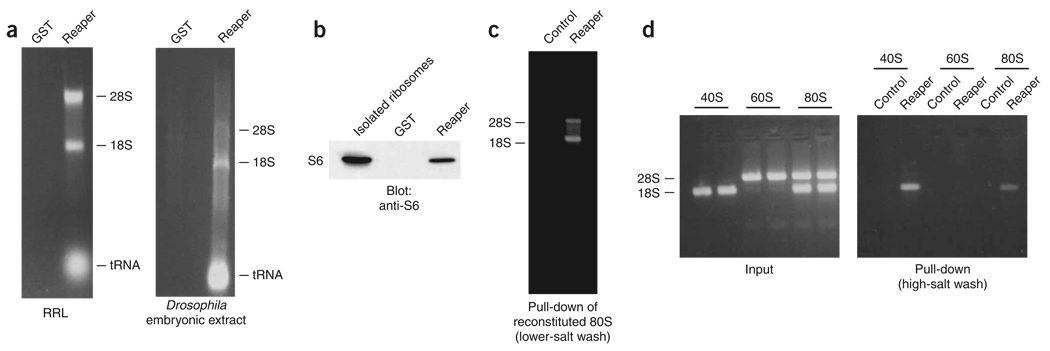
To address more directly the ability of Reaper to interact with the ribosome, we analyzed a number of Reaper-bound factors identified by SDS-PAGE using MS. These analyses revealed that Reaper associated with an array of core ribosomal proteins in RRL, Xenopus egg extracts and Drosophila embryonic extracts (data not shown). To confirm further Reaper’s interaction with the ribosome, biotinylated Reaper pull-downs from RRL and Drosophila embryonic extracts were phenol-extracted and the resulting material was resolved on agarose gels. Biotinylated Reaper, but not control biotinylated GST protein, was able to retrieve both the 18S and 28S rRNAs from RRL and Drosophila embryonic extracts (Fig. 2a). Moreover, immunoblotting of Reaper-bound material for a representative ribosomal protein revealed that Reaper bound sufficiently tightly to ribosomes to retrieve them efficiently from extracts even under stringent washing conditions (Fig. 2b). This ability of Reaper to interact with ribosomes was specific to eukaryotic ribosomes; Reaper was unable to bind prokaryotic ribosomes, but interacted well with eukaryotic ribosomes from RRL, Xenopus egg extracts and Drosophila embryonic extracts (Fig. 2a and data not shown).
Figure 2.
Reaper binds ribosomes through direct interaction with the 40S subunit. (a) Reaper binding to ribosomes. Reaper (or GST)-bound streptavidin beads were used to affinity-deplete RRL (left gel) or Drosophila embryonic extracts (right gel). Bound material was stringently washed, phenol-chloroform extracted and resolved by agarose gel electrophoresis. (b) Reaper binding to tightly associated ribosomal protein S6. RRL was prepared as in a, but bound material was resolved by PAGE and immunoblotted with antibody directed against S6. Input (isolated ribosomes) was also immunoblotted as a control. (c) Reaper binding to 80S complexes. 40S and 60S ribosomal subunits were purified and combined to form 80S complexes, then incubated with biotinylated Reaper, and Reaper-bound material was purified and resolved on denaturing agarose gels (see Methods for details). Addition of Reaper did not cause dissociation of ribosomal subunits. (d) Reaper binding to 80S complexes was assayed as in c, but subunits were either incubated with biotinylated Reaper before 80S reconstitution or stringently washed with high-salt buffer (500 mM KCl) after 80S reconstitution and binding. Stringent high-salt washing conditions resulted in dissociation of 40S and 60S ribosomal subunits, but these same stringent conditions did not affect the 40S-Reaper interaction.
Reaper binds specifically to the 40S subunit
To characterize further the interaction between Reaper and the eukaryotic ribosome, we purified 40S and 60S ribosomal subunits from HeLa cells by high-salt sucrose-gradient centrifugation to yield highly pure, dissociated ribosomal subunits. These subunits are devoid of loosely associated proteins such as initiation and elongation factors, and they contain only the ribosomal core (rRNA and tightly bound ribosomal proteins)30. Biotinylated Reaper was incubated with the purified subunits and stringently washed, and the bound material was then resolved on denaturing agarose gels. We observed that Reaper associated with single 80S ribosomes (monosomes), consistent with our previous data (Fig. 2c). Furthermore, Reaper interacted tightly with the purified 40S ribosomal subunit, but not with the isolated 60S ribosomal subunit (Fig. 2d). Stringent high-salt washing conditions (0.5 M KCl) known to cause 40S–60S dissociation31 did not affect 40S-Reaper interaction (Fig. 2d). These observations underscore the strength and specificity of the Reaper–40S subunit interaction. Furthermore, our data suggest that Reaper–40S subunit association does not directly inhibit factor-independent subunit joining, as 80S monosomes were capable of forming in the presence of Reaper (Fig. 2c). The tight association between Reaper and the 40S subunit was further corroborated by sucrose-gradient centrifugation of either RRL supplemented with Reaper or isolated 40S and 60S subunits supplemented with Reaper. In both cases, Reaper cosedimented with the 40S subunit (data not shown). Together, our data show that Reaper directly binds ribosomes by interacting with the 40S ribosomal core.
Reaper inhibits translation initiation
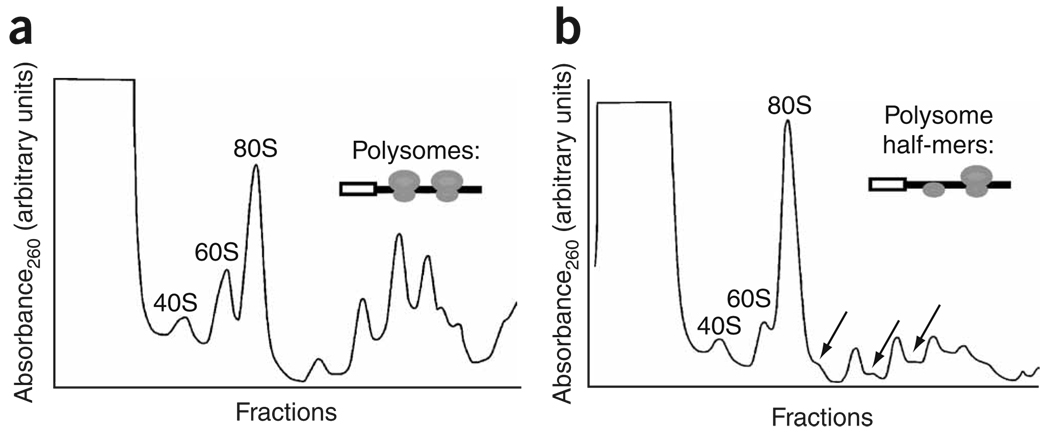
The inhibitory activity that Reaper induces upon binding the 40S subunit could be exerted at the level of either initiation or elongation. To distinguish between these two possibilities, ribosomal species from control RRL or RRL supplemented with Reaper were resolved by sucrose-gradient centrifugation (Fig. 3 and Supplementary Fig. 2 online). These analyses revealed that addition of Reaper resulted in an accumulation of 80S monosomes and a decrease in the number of engaged translating ribosomes (polysome runoff) (Fig. 3b and Supplementary Fig. 2 and Supplementary Fig 3 online). These findings are consistent with a severe reduction in translation initiation.
Figure 3.
Reaper inhibits translation initiation and induces accumulation of 48S half-mers. (a) Sedimentation profile of normally translating RRL resolved by sucrose-gradient centrifugation. (b) Sedimentation as in a, except that RRL was supplemented with Reaper peptide (final concentration of 40 µM). Note the accumulation of 48S half-mers (arrows) and 80S monosomes.
Reaper does not affect 43S or 48S complex formation
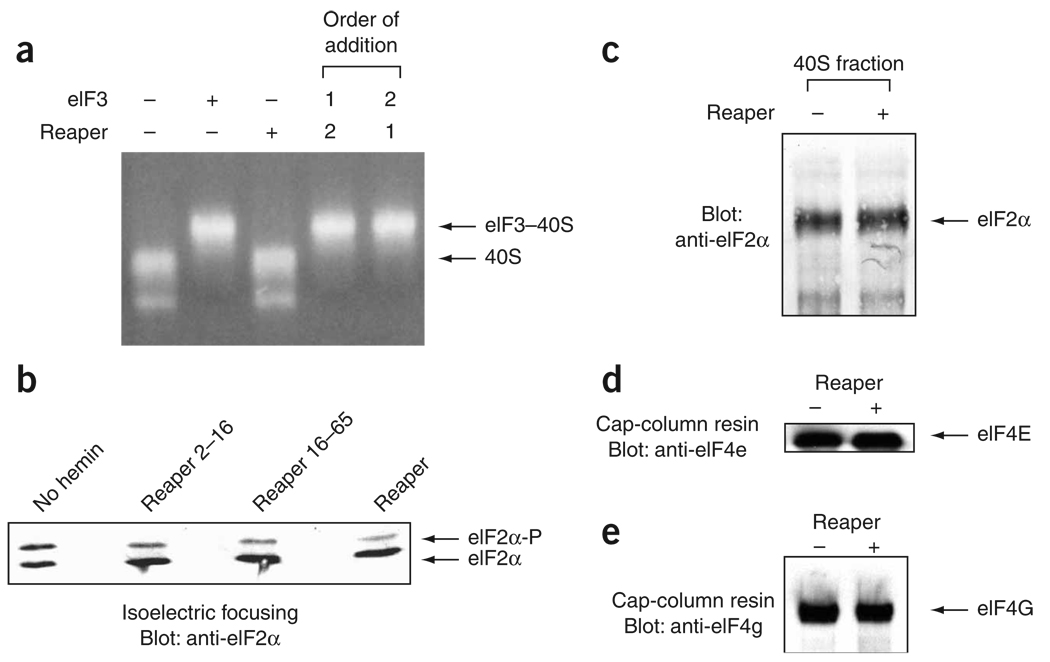
To identify the stage at which Reaper inhibits translation initiation, we assessed whether Reaper affects early initiation events such as formation of the 43S complex. According to current models of initiation, 43S complex formation involves interaction of the eIF3 protein complex with the ‘naked’ 40S subunit. To determine whether Reaper affects the interaction of eIF3 with the 40S subunit, functional eIF3 complex was purified from HeLa cell cytoplasmic extracts and allowed to interact with purified 40S subunits in the presence or absence of purified Reaper. eIF3–40S complex formation was then assayed by resolving the complex on nondenaturing agarose gels. Reaper did not affect the eIF3-induced gel shift of the 40S subunits (Fig. 4a), suggesting that it does not disrupt formation of the eIF3–40S complex by interacting with the 40S ribosomal subunit. eIF3–40S complex formation in the presence of Reaper was also assayed by sucrose-gradient centrifugation of the complex followed by immunoblotting of ribosomal fractions for the presence of eIF3. These assays also demonstrated that Reaper did not affect the association between eIF3 and the 40S ribosomal subunit (data not shown).
Figure 4.
Reaper does not affect eIF2α phosphorylation or 43S complex formation. (a) Purified 40S subunits and purified eIF3 complex were allowed to interact in the presence or absence of Reaper and resolved on nondenaturing agarose gels. 18S rRNA was visualized by ethidium bromide staining. In the last two lanes (order of addition), either Reaper or eIF3 complex was preincubated for 10 min with the 40S subunit as indicated by the number 1, and the remaining component was then added (number 2). (b) Phosphorylation of eIF2α in the presence or absence of Reaper. Translating RRL not supplemented with hemin, or supplemented with hemin and either a control peptide (Reaper2–16, final concentration of 325 µM) or active Reaper peptide (Reaper16–65 or full-length Reaper, final concentration of 40 µM) was resolved by isoelectric focusing and immunoblotted with antibodies directed against eIF2α to detect eIF2α phosphorylation. Not supplementing RRL with hemin results in activation of HRI kinase, which phosphorylates eIF2α and inhibits translation. (c) Association of eIF2α with 40S subunits in the presence or absence of Reaper. Translating RRL with or without Reaper (final concentration of 40 µM) was resolved by sucrose-gradient centrifugation and fractions were collected. Fractions containing the 40S complex were methanol precipitated, resolved by SDS-PAGE and assayed by immunoblotting using antibody directed against eIF2α. (d,e) Association of eIF4G and eIF4E with the cap structure in the presence or absence of Reaper. Translating RRL extracts with or without Reaper (final concentration of 40 µM) were supplemented and incubated with cap resin (7-methylGpppG resin). Bead-bound material was washed with buffer, resolved by SDS-PAGE and immunoblotted with antibodies directed against eIF4E (d) or eIF4G (e). Free 7-methylGpppG could compete with bound initiation factors from the cap resin (not shown).
Another core component of the 43S complex is eIF2. Phosphorylation of the eIF2 regulatory subunit, eIF2α, is a common regulatory mechanism activated by cells in response to a variety of different stresses32. However, Reaper did not promote increased eIF2α phosphorylation, as determined by isoelectric focusing and immunoblotting using antibodies directed against eIF2α (Fig. 4b and Supplementary Fig. 4 online). These data were verified by immunoblotting using antibodies directed against phospho-eIF2α (data not shown and Supplementary Fig. 4).
To assess further potential effects of Reaper on 43S complex formation, we examined binding of the eIF2–GTP–Met-tRNAinitiator ternary complex (TC) to the 40S subunit. The ribosomal species in translating RRL were resolved by sucrose-gradient centrifugation, and the fraction containing the 40S subunit was probed for the presence of eIF2 by immunoblotting. Reaper did not reduce the amount of eIF2 associated with 40S ribosomal subunits, which suggests that it did not disrupt formation of the 40S–TC complex (Fig. 4c and Supplementary Fig. 4). Collectively, these data suggest that Reaper does not inhibit translation at the stage of 43S complex formation.
During translation initiation of capped mRNAs, the formation of the 43S complex is followed by its association with the eIF4F–mRNA to form a 48S complex5. To determine whether 48S complex formation was compromised by Reaper, we again resolved translating RRL by sucrose-gradient sedimentation. Addition of Reaper to translating RRL led to an accumulation of 80S monosomes, as mentioned above. Addition of Reaper to translating RRL also led to an increase in the number of polysome-associated 48S complexes, visualized as polysome ‘half-mers’33 (Fig. 3b and Supplementary Fig. 2). The presence of these 48S half-mers suggests that Reaper allowed the formation of some 48S complexes and, in these cases, stalled initiation before 60S binding. Moreover, cap-binding studies showed that Reaper did not prevent the association of eIF4G and eIF4E factors with the cap structure (Fig. 4d,e). Together, our data demonstrate that Reaper exerts its effect downstream of 43S binding to the mRNA but, as shown by the accumulation of half-mers, upstream of 60S subunit joining and translation elongation.
Inefficient AUG recognition by Reaper-induced 48S half-mers
Three general models could potentially explain Reaper-induced accumulation of 48S half-mers, which contain nontranslating 40S subunits that have not bound 60S subunits and entered the elongation cycle: (i) inefficient scanning, (ii) inefficient recognition of the start codon or (iii) inefficient ribosomal subunit joining after start-codon recognition.
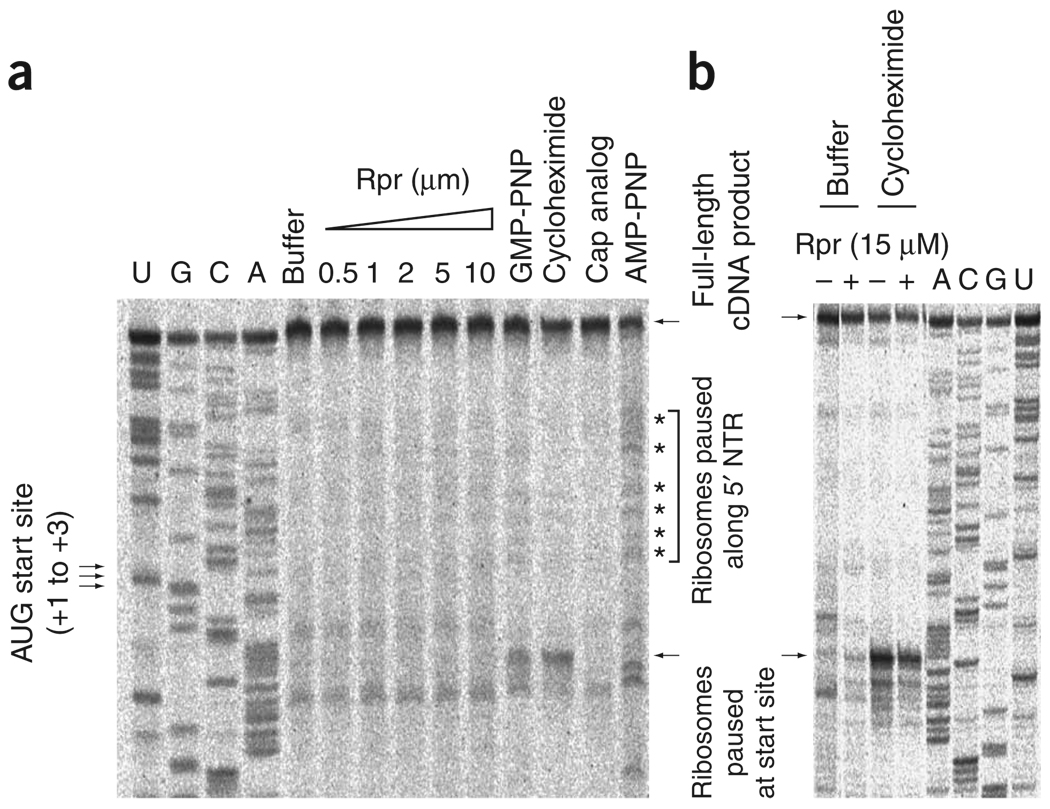
To differentiate between the three possibilities for the observed half-mers, we performed toeprinting analyses on GLA, a cap-dependent mRNA. GLA is a monocistronic mRNA consisting of the globin 5′ untranslated region (UTR) and a luciferase open reading frame, followed by a spacer and a poly(A) tail. Toeprinting allows the positions of stalled ribosomes on the mRNA chain to be identified34,35. We hypothesized that prevention of scanning would lead to an accumulation of 48S complexes in the 5′ UTR, whereas inefficient 80S formation would lead to stalling of the 48S complex at the start codon35. By contrast, inefficient recognition of the start codon would not result in an accumulation of ribosomes stalled in the 5′ UTR or at the start codon.
We used pharmacological agents to provide controls for the different possible scenarios outlined above. AMP-PNP, a nonhydrolyzable analog of ATP, was used to inhibit the ATPase activity of the RNA helicase eIF4A, which is known to promote scanning of the 48S complex36. Addition of AMP-PNP to the translation extract resulted in an increase in the number of ribosomal complexes stalled between the cap and the AUG start codon (Fig. 5a). Addition of GMP-PNP, a nonhydrolyzable analog of GTP that blocks GTP hydrolysis by the eIF2 TC, or addition of cycloheximide, which inhibits the first elongation step, increased the number of ribosomes stalled at the AUG start site, as expected35. When inhibitory concentrations of Reaper were added to translation extracts followed by addition of the GLA mRNA, there was an appreciable increase in ribosomal pausing at the start site or between the cap and the start site (Fig. 5). This was the case even at concentrations of Reaper at which translation was reduced to less than 10% of the level in the absence of Reaper and at which a dramatic accumulation of 80S monosomes and 48S half-mers was observed (Fig. 5 and Supplementary Fig. 2). Therefore, unlike with GMP-PNP or cycloheximide, which stall substantial numbers of 48S and 80S complexes, respectively, at the AUG start site, 48S complexes observed in the presence of Reaper are not stalled at the AUG start site or in the 5′ UTR.
Figure 5.
Reaper-induced 48S half-mers inefficiently recognize the AUG start site. (a) Toeprinting analyses on the GLA mRNA supplemented with buffer, indicated concentrations of Reaper (Rpr) or pharmacological translation inhibitors. Arrows at left, positions of initiation AUG codons; asterisks at right, positions of toeprinting bands. A dideoxynucleotide sequence generated with the same primer was run in parallel (first four rows). The ribosomal pause sites in the Reaper lanes are not appreciably stronger than those present in the buffer control, but are appreciably weaker than those observed for any of the positive controls. (b) Toeprinting as in a, except that Reaper was added at 15 µM (final concentration) in the presence or absence of cycloheximide.
As the 48S complexes were not paused at the start site or 5′ UTR, it is possible that Reaper affected them by promoting scanning past the initiator AUG, perhaps by altering start-site selection or factor-mediated subunit joining. If these steps were affected by Reaper, one would expect addition of Reaper in the presence of cycloheximide to result in a decreased cycloheximide-induced pausing at the start site, as Reaper would promote scanning before the cycloheximide inhibitory step (first elongation cycle). To test this possibility, toeprints were performed as described earlier, but with simultaneous addition of both cycloheximide and Reaper. Notably, addition of Reaper decreased the pausing induced by cycloheximide (Fig. 5b), consistent with the possibility that it promotes accumulation of 48S by promoting scanning past the initiator AUG.
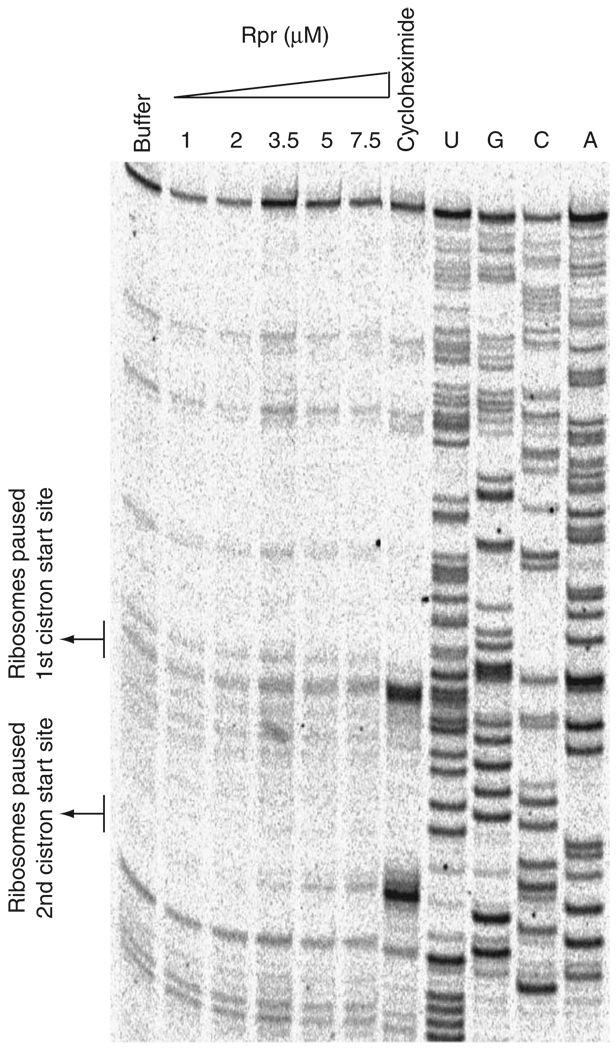
To test more directly whether Reaper affected the capacity of the 48S complex to recognize and pause at the correct AUG start site, we performed toeprinting analyses on BUN-S mRNA, a bicistronic mRNA. BUN-S mRNA consists of the Bunyamwera viral mRNA that encodes, in two overlapping reading frames, the nucleocapsid protein and the nonstructural small (NSs) protein37.
Notably, at lower Reaper concentrations we observed a titra-table increase in Reaper-induced pausing at the second cistron’s start site (Fig. 6). At higher Reaper concentrations, increased pausing was observed for both start sites (data not shown). As no Reaper-induced pausing was observed for the monocistronic GLA mRNA, but pausing was observed at the second start site of the bicistronic BUN-S mRNA even at low Reaper concentrations, it is likely that Reaper’s effect on the 48S complex varies depending on the nature of the mRNA, the position of the AUG start site on the mRNA and the Kozak context of the start site. Together, our toeprinting data on both the GLA and the BUN-S mRNA suggest that Reaper’s interaction with the 40S subunits affects recognition of the initiator AUG by the 48S complex.
Figure 6.
Reaper induces differential pausing in bicistronic mRNA messages. Shown are toeprinting analyses on the BUN-S bicistronic mRNA supplemented with buffer, varying concentrations of Reaper (Rpr) or cycloheximide. A dideoxynucleotide sequence generated with the same primer was run in parallel (last four rows). Note a titratable increase in ribosomal pausing at the second start site upon addition of increasing amounts of Reaper.
Translation inhibition by Reaper is mRNA-dependent
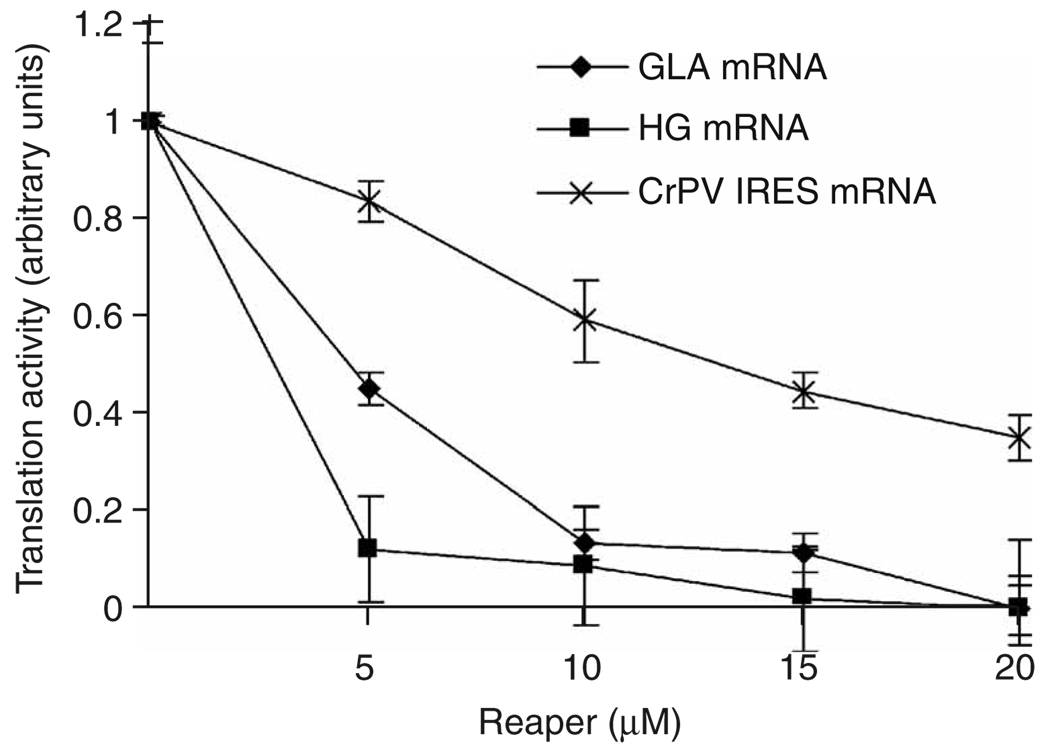
As Reaper inhibits initiation rather than elongation, there is the potential for selective inhibition of translation for particular mRNAs. The toeprinting results described above suggest that Reaper may compromise the ability of the scanning 48S complex to recognize the start codon, and that this capacity is dependent upon the identity of the mRNA. Transcripts driven by certain IRESs do not require scanning, but instead bind the ribosome directly at the start codon38. Therefore, it was plausible that translation of certain IRES-driven mRNAs might be inhibited less effectively by Reaper as compared to translation of cap-dependent messages. To test this, we compared the effects of Reaper on GLA and HG (Hsp70 5′-terminal fragment and β-globin coding sequence, previously described35) cap-dependent messages with its effect on the cricket paralysis virus (CrPV) IRES. This IRES was chosen because it has been extensively characterized and the mechanism by which it allows cap-independent translation is well understood. The CrPV IRES has a long, highly structured region in its 5′ UTR, which prevents initiation through a conventional scanning mechanism39–44. Instead, it directly binds and recruits the 40S ribosomal subunit in the absence of any initiation factors and places it in position to initiate translation without ribosomal scanning7,39,40,43. Thus, if Reaper affected AUG recognition by the scanning ribosome, we hypothesized that the CrPV IRES might escape inhibition. We observed that at concentrations of Reaper under which cap-dependent translation was almost completely inhibited for GLA and HG, substantial translation could still be observed from a CrPV IRES–driven mRNA (Fig. 7).
Figure 7.
Reaper induces differential inhibition of translation in an mRNA-dependent manner. RRLs were supplemented with saturating levels of either GLA mRNA, HG mRNA or CrPV-EGFP mRNA. In vitro ranslation in the absence or presence of Reaper at the indicated concentrations was performed and quantitated as described in Methods. All reactions were carried out in triplicate. The degree of inhibition by Reaper at a given concentration is dependent on the identity of the mRNA, with CrPV-IRES being expressed at Reaper concentrations that completely inhibit expression of cap-dependent GLA and HG mRNAs.
Collectively, these results indicate that direct binding and regulation of ribosome function by Reaper can modulate the protein expression program to allow the preferential expression of certain cap-independent messages while inhibiting general, cap-dependent protein synthesis.
DISCUSSION
Cellular translation is inhibited under many different physiological and pathological circumstances1. Typically, this inhibition results from the covalent modification of translation initiation factors4,5. We have found that Reaper regulates translation initiation through a novel mechanism involving direct binding to the eukaryotic small ribosomal subunit (40S). This binding alters the ribosome by promoting the formation of 48S half-mers defective in correct recognition of the initiator AUG.
Reaper’s interaction with the 40S ribosomal core could potentially be mediated by direct binding to the 18S rRNA or to one of the tightly associated ribosomal proteins. This binding affected neither translation initiation events leading to formation of 48S complexes nor association of regulatory initiation factors, such as eIF2 and eIF3, with the small ribosomal subunit. Moreover, the presence of half-mers in polysome profiles suggests that Reaper-associated 43S complexes are capable of recognizing and binding the mRNA.
Toeprinting analysis on monocistronic GLA messages and bicistronic BUN-S messages suggests that Reaper affects recognition of the initiator AUG by the half-mer. This could result from Reaper directly altering start-site selection or from Reaper affecting subunit joining. It is unlikely that Reaper directly inhibits subunit association, as Reaper interacted with preformed 80S monosomes without causing subunit dissociation (Fig. 2a,c and Supplementary Fig. 2 and Supplementary Fig 3). Reaper also cosedimented with 80S monosomes and polysomes in translating RRL (data not shown). In addition, purified 40S and 60S subunits preincubated with Reaper still formed 80S ribosomes (data not shown). However, Reaper’s interaction with the 48S complex might possibly affect the function of a regulatory factor, such as eIF5B, crucial for correct 60S joining and 80S activation, leading to an accumulation of orphaned 48S complexes. Alternatively, Reaper’s association with the 48S complex could directly disrupt recognition of the initiator codon by the ribosome, perhaps by sterically hindering their interaction or altering 48S conformation. In both models, as Reaper does not affect the association of rate-limiting eIFs associated with the 48S complex, this mechanism could lead to general translation inhibition through sequestration of stoichiometrically limiting initiation factors associated with the nonproductive, Reaper-bound 48S complex. The scarcity of free initiation factors could then contribute to the pool of inactive 80S monosomes that forms in the presence of Reaper.
Although cap-dependent mRNAs were inhibited effectively by Reaper, a CrPV IRES–driven message was inhibited much less effectively. This IRES directly binds and recruits the 40S subunit in an end-independent manner and without any initiation factors. These data suggest that Reaper’s interaction with the 48S complex can disrupt translation in an mRNA-dependent manner. Notably, not all IRES elements were resistant to Reaper-mediated inhibition: we observed that the EMCV IRES was susceptible to inhibition by Reaper (data not shown). These data are consistent with our toeprinting data, which showed that Reaper’s 48S association differentially affected its capacity to pause at a given AUG depending on the identity of the mRNA, the position and context of the AUG and the concentration of Reaper used (Fig. 5 and Fig 6). Together, the IRES expression and toeprinting data suggest that Reaper’s interaction with the 48S complex can disrupt its function in an mRNA-dependent manner.
The fact that CrPV IRES–mediated translation was not affected (particularly at lower Reaper concentrations) suggests that Reaper-bound 40S subunits can still form functional 80S complexes and sustain protein synthesis, given the proper context. Our observations raise the intriguing possibility that Reaper may promote differential regulation of mRNA expression. The fact that certain end-independent mRNAs are less sensitive to Reaper’s inhibition allows for scenarios in which specific cellular mRNAs could be expressed even in the presence of Reaper. It is plausible that Reaper allows the selective translation of novel open reading frames to augment the apoptotic program. Several apoptotic regulators, including Reaper, have been shown to be translated from IRES elements45–49, raising the possibility that their translation may be maintained even when cap-dependent translation is impaired.
Notably, Drosophila Grim, another regulator of apoptosis with sequence similarity to Reaper, has also been shown to inhibit translation and induce cell death27. Furthermore, a human apoptotic inducer related in function to Reaper, Smac (also called Diablo), was recently shown to globally inhibit protein expression50. This activity could be linked with the ability of Smac to sensitize cells to apoptosis and inhibit clonogenic tumor growth. Apoptotic regulators such as Grim and Smac, like Reaper, could potentially alter the protein expression program to allow for rapid responses to changing physiological conditions and the coordinated expression of regulatory mRNAs in the determination of cell fate.
Originally, our identification of Reaper as a regulator of translation was aided by the fact that Reaper bears marked sequence homology to a group of translational inhibitory proteins found in the Bunyamwera genus of Bunyaviruses25. Bunyaviruses cause encephalitis and hemorrhagic fevers in humans and are the major cause of pediatric encephalitis in North America51,52. We have found that NSs protein from San Angelo virus binds the ribosome and promotes the accumulation of half-mers detectable on sucrose gradients (data not shown and Supplementary Fig. 2). These observations strengthen the notion that the Reaper-NSs sequence homology reflects a shared mechanism of translational inhibition in addition to their similarities in promotion of apoptosis25. Our observations also suggest an intriguing context in which altered start site selection might be crucial. The NSs protein is present in the viral genome as a second open reading frame embedded within a larger reading frame. Therefore, it is plausible that NSs synthesis, initially low early in viral infection, is upregulated later in infection by the ability of the initially synthesized NSs protein to facilitate bypass of the first AUG codon in favor of its own, internally located AUG53. Similar regulatory loops have been observed in prokaryotes, where bacterial IF3, which initiates from a non-AUG codon, alters its own translation by affecting the fidelity of initiation codon selection54. Alterations in start codon selection could also be used in eukaryotes for proper temporal coordination of the expression of viral proteins or, in the case of Reaper, for alterations in the gene expression program for determining cell fate.
METHODS
Affinity depletion and biochemical fractionation
Rabbit reticulocyte lysates with endogenous mRNAs (from here on referred to as ‘untreated RRL’) were generated as previously described29. Before use, lysates were supplemented with translation mix (final concentrations: 10 mM creatine phosphate, 50 µg ml−1 creatine phosphokinase, 2 mM DTT, 50 µg ml−1 calf liver tRNA, 86 mM potassium acetate, 0.6 mM magnesium acetate and 40 µM hemin).
The Reaper16–65 fragment was generated as a biotin-tagged synthetic peptide by B. Kaplan (City of Hope, Beckman Research Institute). Full-length Reaper and Reaper2–16 peptides were also generated by B. Kaplan. The NSs40–88 peptide fragment was generated by Anaspec. For affinity depletions, biotinylated Reaper peptide was resuspended in DMSO at 25 mg ml−1, and 4 µl of peptide was added to 200 µl of streptavidin sepharose beads (Amersham Pharmacia), allowed to incubate for 1 h and washed extensively (five times) with 1 ml of PBS buffer. For affinity depletions, 25 µl of packed beads were incubated with 50 µl of untreated RRL extract for 1 h at 4 °C, then removed by centrifugation. Biochemical fractionations of fresh, untreated RRL were conducted as previously described28,29. The biochemical fractions were then tested for their capacity to recover the affinity-depleted activity by adding 30% of any given fractionation to the indicated affinity-depleted RRL (that is, 30% of total final volume of the RRL reaction). Translation of RRL endogenous mRNAs was assayed by radiolabeling of all newly synthesized proteins with addition of 1 µCi µl−1 of Redivue l-[35S]methionine (Amersham Pharmacia Biotech) and 0.02 mM of amino acid mixture minus methionine (Promega). Protein synthesis was performed for 25 min at 30 °C and assayed by trichloroacetic acid precipitation, and radioactivity was measured with a scintillation counter. Protein-bound beads were also resolved by SDS-PAGE and silver-stained to visualize Reaper-interacting proteins in the RRL. The silver-stained bands were then analyzed by MALDI at the Duke University Proteomic Facility.
Ribosomal pull-downs
Reaper-bound streptavidin sepharose beads were incubated in untreated RRL or Drosophila embryonic extract as described above. Bound material was stringently washed with high-salt buffer (10 mM HEPES (pH 7.4), 500 mM KCl, 2.5 mM MgCl2, 1 mM DTT). Bound material was phenol-chloroform extracted and resolved by agarose gel to analyze rRNA binding. For the S6 immunoblot, bound material was boiled and resolved by SDS-PAGE, and S6 was visualized by immunoblot using an antibody to S6 (Cell Signaling Technologies).
Purification of 40S and 60S subunits and Reaper binding assays
40S and 60S ribosomal subunits were purified from HeLa cells by high-salt sucrose-gradient centrifugation as previously described30. Purified subunits were added to a final concentration of 1 µM and allowed to interact with 3 µM of Reaper peptide for 20 min at 37 °C, then incubated with streptavidin sepharose beads. Bound material was stringently washed in high-salt buffer (20 mM HEPES (pH 7.5), 6 mM magnesium acetate, 500 mM KCl, 2 mM DTT) or low-salt buffer (20 mM HEPES (pH 7.5), 100 mM KCl, 2.5 mM MgCl2, 2 mM DTT), phenol-chloroform extracted, resolved by agarose gel and visualized with ethidium bromide staining.
Sucrose-gradient polysome profiles
Open-top wetable centrifuge polyclear tubes (Seton Scientific) were placed on a sucrose-gradient maker. Molecular biology–grade sucrose (Sigma) was used to generate 40% and 15% (w/v) sucrose solutions, which were used in the sucrose-gradient maker to pour 5-ml sucrose gradients (for high-salt sucrose gradients, solutions were made to a final concentration of 0.5 M KCl). Gradients were allowed to settle at 4 °C for 30 min. 100 µl of untreated reticulocyte lysate was supplemented with hemin, translation mix (as described above) and 0.02 mM of amino acid mixture (Promega) in the presence or absence of Reaper (at indicated concentrations), allowed to translate at 30 °C for 25 min and then gently poured at the top of the gradient. Gradients were then spun in a Sorval 60 A11-650 swinging bucket at 45,000 r.p.m. for 1.25 h (or 3 h for higher-resolution gradients). Gradient fractions were then read in a spectrophotometer at absorbance of 260 nm. For analysis of eIF2α, fractions were collected, methanol-precipitated and resolved by SDS-PAGE, and eIF2 was visualized by immunoblot using an antibody to eIF2α (Cell Signaling Technologies).
eIF3–40S complex formation in the presence of Reaper
eIF3 was purified from HeLa cell extracts as previously described40, added at 1 µM final concentration and allowed to interact with 1 µM of 40S subunit for 10 min at 37 °C in the presence or absence of saturating amounts (3 µM) of Reaper (also incubated for 10 min at 37 °C). The complex was resolved by sucrose-gradient centrifugation and fractions were collected, methanol-precipitated, resolved by SDS-PAGE and immunoblotted against eIF3 (antibody kindly provided by C. Fraser, University of California at Berkeley). The order-of-addition experiments were carried out as described above, but either Reaper or eIF3 was allowed to interact for 10 min at 37 °C before the other component was added and incubated for an additional 10 minutes at 37 °C (as indicated by numbering in Figure 4a). The complex was also resolved in nondenaturing agarose gels (1% agarose gels in THEM buffer, as previously described36) and the 18S rRNA was visualized by ethidium bromide staining.
PAGE and immunoblots
To determine the phosphorylation status of eIF2α, samples of untreated RRL without hemin or with hemin and Reaper peptide fragments were supplemented with translation mix (as described above) and 0.02 mM of amino acid mixture (Promega), allowed to translate for 20 min at 30 °C and then resuspended in VSIEF buffer (9.5 M urea, 5% CHAPS, 50 mM sodium fluoride, 5% β-mercaptoethanol). The samples were then subjected to isoelectric focusing on vertical isoelectric focusing gels. eIF2α was visualized by immunoblotting using an antibody to eIF2α (Cell Signaling Technologies). To determine whether Reaper affected eIF2α phosphorylation in Drosophila S2 cells, 5 × 105 S2 cells were transfected with 7 µg of either control pMT-GFP vector (expressing green fluorescent protein (GFP)) or pMT-RPR-GFP (expressing Rpr-GFP) using Cellfectin reagent (Invitrogen). To prevent apoptosis, Schneider’s Drosophila cell media (Gibco) was supplemented with 50 µM zVAD-fmk (BioMol). Cells were then supplemented with 500 µM of copper sulfate (to induce protein expression off the transfected constructs) and harvested after 12 h. Next, S2 cells were sorted for GFP fluorescence using a BD FacsVantage SE cell sorter. GFP-positive cells were then lysed in hypotonic solution (20 mM HEPES-KOH, (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, 1 mM DTT) for 10 min and passed through a 27-gauge needle with 25 passes. Cell material was then resolved by SDS-PAGE and immunoblotted for eIF2α or phospho-eIF2α, as well as for actin to control for protein loading levels (Cell Signaling Technologies). To determine whether Reaper affected eIF4E and eIF4G association with the cap structure, untreated RRL with hemin were supplemented with translation mix (as described above), 0.02 mM of amino acid mixture (Promega) and, where indicated, Reaper to a final concentration of 40 µM. Extracts were incubated for 10 min at 30 °C, supplemented with 20 µl of 7-methylGpppG resin (Amersham-Pharmacia) and incubated for an additional 10 min at 30 °C. Bead-bound material was collected by centrifugation, washed at least three times in buffer (10 mM HEPES (pH 7.7), 50 mM KCl, 2.5 mM MgCl2, 1 mM DTT), resolved by PAGE and immunoblotted using an antibody to eIF4E (Cell Signaling Technologies) or eIF4G (kindly provided by R. Rhodes, Louisiana State University).
In vitro translation
In vitro translation was performed in Flexi RRL (Promega) supplemented with all amino acids except methionine (20 µM) and with radiolabeled methionine (20 µCi), creatine phosphate, KCl or potassium acetate to 87.5 mM, RNase inhibitor (40 u) and Reaper (5–20 µM). mRNA was added to saturating levels (10 µg ml−1 for GLA, 40 µg ml−1 for CrPV–enhanced GFP and 30 µg ml−1 for HG). Reactions were allowed to proceed at 30 °C for 60 min before being stopped on ice. All reactions were carried out in triplicate. Translation was quantified by using nitrocellulose filter binding followed by scintillation counting of incorporated [35S]methionine. Scintillation counts corresponded to production of full-length protein, as confirmed by autoradiographs of SDS PAGE gels of the translation reactions (data not shown). To plot the data, translational activity was normalized to the activity in the absence of Reaper (0 µM Reaper) for each mRNA after it was averaged (this allowed calculation of standard deviation for ‘0 µM Reaper’). Expression levels of mRNAs were compared and normalized only to their own expression prior to addition of Reaper (0 µM Reaper); they were not compared or normalized to one another, to avoid apparent and artificial increases owing to differences among the expression levels of the mRNA classes. This allowed us to illustrate the titratable effect of Reaper on the expression of each mRNA class.
GLA and BUN-S toeprinting
For each set of reactions, an RRL mix was made up containing the following per reaction: 11.55 µl Flexi RRL (Promega), 0.35 µl RNasin RNase inhibitor (Promega), 0.578 µl complete amino acid mix (Promega), 0.49 µl 1.25 M KCl and 0.035 µl 1 M DTT. To this mix was added 2 µl buffer RB (10 mM HEPES (pH 7.5), 50 mM KCl, 2 mM DTT) or 2 µl of Reaper in buffer RB at ten times the desired final concentration, and, where indicated, 2 µl of either 4 mM GMP-PNP or 10 g l−1 cycloheximide. The reactions were made up to a total of 17 µl with water and incubated at 30 °C for 5 min. Next, 3 µl of GLA or BUN-S mRNA at 67 µg ml−1 was added and the reactions were incubated for an additional 15 min at 30 °C. RT mix (3 µl) was added, which contained 1.84 µl of RT elongation mix (10 mM Tris (pH 8.3), 50 mM MgCl2, 10 mM each dNTP, 74.3 mM KCl, 2 mM DTT), 0.16 µl of 20 U µl−1 AMV reverse transcriptase and 1 µl of the 33P-labeled primer at 3.2 µM. The reactions were incubated for 15 min and then placed on ice. The elongated primers were resolved on a 10% sequencing gel and the bands visualized by autoradiography, scanned in a Storm PhosphorImager scanner (Molecular Dynamics) and quantified using ImageQuant.
Supplementary Material
ACKNOWLEDGMENTS
We thank R.M. Elliott, S.S. Margolis, J.E. Irazoqui, K. Silva, T. Prince, A. Vila-Sanjurjo, S. Paranjape, B. Kaplan, E. Harris, R. Rhodes, I.N. Shatsky, E. Nogales and J. Doudna for helpful discussions and generous sharing of advice and reagents. We thank in particular C. Fraser (University of California at Berkeley) for advice and kindly providing purified eIF3 complex and the US National Cell Culture Center for supplying HeLa cells. This work was supported by US National Institutes of Health grants to S.K. (RO1 GM61919) and J.C. (R01 GM65050). R.M. was supported by the Oklahoma Agricultural Experiment Station (Project 1975). C.L.S. was supported by a Howard Hughes Medical Institute Predoctoral Fellowship. D.C.-R. was supported by a US National Institutes of Health Minority Supplement to grant R01 GM61919 and by the Gates Millennium Scholarship. D.C.-R. is a Damon Runyon Fellow.
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 2.Marash L, Kimchi A. DAP5 and IRES-mediated translation during programmed cell death. Cell Death Differ. 2005;12:554–562. doi: 10.1038/sj.cdd.4401609. [DOI] [PubMed] [Google Scholar]
- 3.Bushell M, et al. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ. 2000;7:628–636. doi: 10.1038/sj.cdd.4400699. [DOI] [PubMed] [Google Scholar]
- 4.Marissen WE, Gradi A, Sonenberg N, Lloyd RE. Cleavage of eukaryotic translation initiation factor 4GII correlates with translation inhibition during apoptosis. Cell Death Differ. 2000;7:1234–1243. doi: 10.1038/sj.cdd.4400750. [DOI] [PubMed] [Google Scholar]
- 5.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 6.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 7.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 8.Holcik M. Targeting translation for treatment of cancer–a novel role for IRES? Curr. Cancer Drug Targets. 2004;4:299–311. doi: 10.2174/1568009043333005. [DOI] [PubMed] [Google Scholar]
- 9.Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell. 2002;1:22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens MJ. Translational control in virus-infected cells: models for cellular stress responses. Semin. Cell Dev. Biol. 2005;16:13–20. doi: 10.1016/j.semcdb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pestova TV, Hellen CU. Functions of eukaryotic factors in initiation of translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:389–396. doi: 10.1101/sqb.2001.66.389. [DOI] [PubMed] [Google Scholar]
- 13.Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asano K, et al. A multifactor complex of eIF1, eIF2, eIF3, eIF5, and tRNA(i)Met promotes initiation complex assembly and couples GTP hydrolysis to AUG recognition. Cold Spring Harb. Symp. Quant. Biol. 2001;66:403–415. doi: 10.1101/sqb.2001.66.403. [DOI] [PubMed] [Google Scholar]
- 15.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 16.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez J, et al. Regulation of internal ribosome entry site-mediated translation by eukaryotic initiation factor-2alpha phosphorylation and translation of a small upstream open reading frame. J. Biol. Chem. 2002;277:2050–2058. doi: 10.1074/jbc.M109199200. [DOI] [PubMed] [Google Scholar]
- 18.Schneider R, et al. New ways of initiating translation in eukaryotes. Mol. Cell. Biol. 2001;21:8238–8246. doi: 10.1128/MCB.21.23.8238-8246.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holcik M, Gordon BW, Korneluk RG. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 2003;23:280–288. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 21.Pickering BM, Mitchell SA, Spriggs KA, Stoneley M, Willis AE. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- 23.Narayanan S, Surendranath K, Bora N, Surolia A, Karande AA. Ribosome inactivating proteins and apoptosis. FEBS Lett. 2005;579:1324–1331. doi: 10.1016/j.febslet.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 24.Tait SW, Werner AB, de Vries E, Borst J. Mechanism of action of Drosophila Reaper in mammalian cells: Reaper globally inhibits protein synthesis and induces apoptosis independent of mitochondrial permeability. Cell Death Differ. 2004;11:800–811. doi: 10.1038/sj.cdd.4401410. [DOI] [PubMed] [Google Scholar]
- 25.Colon-Ramos DA, et al. Inhibition of translation and induction of apoptosis by Bunyaviral nonstructural proteins bearing sequence similarity to reaper. Mol. Biol. Cell. 2003;14:4162–4172. doi: 10.1091/mbc.E03-03-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat. Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo SJ, et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- 28.Voorma HO, Thomas A, Goumans H, Amesz H, van der Mast C. Isolation and purification of initiation factors of protein synthesis from rabbit reticulocyte lysate. Methods Enzymol. 1979;60:124–135. doi: 10.1016/s0076-6879(79)60012-5. [DOI] [PubMed] [Google Scholar]
- 29.Safer B, Jagus R, Kemper WM. Analysis of initiation factor function in highly fractionated and unfractionated reticulocyte lysate systems. Methods Enzymol. 1979;60:61–87. doi: 10.1016/s0076-6879(79)60008-3. [DOI] [PubMed] [Google Scholar]
- 30.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 31.Merrick WC. Assays for eukaryotic protein synthesis. Methods Enzymol. 1979;60:108–123. doi: 10.1016/s0076-6879(79)60011-3. [DOI] [PubMed] [Google Scholar]
- 32.Clemens MJ. Initiation factor eIF2 alpha phosphorylation in stress responses and apoptosis. Prog. Mol. Subcell. Biol. 2001;27:57–89. doi: 10.1007/978-3-662-09889-9_3. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen KH, et al. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozak M. Primer extension analysis of eukaryotic ribosome-mRNA complexes. Nucleic Acids Res. 1998;26:4853–4859. doi: 10.1093/nar/26.21.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dmitriev SE, Pisarev AV, Rubtsova MP, Dunaevsky YE, Shatsky IN. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 2003;533:99–104. doi: 10.1016/s0014-5793(02)03776-6. [DOI] [PubMed] [Google Scholar]
- 36.Korneeva NL, First EA, Benoit CA, Rhoads RE. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 2005;280:1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- 37.Bridgen A, Elliott RM. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc. Natl. Acad. Sci. USA. 1996;93:15400–15404. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 40.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 41.Pestova TV, Lomakin IB, Hellen CU. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolupaeva VG, Pestova TV, Hellen CU. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellen CU, Pestova TV. Translation of hepatitis C virus RNA. J. Viral Hepat. 1999;6:79–87. doi: 10.1046/j.1365-2893.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 44.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. USA. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell. Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henis-Korenblit S, et al. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl. Acad. Sci. USA. 2002;99:5400–5405. doi: 10.1073/pnas.082102499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 48.Sherrill KW, Byrd MP, Van Eden ME, Lloyd RE. BCL-2 translation is mediated via internal ribosome entry during cell stress. J. Biol. Chem. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- 49.Hernandez G, Vazquez-Pianzola P, Sierra JM, Rivera-Pomar R. Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA. 2004;10:1783–1797. doi: 10.1261/rna.7154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogler M, et al. Inhibition of clonogenic tumor growth: a novel function of Smac contributing to its antitumor activity. Oncogene. 2005;24:7190–7202. doi: 10.1038/sj.onc.1208876. [DOI] [PubMed] [Google Scholar]
- 51.Elliott RM. Emerging viruses: the Bunyaviridae. Mol. Med. 1997;3:572–577. [PMC free article] [PubMed] [Google Scholar]
- 52.Gonzalez-Scarano F, Jacoby D, Griot C, Nathanson N. Genetics, infectivity and virulence of California serogroup viruses. Virus Res. 1992;24:123–135. doi: 10.1016/0168-1702(92)90001-p. [DOI] [PubMed] [Google Scholar]
- 53.Fuller F, Bhown AS, Bishop DH. Bunyavirus nucleoprotein, N, and a nonstructural protein, NSS, are coded by overlapping reading frames in the S RNA. J. Gen. Virol. 1983;64:1705–1714. doi: 10.1099/0022-1317-64-8-1705. [DOI] [PubMed] [Google Scholar]
- 54.Sacerdot C, et al. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 1996;21:331–346. doi: 10.1046/j.1365-2958.1996.6361359.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.